Blocking IbmiR319a Impacts Plant Architecture and Reduces Drought Tolerance in Sweet Potato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Plasmid and Sweet Potato Genetic Transformation
2.3. Vector Construction and Arabidopsis Transformation
2.4. RNA Isolation and qRT–PCR Analysis
2.5. Microscopic Observations
2.6. Lignin Deposition Experiment and Lignin Content Measurement
2.7. Measurement of Breaking Force
2.8. Analysis of Drought Tolerance
2.9. Gene Expression Analysis
2.10. Transcriptome Analysis
2.11. Statistical Analysis
3. Results
3.1. Identification and Characterization of IbmiR319 in Sweet Potato
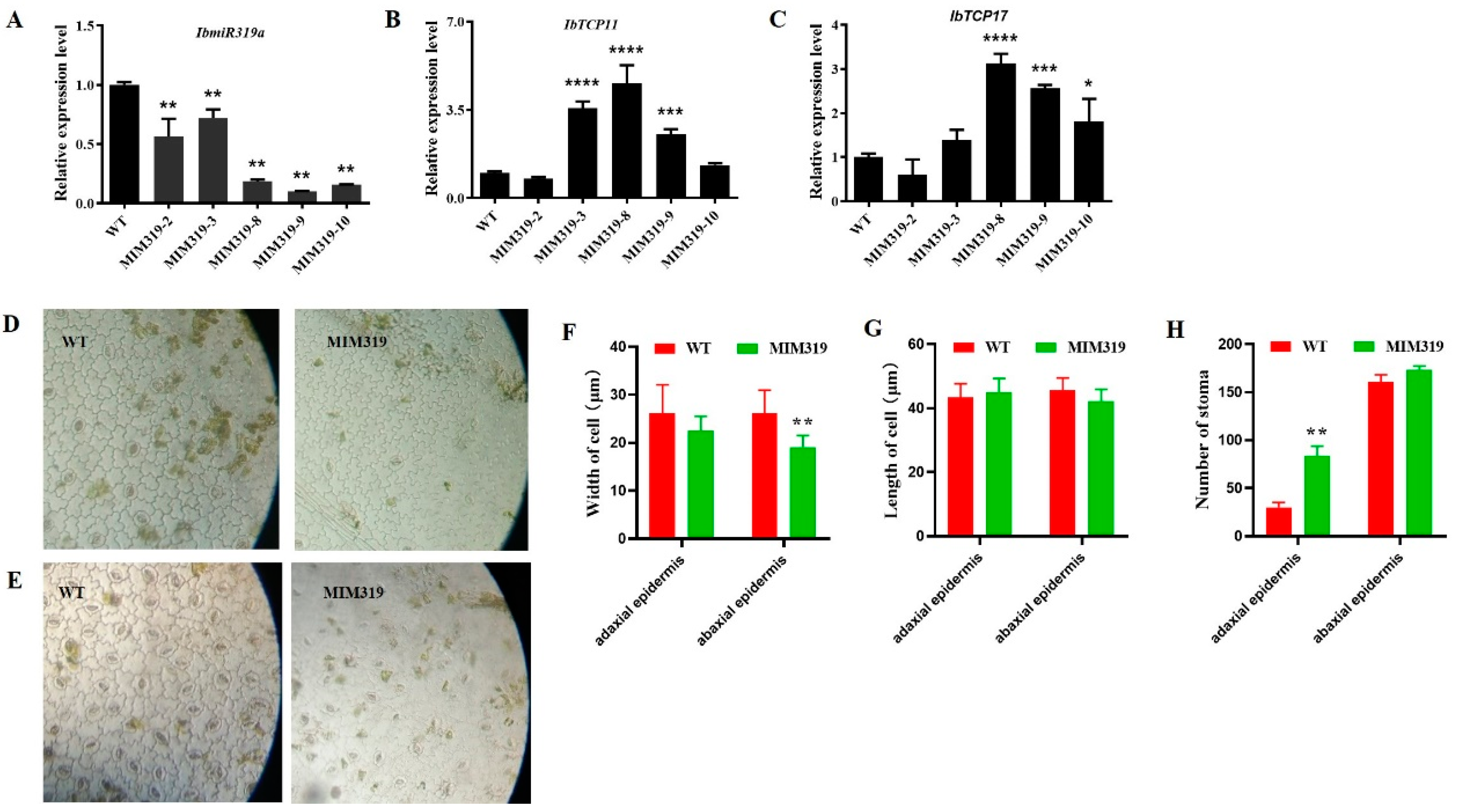
3.2. Generation of Transgenic Sweet Potato Plants with miR319a Blocked
3.3. Expression Level of the Putative IbmiR319a Target Genes Was Upregulated in MIM319 Transgenic Sweet Potato
3.4. Blocking IbmiR319a in Sweet Potato Caused Pleiotropic Phenotype Changes
3.5. RNA-Seq Analysis of Transgenic Sweet Potato Plants with Blocked IbmiR319a
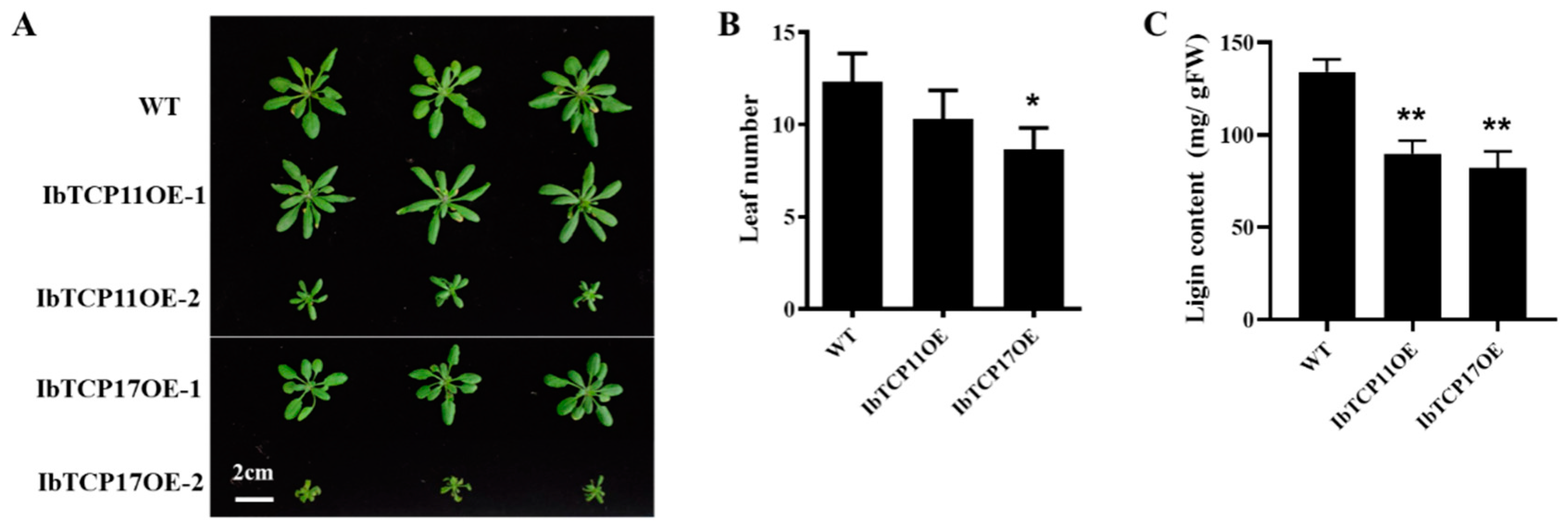
3.6. Transgenic Arabidopsis Plants Overexpressing IbTCP11 and IbTCP17 Also Had a Narrow and Small Leaf Phenotype and Decreased Lignin Content
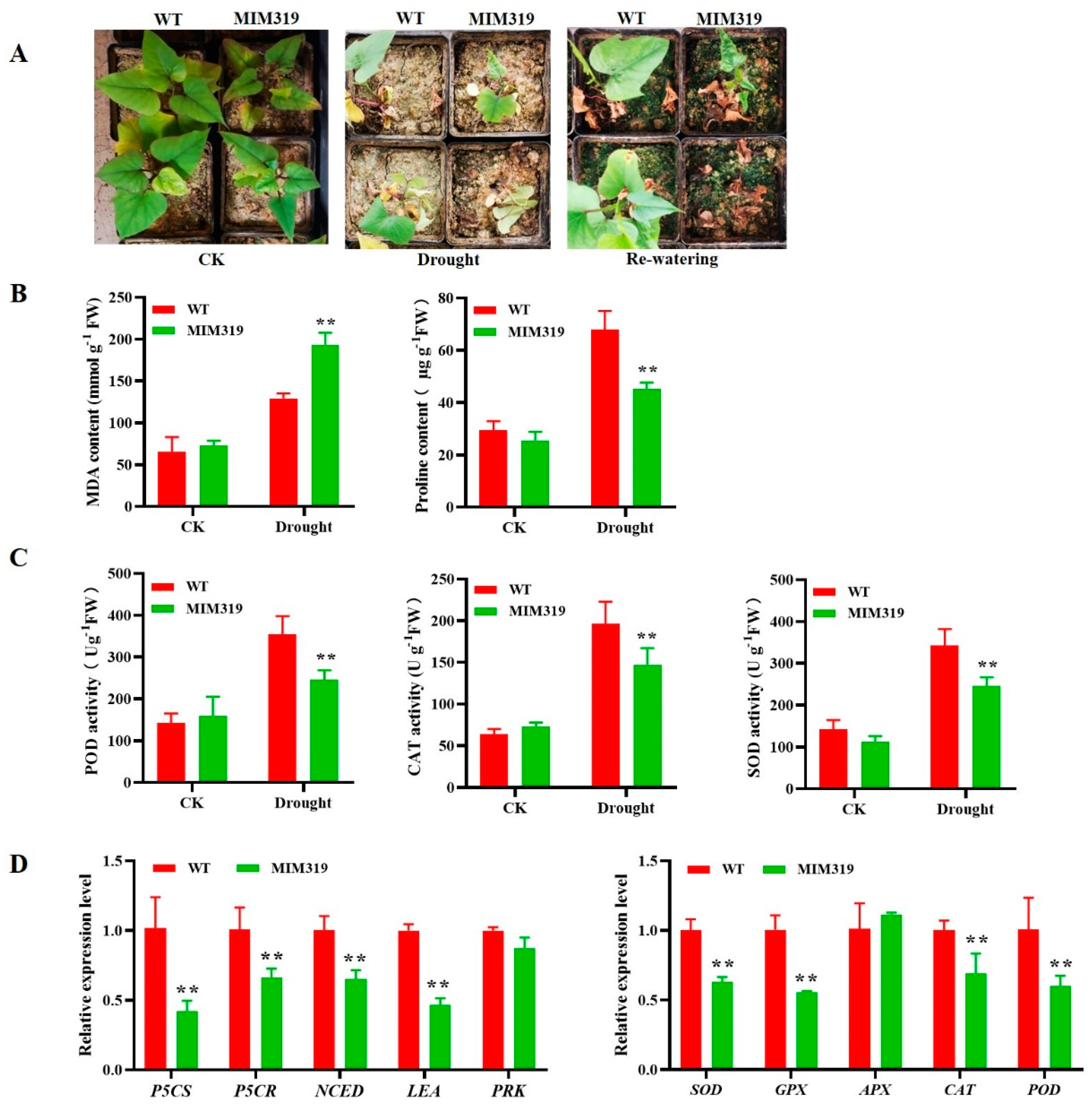
3.7. Blocking IbmiR319a in Sweet Potato Resulted in Decreased Drought Tolerance
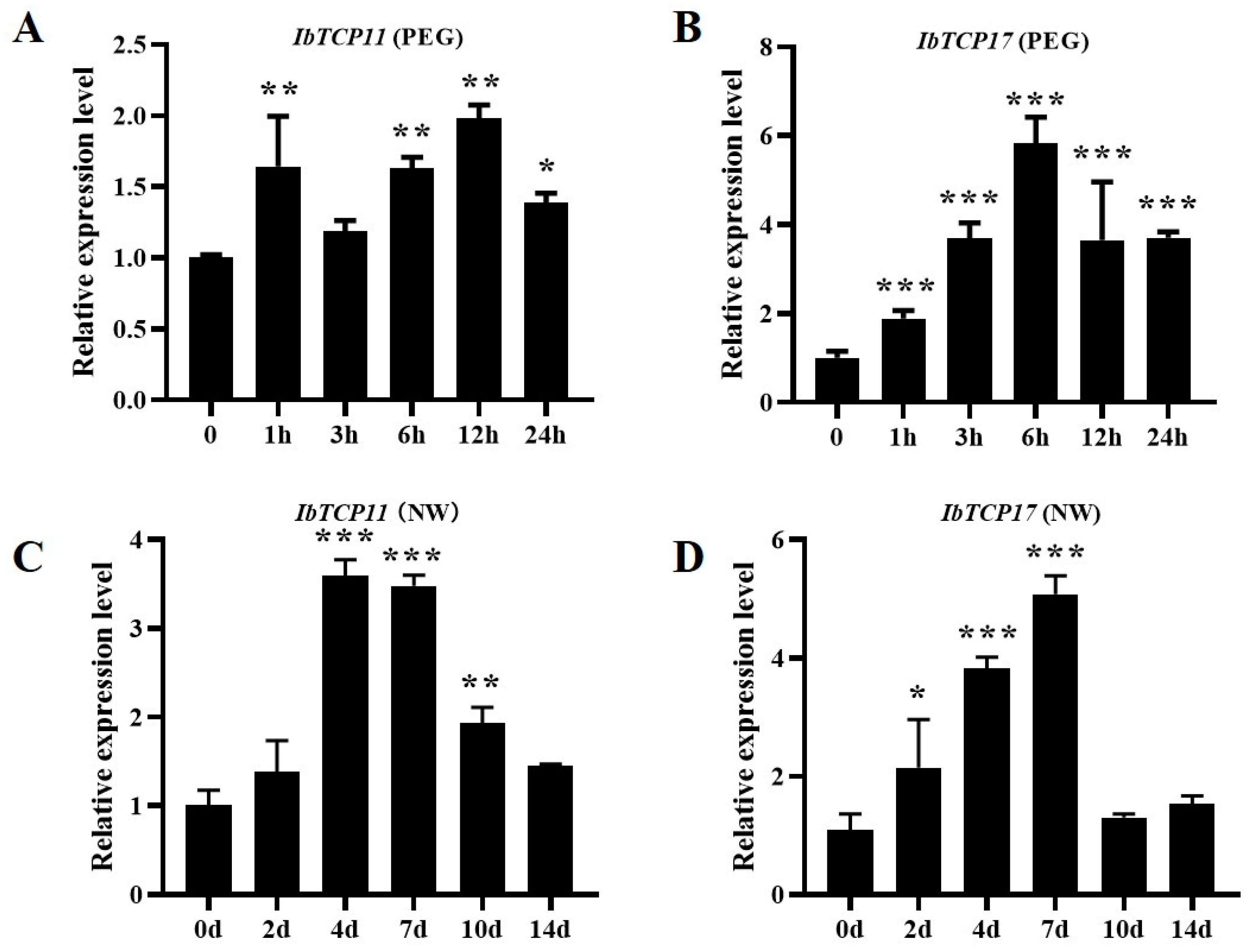
3.8. IbmiR319a Functions in Plant Tolerance of Drought Stress Possibly by Inhibiting IbTCP11/17 Expression in Sweet Potato
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Pearce, R.S. Plant freezing and damage. Ann. Bot. 2001, 87, 417–424. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, A.; Li, H.; Yu, J.; Jiang, J.; Tang, Z.; Ma, D.; Zhang, B.; Han, Y.; Li, Z. High throughput deep sequencing reveals the important roles of microRNAs during sweetpotato storage at chilling temperature. Sci. Rep. 2017, 7, 16578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.Q.; Li, G.; Yu, J.; Cai, J.; Dong, T.T.; Sun, J.; Xu, T.; Li, Z.Y.; Pan, S.Y.; Ma, D.F.; et al. Isolation, Expression Analysis, and Function Evaluation of 12 Novel Stress-Responsive Genes of NAC Transcription Factors in Sweetpotato. Crop Sci. 2018, 58, 1328–1341. [Google Scholar] [CrossRef]
