The Psychoemotional Stress-Induced Changes in the Abundance of SatIII (1q12) and Telomere Repeats, but Not Ribosomal DNA, in Human Leukocytes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population Samples
2.1.1. The SZ Group
2.1.2. The HC Group
2.1.3. The St Group
2.1.4. The Patient’s Consents to the Analyses Performed
2.2. DNA Isolation from Leukocytes
2.3. Nonradioactive Quantitative Hybridization (NQH)
2.3.1. DNA Probe for SatIII Quantification (f-SatIII)
2.3.2. DNA Probe for rDNA Quantification
2.3.3. DNA Probe for Telomere Repeat Quantification
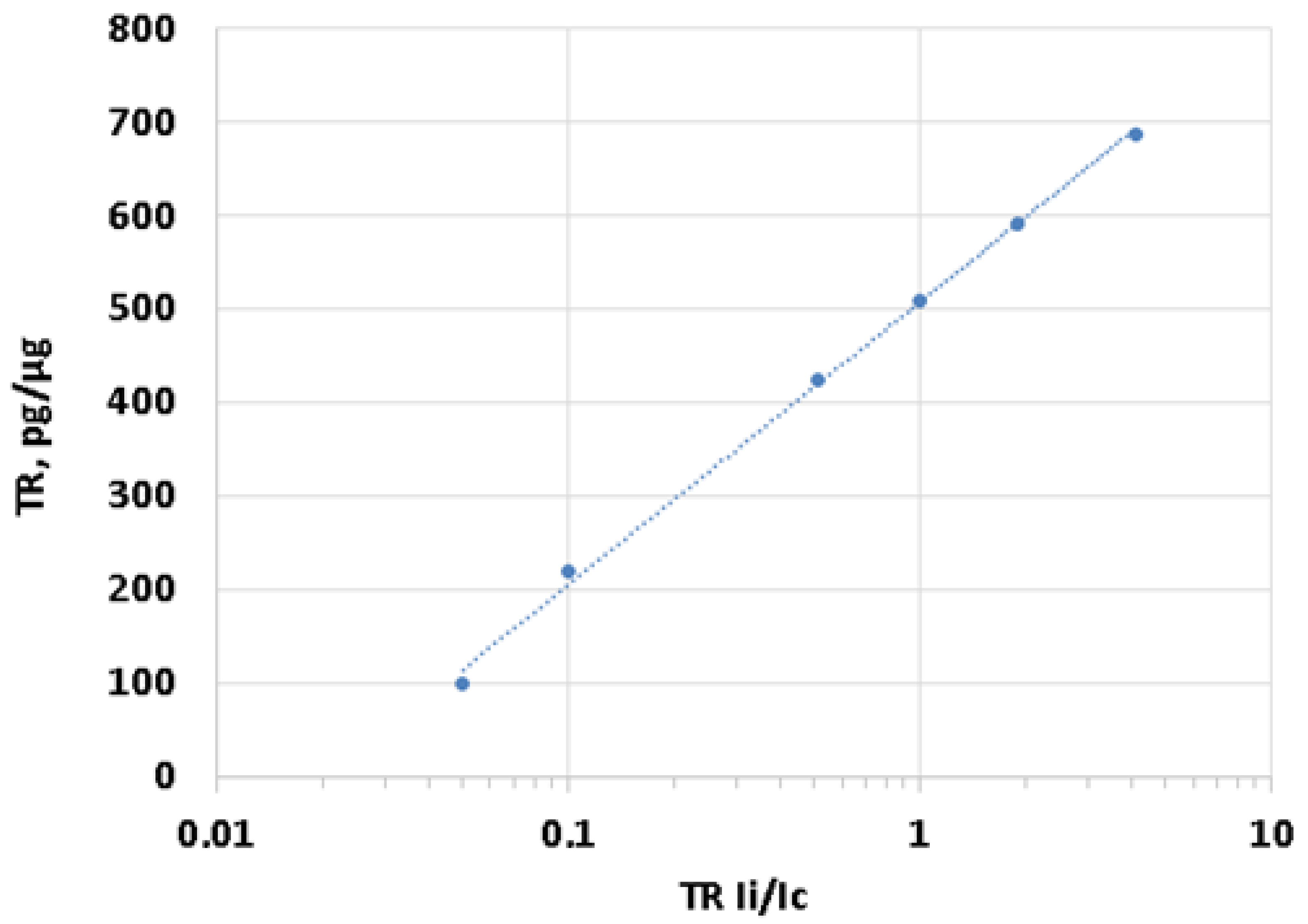
2.3.4. Standard Calibration Curve for TR Quantification
2.4. Flow Cytometry Assay (FCA)
2.5. Statistical Analyses
3. Results
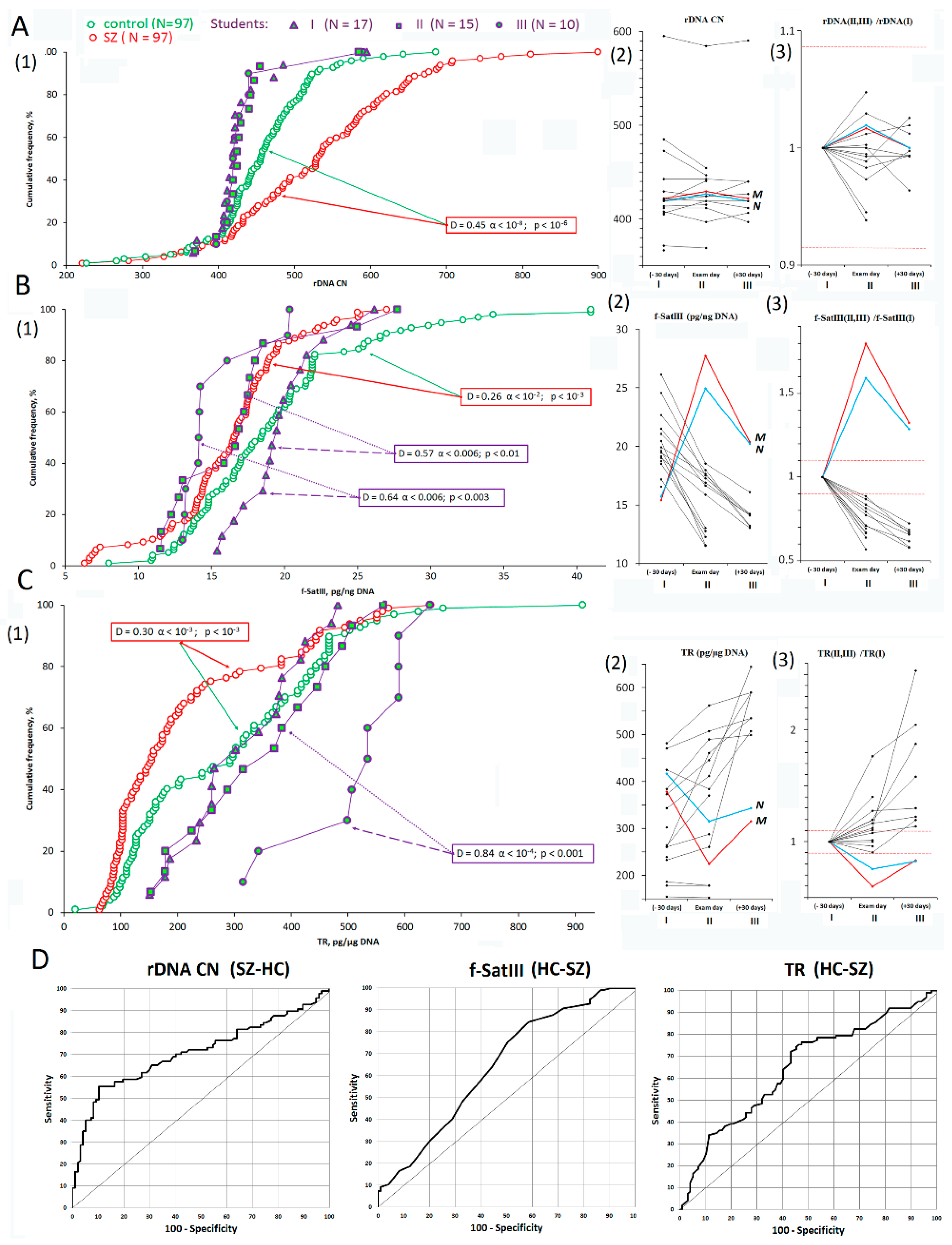
3.1. rDNA, f-SatIII, and TR CNVs under Stress
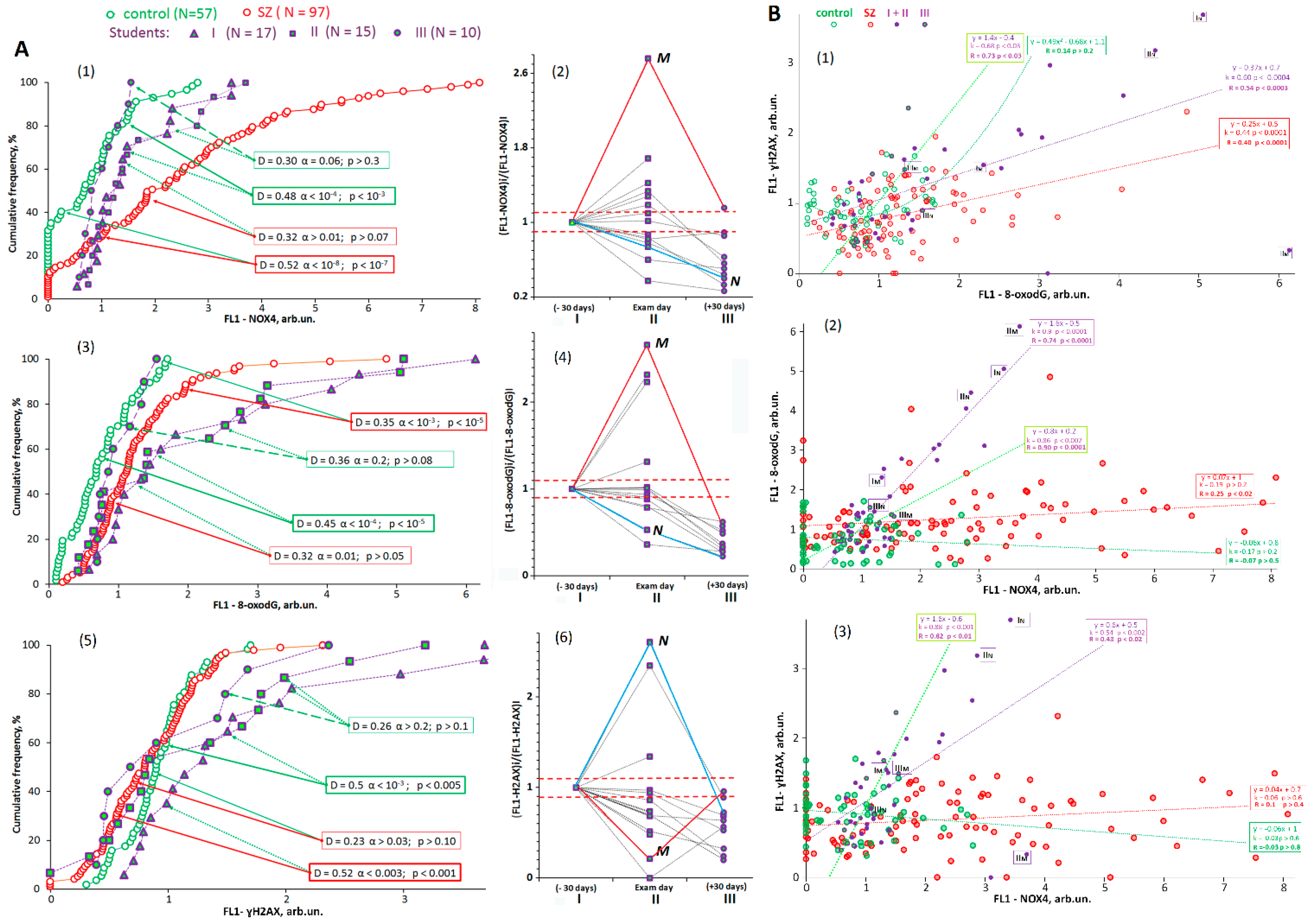
3.2. Oxidative Stress Markers in Lymphocytes from the Different Groups Studied
3.3. Relationships between rDNA CNs and Oxidative Stress Markers
3.4. Relationship between Oxidative Stress Markers and Abundance of f-SatIII and TR
3.5. Changes in the Levels of Proteins Involved in the Exam Stress Response in Students
4. Discussion
4.1. Psychoemotional Stress Induces Oxidative Stress in the Human Body
4.2. Cellular Response to Oxidative Stress in Students
4.3. Cellular Response to Oxidative Stress in SZ Patients
4.4. Effect of rDNA Repeat Abundance on f-SatIII and TR CNVs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Koning, A.P.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monlong, J.; Cossette, P.; Meloche, C.; Rouleau, G.; Girard, S.L.; Bourque, G. Human copy number variants are enriched in regions of low mappability. Nucleic Acids Res. 2018, 46, 7236–7249. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.L.; Perry, G.H.; Feuk, L.; Redon, R.; McCarroll, S.A.; Altshuler, D.M.; Aburatani, H.; Jones, K.W.; Tyler-Smith, C.; Hurles, M.E.; et al. Copy number variation: New insights in genome diversity. Genome Res. 2006, 16, 949–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henrichsen, C.N.; Chaignat, E.; Reymond, A. Copy number variants, diseases and gene expression. Hum. Mol. Genet. 2009, 18, R1–R8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrad, D.F.; Pinto, D.; Redon, R.; Feuk, L.; Gokcumen, O.; Zhang, Y.; Aerts, J.; Andrews, T.D.; Barnes, C.; Campbel, P.; et al. Origins and functional impact of copy number variation in the human genome. Nature 2010, 464, 704–712. [Google Scholar] [CrossRef] [Green Version]
- Brahmachary, M.; Guilmatre, A.; Quilez, J.; Hasson, D.; Borel, C.; Warburton, P.; Sharp, A.J. Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet. 2014, 10, e1004418. [Google Scholar] [CrossRef] [Green Version]
