Comparative Genome Analysis of Two Bacillus pumilus Strains Producing High Level of Extracellular Hydrolases
Abstract
:1. Introduction
2. Materials and Methods
2.1. B. pumilus Strains and Total DNA Extraction
2.2. Genome Sequencing and Assembly
2.3. Genome Annotation and Comparative Analysis
2.4. Plasmid and Phage Regions Prediction
2.5. Classification of Proteases Family
2.6. Variants Calling
3. Results and Discussion
3.1. Genome Assembly of B. pumilus Strains 7P and 3-19
3.2. Phylogenetic Classification of B. pumilus Strains 7P and 3-19
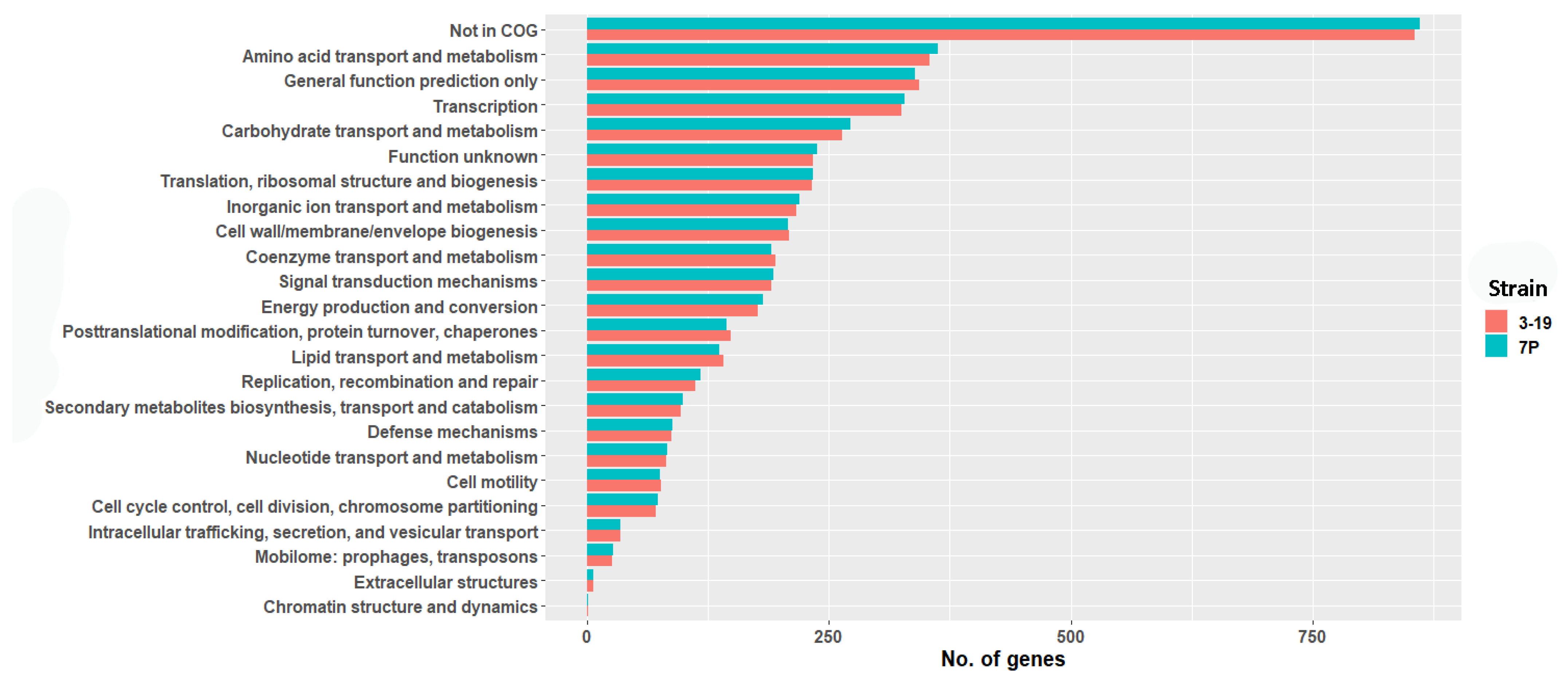
3.3. Functional Prediction of the Annotated Genes
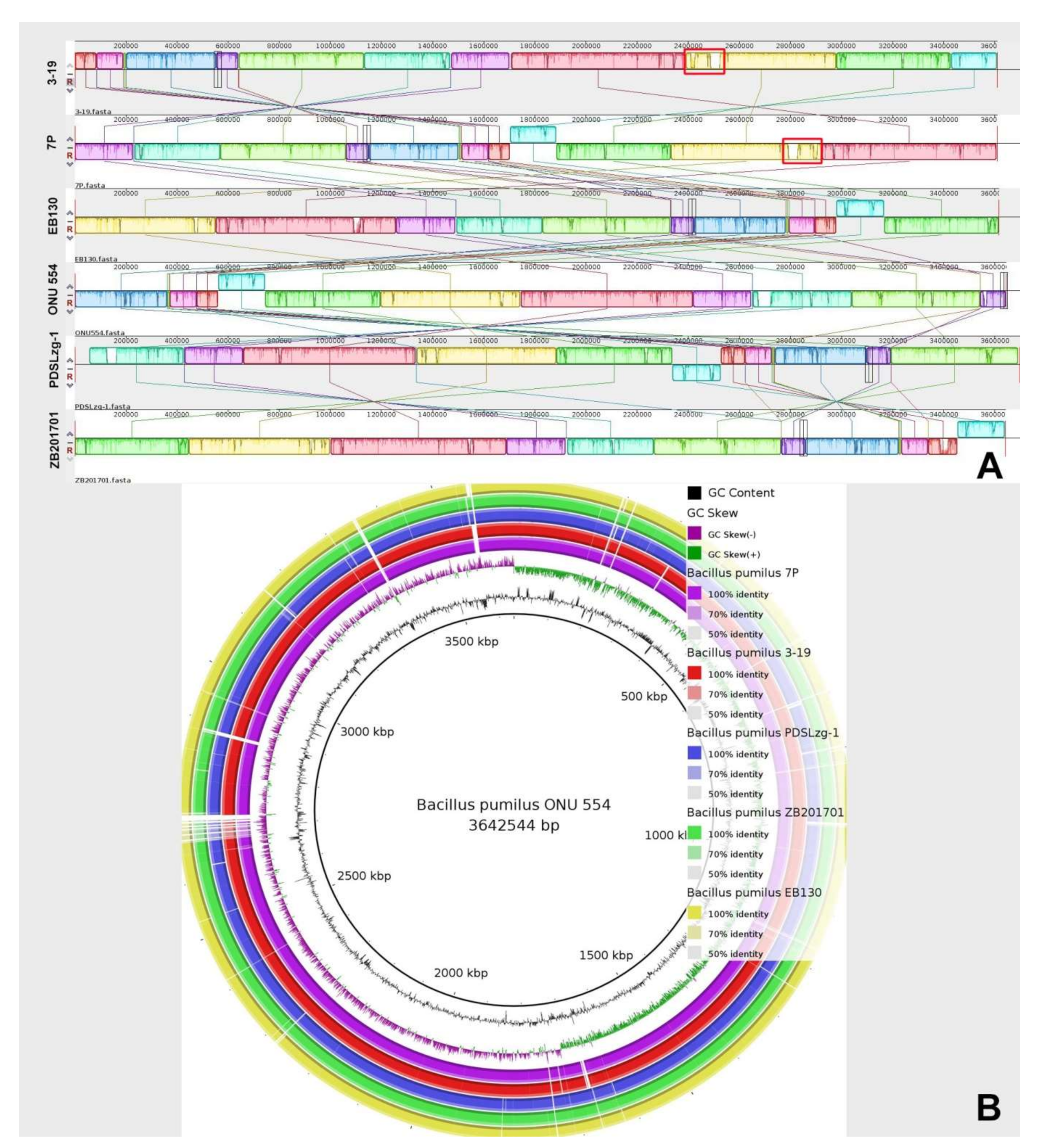
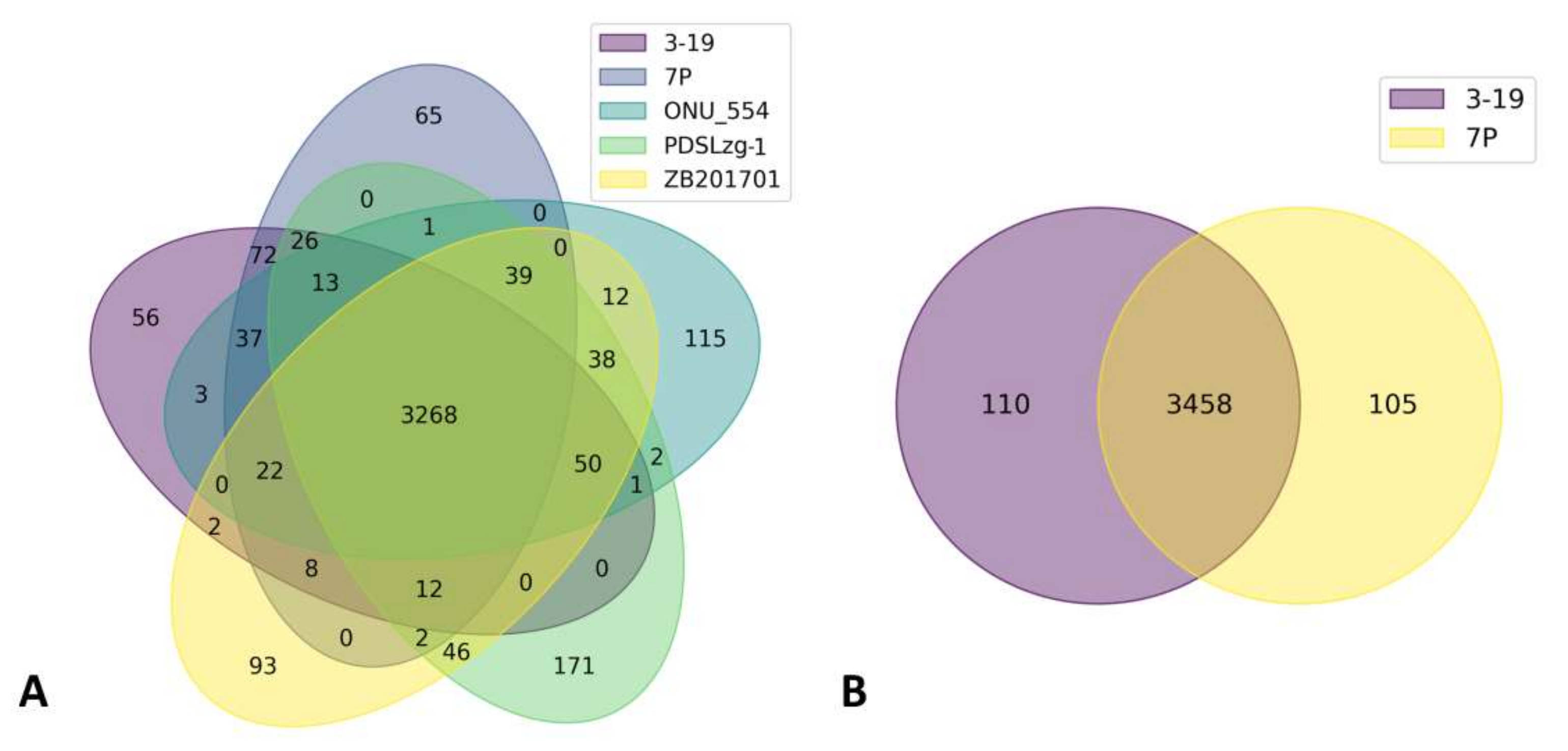
3.4. Comparative Genomic Analysis of B. pumilus Strains
3.5. Genome-Based Identification of Prophage Regions
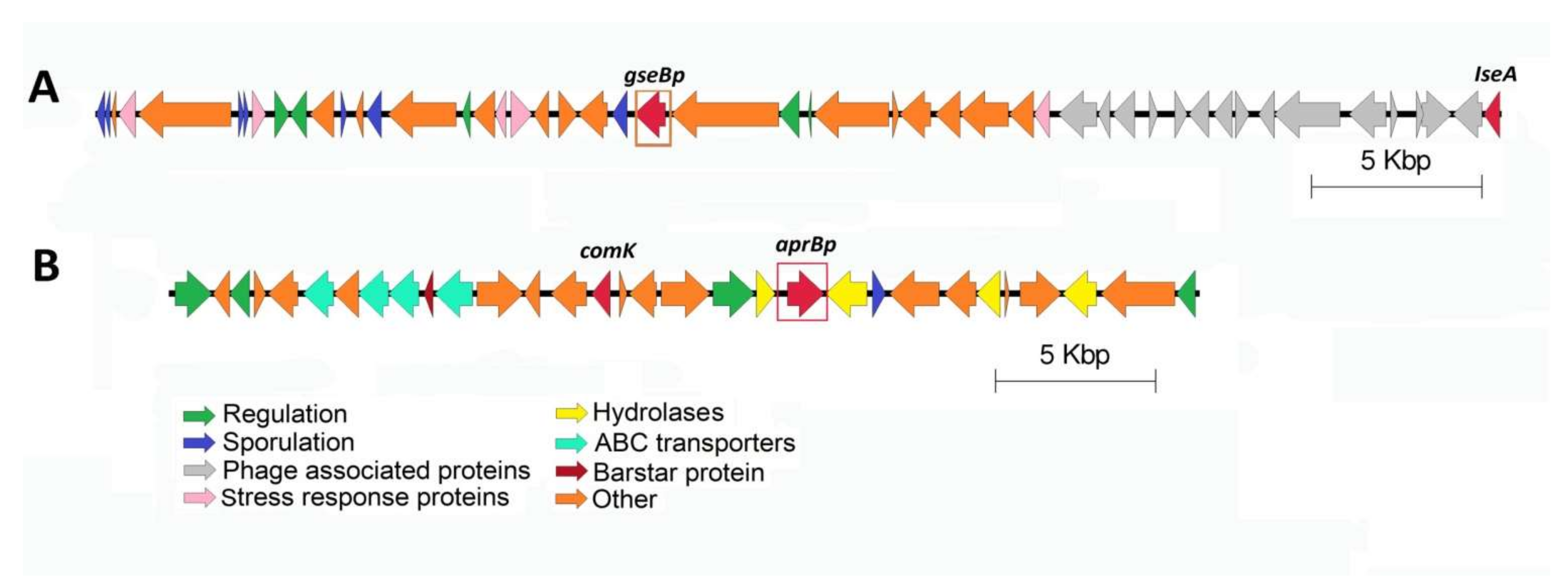
3.6. Genomic Analysis of B. pumilus Proteases and Protease Inhibitors
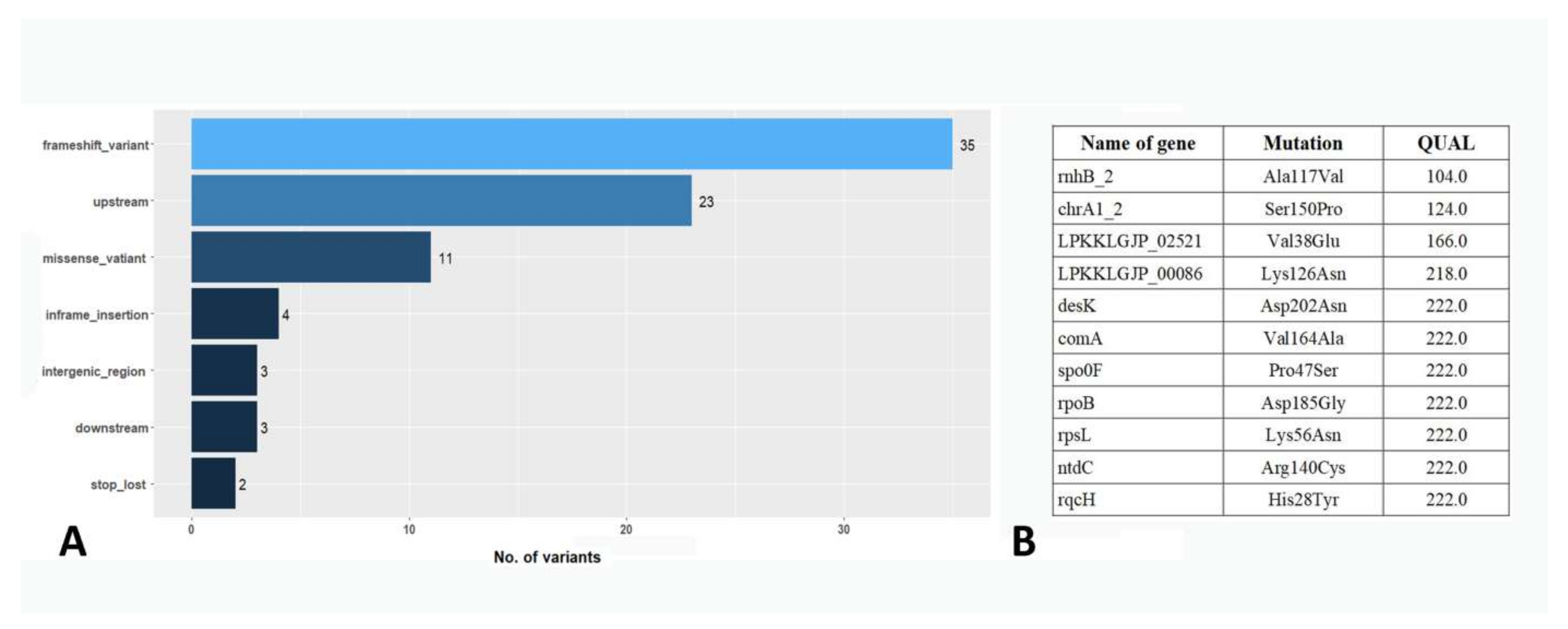
3.7. Analysis of Single-Nucleotide Polymorphisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erkmen, O.; Bozoglu, T.F. (Eds.) Foodborne intoxications. In Food Microbiology Principles