- Burke, M.B.; Lobell, D.B.; Guarino, L. Shifts in African crop climates by 2050, and the implications for crop improvement and genetic resources conservation. Glob. Environ. Chang. 2009, 19, 317–325. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2014, 161, 1375–1391. [Google Scholar] [CrossRef] [Green Version]
- Lin, K.H.; Chiang, C.M.; Lai, Y.C.; You, S.H.; Lo, H.F. Identification of Chilling-inducible Genes in Sweet potato by Suppression Subtractive Hybridization. Res. J. Biotechol. 2011, 6, 37–43. [Google Scholar]
- Sato, Y.; Murakami, T.; Funatsuki, H.; Matsuba, S.; Saruyama, H.; Tanida, M. Heat shock-mediated APX gene expression and protection against chilling injury in rice seedlings. J. Exp. Bot. 2001, 52, 145. [Google Scholar] [CrossRef]
- Motsa, N.M.; Modi, A.T.; Mabhaudhi, T. Sweet potato (Ipomoea batatas L.) as a drought tolerant and food security crop. S. Afr. J. Sci. 2005, 111, 11–12. [Google Scholar] [CrossRef] [Green Version]
- Mbinda, W.; Dixelius, C.; Oduor, R. Induced Expression of Xerophyta viscosa XvSap1 Gene Enhances Drought Tolerance in Transgenic Sweet Potato. Front. Plant Sci. 2019, 10, 1119. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Zhang, M.; Zhang, H.; Zhang, P. Improved tolerance to various abiotic stresses in transgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PLoS ONE 2012, 7, e37344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, L.; Chen, L.J.; Chen, Y.H.; He, J.L.; Zhang, W.; Gao, Z.L.; Zhang, Y.H. Expression of Arabidopsis HOMEODOMAIN GLABROUS 11 Enhances Tolerance to Drought Stress in Transgenic Sweet Potato Plants. J. Plant Biol. 2011, 55, 151–158. [Google Scholar] [CrossRef]