- Jackson, M.; Marks, L.; May, G.H.W.; Wilson, J.B. The genetic basis of disease. Essays Biochem. 2018, 62, 643–723. [Google Scholar] [CrossRef] [Green Version]
- Warburton, P.E.; Hasson, D.; Guillem, F.; Lescale, C.; Jin, X.; Abrusan, G. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genom. 2008, 9, 533. [Google Scholar] [CrossRef] [Green Version]
- Black, E.M.; Giunta, S. Repetitive fragile sites: Centromere satellite DNA as a source of genome instability in human diseases. Genes 2018, 9, E615. [Google Scholar] [CrossRef] [Green Version]
- Hannan, A.J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 2018, 19, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Veiko, N.N.; Egolina, N.A.; Radzivil, G.G.; Nurbaev, S.D.; Kosyakova, N.V.; Shubaeva, N.O.; Liapunova, N.A. Quantitative analysis of repetitive sequences in human genomic DNA and detection of an elevated ribosomal repeat copy number in patients with schizophrenia (the results of molecular and cytogenetic analysis). Mol. Biol. 2003, 37, 349–357. [Google Scholar] [CrossRef]
- Korzeneva, I.B.; Kostuyk, S.V.; Ershova, E.S.; Skorodumova, E.N.; Zhuravleva, V.F.; Pankratova, G.V.; Volkova, I.V.; Stepanova, E.V.; Porokhovnik, L.N.; Veiko, N.N. Human circulating ribosomal DNA content significantly increases while circulating satellite III (1q12) content decreases under chronic occupational exposure to low-dose gamma-neutron and tritium beta-radiation. Mutat. Res. 2016, 791–792, 49–60. [Google Scholar] [CrossRef]
- Malinovskaya, E.M.; Ershova, E.S.; Golimbet, V.E.; Porokhovnik, L.N.; Lyapunova, N.A.; Kutsev, S.I.; Veiko, N.N.; Kostyuk, S.V. Copy number of human ribosomal genes with aging: Unchanged mean, but narrowed range and decreased variance in elderly group. Front. Genet. 2018, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Ershova, E.S.; Malinovskaya, E.M.; Konkova, M.S.; Veiko, R.V.; Umriukhin, P.E.; Martynov, A.V.; Kutsev, S.I.; Veiko, N.N.; Kostyuk, S.V. Copy number variation of human satellite III (1q12) with aging. Front. Genet. 2019, 10, 704. [Google Scholar] [CrossRef] [Green Version]
- Ershova, E.S.; Agafonova, O.N.; Zakharova, N.V.; Bravve, L.V.; Jestkova, E.M.; Golimbet, V.E.; Lezheiko, T.V.; Morozova, A.Y.; Martynov, A.V.; Veiko, R.V.; et al. Copy number variation of satellite III (1q12) in patients with schizophrenia. Front. Genet. 2019, 10, 1132. [Google Scholar] [CrossRef] [PubMed]
- Ershova, E.S.; Jestkova, E.M.; Martynov, A.V.; Shmarina, G.V.; Umriukhin, P.E.; Bravve, L.V.; Zakharova, N.V.; Kostyuk, G.P.; Saveliev, D.V.; Orlova, M.D.; et al. Accumulation of circulating cell-free CpG-enriched ribosomal DNA fragments on the background of high endonuclease activity of blood plasma in schizophrenic patients. Int. J. Genom. 2019, 2019, 8390585. [Google Scholar] [CrossRef] [Green Version]