into Practice; Volume 1: Microorganisms Related to Foods, Foodborne Diseases and Food Spoilage; John Wiley and Sons, Ltd.: Oxford, UK; Chicester, UK, 2016; pp. 186–207. [Google Scholar]
- Zhang, Z.; Yin, L.; Li, X.; Zhang, C.; Zou, H.; Liu, C.; Wu, Z. Analyses of the Complete Genome Sequence of the Strain Bacillus pumilus ZB201701 Isolated from Rhizosphere Soil of Maize under Drought and Salt Stress. Microbes Environ. 2019, 34, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Ettoumi, B.; Raddadispi, N.; Borin, S.; Daffonchio, D.; Boudabous, A.; Cherif, A. Diversity and phylogeny of culturable spore-forming Bacilli isolated from marine sediments. J. Basic Microbiol. 2009, 49, S13–S23. [Google Scholar] [CrossRef] [PubMed]
- Vaishampayan, P.A.; Rabbow, E.; Horneck, G.; Venkateswaran, K.J. Survival of Bacillus pumilus spores for a prolonged period of time in real space conditions. Astrobiology 2012, 12, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Tewari, A.; Abdullah, S. Bacillus cereus food poisoning: International and Indian perspective. J. Food Sci. Technol. 2015, 52, 2500. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, V.M. You Can’t B. cereus—A Review of Bacillus cereus Strains That Cause Anthrax-Like Disease. Front. Microbiol. 2020, 11, 1731. [Google Scholar] [CrossRef]