- Mbinda, W.; Ombori, O.; Dixelius, C.; Oduor, R. Xerophyta viscosa Aldose Reductase, XvAld1, Enhances Drought Tolerance in Transgenic Sweetpotato. Mol. Biotechnol. 2018, 60, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.M.; Zhu, M.K.; Luo, Y.H.; Liu, Y.J.; Li, R.S.; Kou, M.; Wang, X.; Zhang, Y.G.; Meng, X.Q.; Zheng, Y.L.; et al. A sweet potato cinnamate 4-hydroxylase gene, IbC4H, increases phenolics content and enhances drought tolerance in tobacco. Acta Physiol. Plant 2017, 39, 276. [Google Scholar] [CrossRef]
- Zhai, H.; Wang, F.; Si, Z.; Huo, J.; Xing, L.; An, Y.; He, S.; Liu, Q. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 2016, 14, 592–602. [Google Scholar] [CrossRef]
- Jin, R.; Kim, B.H.; Ji, C.Y.; Kim, H.S.; Li, H.M.; Ma, D.F.; Kwak, S.S. Overexpressing IbCBF3 increases low temperature and drought stress tolerance in transgenic sweetpotato. Plant Physiol. Biochem. 2017, 118, 45–54. [Google Scholar] [CrossRef]
- Wang, B.; Zhai, H.; He, S.Z.; Zhang, H.; Ren, Z.T.; Zhang, D.D.; Liu, Q.C. A vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci. Hortic. 2016, 201, 153–166. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhai, H.; He, S.; Zhu, H.; Gao, S.; Xing, S.; Wei, Z.; Zhao, N.; Liu, Q. The Sweetpotato BTB-TAZ Protein Gene, IbBT4, Enhances Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2020, 11, 877. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A Sweetpotato Auxin Response Factor Gene (IbARF5) Is Involved in Carotenoid Biosynthesis and Salt and Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 1307. [Google Scholar] [CrossRef]
- Kang, C.; Zhai, H.; He, S.; Zhao, N.; Liu, Q. A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2019, 38, 1373–1382. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Qiu, X.; Yang, Y.; Kim, H.S.; Jia, X.; Yu, H.; Kwak, S.S. Sweetpotato bZIP Transcription Factor IbABF4 Confers Tolerance to Multiple Abiotic Stresses. Front. Plant Sci. 2019, 10, 630. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, H.; He, S.; Zhai, H.; Zhao, N.; Xing, S.; Wei, Z.; Liu, Q. A Novel Sweetpotato Transcription Factor Gene IbMYB116 Enhances Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2019, 10, 1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 1, 21–44. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R.S.; Tarver, J.E.; Hiscock, S.J.; Donoghue, P.C. Evolutionary history of plant microRNAs. Trends Plant Sci. 2014, 19, 175–182. [Google Scholar] [CrossRef]