- Konkova, M.S.; Ershova, E.S.; Savinova, E.A.; Malinovskaya, E.M.; Shmarina, G.V.; Martynov, A.V.; Veiko, R.V.; Zakharova, N.V.; Umriukhin, P.; Kostyuk, G.P.; et al. 1Q12 Loci Movement in the Interphase Nucleus Under the Action of ROS Is an Important Component of the Mechanism That Determines Copy Number Variation of Satellite III (1q12) in Health and Schizophrenia. Front. Cell Dev. Biol. 2020, 8, 386. [Google Scholar] [CrossRef]
- Chestkov, I.V.; Jestkova, E.M.; Ershova, E.S.; Golimbet, V.E.; Lezheiko, T.V.; Kolesina, N.Y.; Porokhovnik, L.N.; Lyapunova, N.A.; Izhevskaya, V.L.; Kutsev, S.I.; et al. Abundance of ribosomal RNA gene copies in the genomes of schizophrenia patients. Schizophr. Res. 2018, 197, 305–314. [Google Scholar] [CrossRef]
- Bersaglieri, C.; Santoro, R. Genome Organization in and around the Nucleolus. Cells 2019, 8, 579. [Google Scholar] [CrossRef] [Green Version]
- Symonová, R. Integrative rDNAomics—Importance of the Oldest Repetitive Fraction of the Eukaryote Genome. Genes 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schöfer, C.; Weipoltshammer, K. Nucleolus and chromatin. Histochem. Cell Biol. 2018, 150, 209–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diesch, J.; Hannan, R.D.; Sanij, E. Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease. Cell Biosci. 2014, 4, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McStay, B.; Grummt, I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008, 24, 131–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, B.; Li, H.; Perry, J.M.; Singh, V.P.; Unruh, J.; Yu, Z.; Zakari, M.; McDowell, W.; Li, L.; Gerton, J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017, 13, e1006771. [Google Scholar] [CrossRef] [PubMed]
- Cooke, H.J.; Hindley, J. Cloning of human satellite III DNA: Different components are on different chromosomes. Nucleic Acids Res. 1979, 6, 3177–3197. [Google Scholar] [CrossRef] [Green Version]
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153. [Google Scholar] [CrossRef] [Green Version]
- Turner, K.J.; Vasu, V.; Griffin, D.K. Telomere Biology and Human Phenotype. Cells 2019, 8, 73. [Google Scholar] [CrossRef] [Green Version]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef] [Green Version]
- Shay, J.W. Telomeres and aging. Curr. Opin. Cell Biol. 2018, 52, 1–7. [Google Scholar] [CrossRef]