- Danilova, I.; Sharipova, M. The Practical Potential of Bacilli and Their Enzymes for Industrial Production. Front. Microbiol. 2020, 11, 1782. [Google Scholar] [CrossRef]
- Reyes-cortes, J.L.; Azaola-espinosa, A.; Lozano-aguirre, L.; Ponce-alquicira, E. Physiological and Genomic Analysis of Bacillus pumilus UAMX Isolated from the Gastrointestinal Tract of Overweight Individuals. Microorganisms 2021, 9, 1076. [Google Scholar] [CrossRef]
- Hao, K.; Li, H.; Li, F.; Guo, P. Complete Genome Sequence of Bacillus pumilus PDSLzg-1, a Hydrocarbon-Degrading Bacterium Isolated from Oil-Contaminated Soil in China. Genome Announc. 2016, 4, e01079-16. [Google Scholar] [CrossRef] [Green Version]
- Bulgakova, R.S.; Leshchinskaya, I.B.; Balaban, N.P.; Egorova, G.S. The principal biochemical properties of highly purified Bacillus intermedius extracellular RNAase. Biochemistry 1974, 39, 245–247. [Google Scholar] [CrossRef]
- Koryagina, A.O.; Rudakova, N.L.; Smolentsev, S.Y.; Khadieva, G.F.; Lutfullin, M.T.; Mardanova, A.M.; Sharipova, M.R.; Org, P. Effect of bacterial serine proteinase on growth performance and nutrient digestibility in broilers. Int. J. Res. Pharm. Sci. 2021, 12, 262–273. [Google Scholar] [CrossRef]
- Hosaka, T.; Tamehiro, N.; Chumpolkulwong, N.; Hori-Takemoto, C.; Shirouzu, M.; Yokoyama, S.; Ochi, K. The novel mutation K87E in ribosomal protein S12 enhances protein synthesis activity during the late growth phase in Escherichia coli. Mol. Genet. Genom. 2004, 271, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hosaka, T.; Ochi, K. Dramatic Activation of Antibiotic Production in Streptomyces coelicolor by Cumulative Drug Resistance Mutations. Appl. Environ. Microbiol. 2008, 74, 2834–2840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelenikhin, P.; Pukhovskaya, V.; Garipov, A.; Makeeva, A.; Sokolova, E.; Ilinskaya, O. Obvious and Hidden Reasons of Breast Cancer Cell Sensitivity to Antitumor RNase. Bionanoscience 2016, 6, 528–533. [Google Scholar] [CrossRef]
- Itskovich, E.L.; Liutova, L.I.; Balaban, N.P.; Mardanova, A.M.; Shakirov, E.V.; Sharipova, M.R.; Leshchinskaia, I.B.; Rudenskaia, G.N. Thrombolytic and anticoagulant properties of thiol-dependent proteinase from Bacillus intermedius 3–19. Vopr. Med. Khim. 1998, 44, 288–291. [Google Scholar]