- Axtell, M.J.; Snyder, J.A.; Bartel, D.P. Common Functions for Diverse Small RNAs of Land Plants. Plant Cell 2007, 19, 1750–1769. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Zhu, J.K. Novel and Stress-Regulated MicroRNAs and Other Small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [Green Version]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Schommer, C.; Debernardi, J.M.; Bresso, E.G.; Rodriguez, R.E.; Palatnik, J.F. Repression of Cell Proliferation by miR319-Regulated TCP4. Mol. Plant 2014, 7, 1533–1544. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Li, D.; Mao, D.; Liu, X.; Ji, C.; Li, X.; Zhao, X.; Cheng, Z.; Chen, C.; Zhu, L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 2207–2218. [Google Scholar] [CrossRef]
- Wang, S.T.; Sun, X.L.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.W.; Sun, M.Z.; Duan, X.B.; Zhu, Y.M. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Mi, X.; Zhang, R.; An, Y.; Zhou, Q.; Yang, T.; Xia, X.; Guo, R.; Wang, X.; Wei, C. Integrated analysis of miRNAs and their targets reveals that miR319c/TCP2 regulates apical bud burst in tea plant (Camellia sinensis). Planta 2019, 250, 1111–1129. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.; Ran, L.; Hu, J.; Ye, X.; Xu, D.; Li, J.; Su, H.; Wang, X.; Ren, S.; Luo, K. miR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 2020, 227, 867–883. [Google Scholar] [CrossRef]
- Wang, F.L.; Zheng, T.; Wu, G.T.; Lang, C.X.; Hu, Z.H.; Shi, J.H.; Wei, J.; Chen, J.Q.; Liu, R.H. Overexpression of miR319a Affects the Balance between Mitosis and Endoreduplication in Arabidopsis Leaves. Plant Mol. Biol. Rep. 2015, 33, 2006–2013. [Google Scholar] [CrossRef]