- Solana, C.; Pereira, D.; Tarazona, R. Early Senescence and Leukocyte Telomere Shortening in SCHIZOPHRENIA: A Role for Cytomegalovirus Infection? Brain Sci. 2018, 8, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anitha, A.; Thanseem, I.; Vasu, M.M.; Viswambharan, V.; Poovathinal, S.A. Telomeres in neurological disorders. Adv. Clin. Chem. 2019, 90, 81–132. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Sun, J.; Wang, S.; Milush, J.M.; Baker, C.A.R.; Coccia, M.; Effros, R.B.; Puterman, E.; Blackburn, E.; Prather, A.A.; et al. In vitro proinflammatory gene expression predicts in vivo telomere shortening: A preliminary study. Psychoneuroendocrinology 2018, 96, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Gomes, N.M.; Ryder, O.A.; Houck, M.L.; Charter, S.J.; Walker, W.; Forsyth, N.R.; Austad, S.N.; Venditti, C.; Pagel, M.; Shay, J.W.; et al. Comparative biology of mammalian telomeres: Hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011, 10, 761–768. [Google Scholar] [CrossRef] [Green Version]
- Barnes, R.P.; Fouquerel, E.; Opresko, P.L. The impact of oxidative DNA damage and stress on telomere homeostasis. Mech. Ageing Dev. 2019, 177, 37–45. [Google Scholar] [CrossRef]
- Eskiocak, S.; Gozen, A.S.; Yapar, S.B.; Tavas, F.; Kilic, A.S.; Eskiocak, M. Glutathione and free sulphydryl content of seminal plasma in healthy medical students during and after exam stress. Hum. Reprod. 2005, 20, 2595–2600. [Google Scholar] [CrossRef] [Green Version]
- Afrisham, R.; Sadegh-Nejadi, S.; SoliemaniFar, O.; Kooti, W.; Ashtary-Larky, D.; Alamiri, F.; Aberomand, M.; Najjar-Asl, S.; Khaneh-Keshi, A. Salivary Testosterone Levels Under Psychological Stress and Its Relationship with Rumination and Five Personality Traits in Medical Students. Psychiatry Investig. 2016, 13, 637–643. [Google Scholar] [CrossRef] [Green Version]
- Yusoff, M.S.; Esa, A.R.; Mat Pa, M.N.; Mey, S.C.; Aziz, R.A.; Abdul Rahim, A.F. A longitudinal study of relationships between previous academic achievement, emotional intelligence and personality traits with psychological health of medical students during stressful periods. Educ. Health 2013, 26, 39–47. [Google Scholar] [CrossRef]
- Hammoud, S.; Karam, R.; Mourad, R.; Saad, I.; Kurdi, M. Stress and Heart Rate Variability during University Final Examination among Lebanese Students. Behav. Sci. 2018, 9, 3. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, K.; Vaz, M.; Sucharita, S. A study of stress and autonomic nervous function in first year undergraduate medical students. Indian J. Physiol. Pharmacol. 2006, 50, 257–264. [Google Scholar]
- Gazzaz, Z.J.; Baig, M.; Al Alhendi, B.S.M.; al Suliman, M.M.O.; al Alhendi, A.S.; Al-Grad, M.S.H.; Quraysha, M.A.A. Perceived stress, reasons for and sources of stress among medical students at Rabigh Medical College, King Abdulaziz University, Jeddah, Saudi Arabia. BMC Med. Educ. 2018, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Abdel Rahman, A.G.; Al Hashim, B.N.; Al Hiji, N.K.; Al-Abbad, Z. Stress among medical Saudi students at College of Medicine, King Faisal University. J. Prev. Med. Hyg. 2013, 54, 195–199. [Google Scholar] [PubMed]