- Danilova, Y.V.; Shagimardanova, E.I.; Margulis, A.B.; Toymentseva, A.A.; Balaban, N.P.; Rudakova, N.L.; Rizvanov, A.A.; Sharipova, M.R.; Palotás, A. Bacterial enzymes effectively digest Alzheimer’s β-amyloid peptide. Brain Res. Bull. 2014, 108, 113–117. [Google Scholar] [CrossRef]
- Mitrofanova, O.; Mardanova, A.; Evtugyn, V.; Bogomolnaya, L.; Sharipova, M. Effects of Bacillus Serine Proteases on the Bacterial Biofilms. BioMed Res. Int. 2017, 2017, 8525912. [Google Scholar] [CrossRef] [Green Version]
- Karimi, M.; Behzadian, F.; Rouhaninejad, H.; Yari, S. A Feasibility Study to Evaluate Bacillus subtilis as a Host for Producing Recombinant Human Parathyroid Hormone. Avicenna J. Med. Biotechnol. 2018, 10, 147. [Google Scholar]
- Wright, M.H.; Adelskov, J.; Greene, A.C. Bacterial DNA Extraction Using Individual Enzymes and Phenol/Chloroform Separation. J. Microbiol. Biol. Educ. 2017, 18, 18.2.48. [Google Scholar] [CrossRef] [Green Version]