- Chen, H.; Rosin, F.M.; Prat, S.; Hannapel, D.J. Interacting transcription factors from the three-amino acid loop extension superclass regulate tuber formation. Plant Physiol. 2003, 132, 1391–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Jiang, F.; Wen, J.; Wu, Z. Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum). BMC Plant Biol. 2019, 19, 214. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Yan, J.; Wang, K.; Zhang, W. MiR319-mediated ethylene biosynthesis, signalling and salt stress response in switchgrass. Plant Biotechnol. J. 2019, 17, 2370–2383. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef]
- Yang, J.; Bi, H.P.; Fan, W.J.; Zhang, M.; Wang, H.X.; Zhang, P. Efficient embryogenic suspension culturing and rapid transformation of a range of elite genotypes of sweet potato (Ipomoea batatas [L.] Lam.). Plant Sci. 2011, 181, 701–711. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, R.D.; Jung, H.J.G.; Ralph, J.; Buxton, D.R.; Weimer, P.J. A comparison of the insoluble residues produced by the Klason lignin and acid detergent lignin procedures. J. Sci. Food Agric. 2010, 65, 51–58. [Google Scholar] [CrossRef]
- Abdi, H. The Bonferonni and Šidák Corrections for Multiple Comparisons. Encycl. Meas. Stat. 2007, 1, 1–9. [Google Scholar]
- Ren, L.; Wu, H.; Zhang, T.; Ge, X.; Wang, T.; Zhou, W.; Zhang, L.; Ma, D.; Wang, A. Genome-Wide Identification of TCP Transcription Factors Family in Sweet Potato Reveals Significant Roles of miR319-Targeted TCPs in Leaf Anatomical Morphology. Front. Plant Sci. 2021, 12, 686698. [Google Scholar] [CrossRef]
- Lee, Y.K.; Kim, G.T.; Kim, I.J.; Park, J.; Kwak, S.S.; Choi, G.; Chung, W.I. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 2006, 133, 4305–4314. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.K.; Kim, I.J. Functional conservation of Arabidopsis LNG1 in tobacco relating to leaf shape change by increasing longitudinal cell elongation by overexpression. Genes Genom. 2018, 40, 1053–1062. [Google Scholar] [CrossRef]
- Zou, J.J.; Zheng, Z.Y.; Xue, S.; Li, H.H.; Wang, Y.R.; Le, J. The role of Arabidopsis Actin-Related Protein 3 in amyloplast sedimentation and polar auxin transport in root gravitropism. J. Exp. Bot. 2016, 67, 5325–5337. [Google Scholar] [CrossRef] [Green Version]