- Gupta, S.; Choudhury, S.; Das, M.; Mondol, A.; Pradhan, R. Factors causing stress among students of a medical college in Kolkata, India. Educ. Health 2015, 28, 92–95. [Google Scholar] [CrossRef] [PubMed]
- Nimkuntod, P.; Uengarpon, N.; Benjaoran, F.; Pinwanna, K.; Ratanakeereepun, K.; Tongdee, P. Psychometric Properties of Depression Anxiety and Stress in Preclinical Medical Students. J. Med. Assoc. Thai. 2016, 99 (Suppl. S7), S111–S117. [Google Scholar]
- Dyrbye, L.N.; Thomas, M.R.; Shanafelt, T.D. Systematic review of depression, anxiety, and other indicators of psychological distress among U.S. and Canadian medical students. Acad. Med. 2006, 81, 354–373. [Google Scholar] [CrossRef]
- Brazeau, C.M.; Shanafelt, T.; Durning, S.J.; Massie, F.S.; Eacker, A.; Moutier, C.; Satele, D.V.; Sloan, J.A.; Dyrbye, L.N. Distress among matriculating medical students relative to the general population. Acad. Med. 2014, 89, 1520–1525. [Google Scholar] [CrossRef]
- Hope, V.; Henderson, M. Medical student depression, anxiety and distress outside North America: A systematic review. Med. Educ. 2014, 48, 963–979. [Google Scholar] [CrossRef]
- Howes, O.D.; Murray, R.M. Schizophrenia: An integrated socio developmental-cognitive model. Lancet 2014, 383, 1677–1687. [Google Scholar] [CrossRef] [Green Version]
- Boyum, A. Separation of leukocytes from blood and bone marrow. Scand. J. Clin. Lab. Investig. 1968, 21, 90–109. [Google Scholar]
- Guo, S.; Chen, X. The human Nox4: Gene, structure, physiological function and pathological significance. J. Drug Target 2015, 23, 888–896. [Google Scholar] [CrossRef]
- Shigenaga, M.K.; Ames, B.N. Assays for 8-hydroxy-2’-deoxyguanosine: A biomarker of in vivo oxidative DNA damage. Free Radic. Biol. Med. 1991, 10, 211–216. [Google Scholar] [CrossRef]
- Pilger, A.; Rüdiger, H.W. 8-Hydroxy-2’-deoxyguanosine as a marker of oxidative DNA damage related to occupational and environmental exposures. Int. Arch. Occup. Environ. Health. 2006, 80, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Valdiglesias, V.; Giunta, S.; Fenech, M.; Neri, M.; Bonassi, S. γH2AX as a marker of DNA double strand breaks and genomic instability in human population studies. Mutat. Res. 2013, 753, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Veiko, N.N.; Terekhov, S.V.; Shubaeva, N.O.; Simirnova, T.D.; Ivanova, S.M.; Egolina, N.A.; Tsvetkova, T.G.; Spitkovskiĭ, D.M.; Liapunova, N.A. Early and late responses to oxidative stress in human dermal fibroblasts of healthy donors and rheumatoid arthritis patients. Relationship between the cell death rate and the genomic dosage of active ribosomal genes. Mol. Biol. 2005, 39, 234–243. [Google Scholar] [CrossRef]
- Porokhovnik, L.N.; Passekov, V.P.; Gorbachevskaya, N.L.; Sorokin, A.B.; Veiko, N.N.; Lyapunova, N.A. Active ribosomal genes, translational homeostasis and oxidative stress in the pathogenesis of schizophrenia and autism. Psychiatr. Genet. 2015, 25, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Cardno, A.G.; Marshall, E.J.; Coid, B.; Macdonald, A.M.; Ribchester, T.R.; Davies, N.J.; Venturi, P.; Jones, L.A.; Lewis, S.W.; Sham, P.C. Heritability estimates for psychotic disorders: The Maudsley twin psychosis series. Arch. Gen. Psychiatry 1999, 56, 162–168. [Google Scholar] [CrossRef]
- Emiliani, F.E.; Sedlak, T.W.; Sawa, A. Oxidative stress and schizophrenia: Recent breakthroughs from an old story. Curr. Opin. Psychiatry 2014, 27, 185–190. [Google Scholar] [CrossRef] [Green Version]
- Sertan Copoglu, U.; Virit, O.; Hanifi Kokacya, M.; Orkmez, M.; Bulbul, F.; Binnur Erbagci, A.; Semiz, M.; Alpak, G.; Unal, A.; Ari, M.; et al. Increased oxidative stress and oxidative DNA damage in non-remission schizophrenia patients. Psychiatry Res. 2015, 229, 200–205. [Google Scholar] [CrossRef]
- Smaga, I.; Niedzielska, E.; Gawlik, M.; Moniczewski, A.; Krzek, J.; Przegaliński, E.; Pera, J.; Filip, M. Oxidative stress as an etiological factor and a potential treatment target of psychiatric disorders. Part 2. Depression, anxiety, schizophrenia and autism. Pharmacol. Rep. 2015, 67, 569–580. [Google Scholar] [CrossRef]
- Hardingham, G.E.; Do, K.Q. Linking early-life NMDAR hypofunction and oxidative stress in schizophrenia pathogenesis. Nat. Rev. Neurosci. 2016, 17, 125–134. [Google Scholar] [CrossRef]
- Koga, M.; Serritella, A.V.; Sawa, A.; Sedlak, T.W. Implications for reactive oxygen species in schizophrenia pathogenesis. Schizophr. Res. 2016, 176, 52–71. [Google Scholar] [CrossRef] [PubMed]
- Barron, H.; Hafizi, S.; Andreazza, A.C.; Mizrahi, R. Neuroinflammation and oxidative stress in psychosis and psychosis risk. Int. J. Mol. Sci. 2017, 18, E651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maas, D.A.; Vallès, A.; Martens, G.J.M. Oxidative stress, prefrontal cortex hypomyelination and cognitive symptoms in schizophrenia. Transl. Psychiatry 2017, 7, e1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.; Sharma, D.; Kalia, K.; Tiwari, V. Crosstalk between endoplasmic reticulum stress and oxidative stress in schizophrenia: The dawn of new therapeutic approaches. Neurosci. Biobehav. Rev. 2017, 83, 589–603. [Google Scholar] [CrossRef]