- Stortenbeker, N.; Bemer, M. The SAUR gene family: The plant’s toolbox for adaptation of growth and development. J. Exp. Bot. 2019, 70, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, N.; Zhang, F.; Yu, R.; Chen, H.; Deng, X.W.; Wei, N. SAUR17 and SAUR50 Differentially Regulate PP2C-D1 during Apical Hook Development and Cotyledon Opening in Arabidopsis. Plant Cell 2002, 32, 3792–3811. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, J.; Zhang, P.; He, S. Root and Tuber Crops; Encyclopedia of Agriculture and Food Systems; Elsevier: Amsterdam, The Netherlands, 2014; pp. 46–61. [Google Scholar]
- Weng, S.T.; Kuo, Y.W.; King, Y.C.; Lin, H.H.; Tu, P.Y.; Tung, K.S.; Jeng, S.T. Regulation of micoRNA2111 and its target IbFBK in sweet potato on wounding. Plant Sci. 2020, 292, 110391. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S.; Lin, C.C.; Lin, H.H.; Chen, Y.C.; Jeng, S.T. MicroR828 regulates lignin and H2O2 accumulation in sweet potato on wounding. New Phytol. 2012, 196, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.W.; Lin, J.S.; Li, Y.C.; Jhu, M.Y.; King, Y.C.; Jeng, S.T. MicroR408 regulates defense response upon wounding in sweet potato. J. Exp. Bot. 2019, 70, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.F.; Zhao, B.; Huang, C.C.; Chen, Z.W.; Zhao, T.; Liu, H.R.; Hu, G.J.; Shangguan, X.X.; Shan, C.M.; Wang, L.J.; et al. The miR319-Targeted GhTCP4 Promotes the Transition from Cell Elongation to Wall Thickening in Cotton Fiber. Mol. Plant 2020, 13, 1063–1077. [Google Scholar] [CrossRef]
- Xie, Q.; Liu, X.; Zhang, Y.; Tang, J.; Yin, D.; Fan, B.; Zhu, L.; Han, L.; Song, G.; Li, D. Identification and Characterization of microRNA319a and Its Putative Target Gene, PvPCF5, in the Bioenergy Grass Switchgrass (Panicum virgatum). Front. Plant Sci. 2017, 8, 396. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Zhao, X.; Kong, F.; Zuo, Z.; Liu, X. TCP2 positively regulates HY5/HYH and photomorphogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 775–785. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Zhou, X.; Chen, J.; Yin, L.; Zeng, Z.; Xiang, J.; Liu, S. Identification of a consensus DNA-binding site for the TCP domain transcription factor TCP2 and its important roles in the growth and development of Arabidopsis. Mol. Biol. Rep. 2021, 48, 2223–2233. [Google Scholar] [CrossRef]
- Sun, X.; Wang, C.; Xiang, N.; Li, X.; Yang, S.; Du, J.; Yang, Y.; Yang, Y. Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef] [Green Version]
- Vadde, B.V.L.; Challa, K.R.; Sunkara, P.; Hegde, A.S.; Nath, U. The TCP4 Transcription Factor Directly Activates TRICHOMELESS1 and 2 and Suppresses Trichome Initiation. Plant Physiol. 2019, 181, 1587–1599. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Sun, N.; Yang, J.; Deng, Z.; Lan, J.; Qin, G.; He, H.; Deng, X.W.; Irish, V.F.; Chen, H.; et al. The Transcription Factors TCP4 and PIF3 Antagonistically Regulate Organ-Specific Light Induction of SAUR Genes to Modulate Cotyledon Opening during De-Etiolation in Arabidopsis. Plant Cell 2019, 31, 1155–1170. [Google Scholar] [CrossRef] [Green Version]
- Tsukaya, H. Leaf shape: Genetic controls and environmental factors. Int. J. Dev. Biol. 2005, 49, 547–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, S.; Takano, S.; Sato, M.; Furukawa, K.; Nagasawa, H.; Yoshikawa, S.; Kasuga, J.; Tokuji, Y.; Yazaki, K.; Nakazono, M.; et al. Rice Stomatal Closure Requires Guard Cell Plasma Membrane ATP-Binding Cassette Transporter RCN1/OsABCG5. Mol. Plant 2016, 9, 417–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, J.C.; Bonine, C.A.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and Biotic Stresses and Changes in the Lignin Content and Composition in Plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Domingues-Junior, A.P.; Jansen, S.; Choat, B.; Mazzafera, P. Is embolism resistance in plant xylem associated with quantity and characteristics of lignin? Trees 2018, 32, 349–358. [Google Scholar] [CrossRef]