- Seredenina, T.; Sorce, S.; Herrmann, F.R.; Mulone, X.-J.M.; Plastre, O.; Aguzzi, A.; Jaquet, V.; Krause, K.-H. Decreased NOX2 expression in the brain of patients with bipolar disorder: Association with valproic acid prescription and substance abuse. Transl. Psychiatry 2017, 7, e1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enukashvily, N.I.; Donev, R.; Waisertreiger, I.S.; Podgornaya, O.I. Human chromosome 1 satellite 3 DNA is decondensed, demethylated and transcribed in senescent cells and in A431 epithelial carcinoma cells. Cytogenet. Genome. Res. 2007, 118, 42–54. [Google Scholar] [CrossRef]
- Paredes, S.; Maggert, K.A. Ribosomal DNA contributes to global chromatin regulation. Proc. Natl. Acad. Sci. USA 2009, 106, 17829–17834. [Google Scholar] [CrossRef] [Green Version]
- Paredes, S.; Branco, A.T.; Hartl, D.L.; Maggert, K.A.; Lemos, B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011, 7, e1001376. [Google Scholar] [CrossRef] [Green Version]
- Ershova, E.S.; Jestkova, E.M.; Chestkov, I.V.; Porokhovnik, L.N.; Izevskaya, V.L.; Kutsev, S.I.; Veiko, N.N.; Shmarina, G.; Dolgikh, O.; Kostyuk, S.V. Quantification of cell-free DNA in blood plasma and DNA damage degree in lymphocytes to evaluate dysregulation of apoptosis in schizophrenia patients. J. Psychiatr. Res. 2017, 87, 15–22. [Google Scholar] [CrossRef]



| Index | HC | SZ |
|---|---|---|
| n | 97 | 97 |
| Age | 29 ± 19 | 27 ± 15 |
| Age of SZ manifestation | N/A | 22.1 ± 6.1 |
| Gender (men/women) | 50/47 | 65/32 |
| Never smoked (%) | 68 | 59 |
| Smoke more than 20 cigarettes a day (%) | 12 | 19 |
| Index | SZ | HC | St(I) | St(II) | St(III) | |
|---|---|---|---|---|---|---|
| n | 97 | 97 | 17 | 15 | 10 | |
| rDNA CN | Mean | 531 | 456 | 430 | 434 | 437 |
| SD | 117 | 72 | 51 | 47 | 56 | |
| Min | 221 | 226 | 367 | 369 | 397 | |
| Max | 899 | 686 | 595 | 585 | 591 | |
| Median | 529 | 456 | 419 | 424 | 421 | |
| Cv | 0.22 | 0.16 | 0.12 | 0.11 | 0.13 | |
| f-SatIII pg/ng DNA | Mean | 16 | 19 | 20 | 17 | 15 |
| SD | 4 | 6 | 3 | 5 | 3 | |
| Min | 6 | 8 | 15 | 12 | 13 | |
| Max | 27 | 41 | 26 | 28 | 20 | |
| Median | 17 | 18 | 20 | 17 | 14 | |
| Cv | 0.26 | 0.31 | 0.15 | 0.28 | 0.18 | |
| TR pg/µg DNA | Mean | 213 | 291 | 315 | 349 | 515 |
| SD | 149 | 169 | 103 | 132 | 108 | |
| Min | 63 | 20 | 151 | 153 | 315 | |
| Max | 646 | 913 | 482 | 562 | 644 | |
| Median | 155 | 298 | 302 | 370 | 535 | |
| Cv | 0.70 | 0.58 | 0.33 | 0.38 | 0.21 |
| HC (n = 97) vs. SZ (n = 97) | St(I) (n = 17) vs. St(II) (n = 15) | St(I) (n = 17) vs. St(III) (n = 10) | St(II) (n = 15) vs. St(III) (n = 10) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K–S | U | ROC | K–S | U | K–S | U | K–S | U | |||||||
| D | α | p | AUC | p | D | α | p | D | α | p | D | α | p | ||
| 1 | rDNA | −0.45 | 2 × 10−9 | 4 × 10−7 | 0.710 | <0.001 | −0.31 | 0.3 | 0.3 | −0.15 | 0.9 | 0.7 | 0.17 | 0.9 | 0.6 |