- Bang, S.W.; Lee, D.K.; Jung, H.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Suh, J.W.; Kim, J.K. Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 2019, 17, 118–131. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Tang, W.; Wang, C.; Ge, L.; Sun, J.; Qi, X.; He, Z.; Zhou, Y.; Chen, J.; Xu, Z.; et al. SiMYB56 Confers Drought Stress Tolerance in Transgenic Rice by Regulating Lignin Biosynthesis and ABA Signaling Pathway. Front. Plant Sci. 2020, 11, 785. [Google Scholar] [CrossRef]
- Sun, S.C.; Xiong, X.P.; Zhang, X.L.; Feng, H.J.; Zhu, Q.H.; Sun, J.; Li, Y.J. Characterization of the Gh4CL gene family reveals a role of Gh4CL7 in drought tolerance. BMC Plant Biol. 2020, 20, 125. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, Y.; Wang, C.; Zhao, L.; Jin, Y.; Xing, Q.; Li, M.; Lv, T.; Qi, H. Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance. Plant Mol. Biol. 2020, 103, 689–704. [Google Scholar] [CrossRef]
- Zhao, D.; Luan, Y.; Shi, W.; Zhang, X.; Meng, J.; Tao, J. A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging. Plant Sci. 2021, 303, 110765. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, L.; Zhang, T.; Wu, H.; Ge, X.; Wan, H.; Chen, S.; Li, Z.; Ma, D.; Wang, A. Blocking IbmiR319a Impacts Plant Architecture and Reduces Drought Tolerance in Sweet Potato. Genes 2022, 13, 404. https://doi.org/10.3390/genes13030404
Ren L, Zhang T, Wu H, Ge X, Wan H, Chen S, Li Z, Ma D, Wang A. Blocking IbmiR319a Impacts Plant Architecture and Reduces Drought Tolerance in Sweet Potato. Genes. 2022; 13(3):404. https://doi.org/10.3390/genes13030404
Chicago/Turabian StyleRen, Lei, Tingting Zhang, Haixia Wu, Xinyu Ge, Huihui Wan, Shengyong Chen, Zongyun Li, Daifu Ma, and Aimin Wang. 2022. "Blocking IbmiR319a Impacts Plant Architecture and Reduces Drought Tolerance in Sweet Potato" Genes 13, no. 3: 404. https://doi.org/10.3390/genes13030404
APA StyleRen, L., Zhang, T., Wu, H., Ge, X., Wan, H., Chen, S., Li, Z., Ma, D., & Wang, A. (2022). Blocking IbmiR319a Impacts Plant Architecture and Reduces Drought Tolerance in Sweet Potato. Genes, 13(3), 404. https://doi.org/10.3390/genes13030404