| 2 | f-SatIII | 0.26 | 2 × 10−3 | 6 × 10−4 | 0.642 | <0.001 | 0.57 | 0.005 | 0.009 | 0.64 | 0.005 | 0.002 | 0.37 | 0.3 | 0.6 |
| 3 | TR | 0.30 | 4 × 10−4 | 4 × 10−4 | 0.646 | <0.001 | −0.22 | 0.7 | 0.4 | −0.84 | 8 × 10−5 | 4 × 10−4 | −0.70 | 0.002 | 0.003 |
| 4 | NOX4 | −0.52 | 4 × 10−9 | 2 × 10−8 | 0.772 | <0.001 | −0.16 | 0.9 | 0.8 | 0.34 | 0.4 | 0.1 | 0.33 | 0.4 | 0.07 |
| 5 | 8-oxodG | −0.34 | 3 × 10−4 | 5 × 10−6 | 0.720 | <0.001 | −0.29 | 0.4 | 0.4 | 0.36 | 0.3 | 0.3 | 0.43 | 0.1 | 0.06 |
| 6 | H2AX | 0.23 | 0.03 | 0.2 | 0.567 | 0.149 | 0.29 | 0.4 | 0.3 | 0.40 | 0.2 | 0.09 | 0.23 | 0.8 | 0.4 |
| Index | HC | SZ | St(I) | St(II) | St(III) | |
|---|---|---|---|---|---|---|
| n | 57 | 97 | 17 | 15 | 10 | |
| FL-NOX4 | Mean | 0.76 | 2.42 | 1.55 | 1.65 | 1.01 |
| SD | 0.77 | 2.00 | 0.88 | 0.97 | 0.36 | |
| Min | 0.00 | 0.00 | 0.54 | 0.75 | 0.59 | |
| Max | 2.80 | 8.08 | 3.43 | 3.70 | 1.55 | |
| Median | 0.68 | 1.95 | 1.34 | 1.26 | 0.91 | |
| Coef. var. | 1.01 | 0.83 | 0.57 | 0.59 | 0.35 | |
| FL-8-oxodG | Mean | 0.75 | 1.26 | 1.91 | 2.13 | 1.00 |
| SD | 0.47 | 0.76 | 1.51 | 1.64 | 0.32 | |
| Min | 0.10 | 0.19 | 0.42 | 0.57 | 0.69 | |
| Max | 1.70 | 4.85 | 5.05 | 6.13 | 1.55 | |
| Median | 0.68 | 1.12 | 1.40 | 1.45 | 0.89 | |
| Coef. var. | 0.62 | 0.60 | 0.79 | 0.77 | 0.32 | |
| FL-γH2AX | Mean | 0.92 | 0.83 | 1.58 | 1.25 | 1.03 |
| SD | 0.30 | 0.42 | 0.99 | 0.89 | 0.67 | |
| Min | 0.31 | 0.00 | 0.63 | 0.00 | 0.39 | |
| Max | 1.70 | 2.31 | 3.69 | 3.18 | 2.36 | |
| Median | 0.90 | 0.80 | 1.30 | 0.84 | 0.79 | |
| Coef. var. | 0.33 | 0.51 | 0.63 | 0.71 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Umriukhin, P.E.; Ershova, E.S.; Filev, A.D.; Agafonova, O.N.; Martynov, A.V.; Zakharova, N.V.; Veiko, R.V.; Porokhovnik, L.N.; Kostyuk, G.P.; Kutsev, S.I.; et al. The Psychoemotional Stress-Induced Changes in the Abundance of SatIII (1q12) and Telomere Repeats, but Not Ribosomal DNA, in Human Leukocytes. Genes 2022, 13, 343. https://doi.org/10.3390/genes13020343
Umriukhin PE, Ershova ES, Filev AD, Agafonova ON, Martynov AV, Zakharova NV, Veiko RV, Porokhovnik LN, Kostyuk GP, Kutsev SI, et al. The Psychoemotional Stress-Induced Changes in the Abundance of SatIII (1q12) and Telomere Repeats, but Not Ribosomal DNA, in Human Leukocytes. Genes. 2022; 13(2):343. https://doi.org/10.3390/genes13020343
Chicago/Turabian StyleUmriukhin, Pavel E., Elizaveta S. Ershova, Anton D. Filev, Oksana N. Agafonova, Andrey V. Martynov, Natalia V. Zakharova, Roman V. Veiko, Lev N. Porokhovnik, George P. Kostyuk, Sergey I. Kutsev, and et al. 2022. "The Psychoemotional Stress-Induced Changes in the Abundance of SatIII (1q12) and Telomere Repeats, but Not Ribosomal DNA, in Human Leukocytes" Genes 13, no. 2: 343. https://doi.org/10.3390/genes13020343