- Shagimardanova, E.I.; Toymentseva, A.A.; Balaban, N.P.; Mardanova, A.M.; Danilova, Y.V.; Gusev, O.A.; Kostryukova, E.; Karpova, I.; Manolov, A.; Alexeev, D.; et al. Draft Genome Sequence of Bacillus pumilus 7P, Isolated from the Soil of the Tatarstan Republic, Russia. Genome Announc. 2014, 2, e00599-14. [Google Scholar] [CrossRef] [Green Version]
- Ulyanova, V.; Shah Mahmud, R.; Dudkina, E.; Vershinina, V.; Ilinskaya, O. Draft Whole Genome Sequence of Bacillus pumilus Strain 3–19, a Chemical Mutant Overproducing Extracellular Ribonuclease. Genome Announc. 2014, 2, e00724-14. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.M.A.; Markowitz, V.M.; Chu, K.; Palaniappan, K.; Szeto, E.; Pillay, M.; Ratner, A.; Huang, J.; Andersen, E.; Huntemann, M.; et al. IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017, 45, D507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Lechner, M.; Findeiß, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Bateman, A. How to use the MEROPS database and website to help understand peptidase specificity. Protein Sci. 2021, 30, 83. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; O Pollard, M.; Whitwham, A.; Keane, T.; A McCarthy, S.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Danilova, I.V.; Toymentseva, A.A.; Baranova, D.S.; Sharipova, M.R. The Genetic Mechanism of Resistance to Antibiotics in Bacillus pumilus 3-19 Strain. BioNanoScience 2016, 7, 88–91. [Google Scholar] [CrossRef]
- Sharipova, M.R.; Toymentseva, A.A.; Sabirova, A.R.; Mukhametzyanova, A.D.; Akhmetova, A.I.; Mardanova, A.M.; Balaban, N.P. A new phylogenetic position of strain Bacillus intermedius 3–19. Microbiology 2011, 80, 432–435. [Google Scholar] [CrossRef]
- Fu, X.; Gong, L.; Liu, Y.; Lai, Q.; Li, G.; Shao, Z. Bacillus pumilus Group Comparative Genomics: Toward Pangenome Features, Diversity, and Marine Environmental Adaptation. Front. Microbiol. 2021, 12, 571212. [Google Scholar] [CrossRef]
- Benardini, J.N.; Sawyer, J.; Venkateswaran, K.; Nicholson, W.L. Spore UV and acceleration resistance of endolithic Bacillus pumilus and Bacillus subtilis isolates obtained from Sonoran desert basalt: Implications for lithopanspermia. Astrobiology 2003, 3, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; de Souza, R.F.; Anantharaman, V.; Iyer, L.M.; Aravind, L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 2012, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaundal, S.; Deep, A.; Kaur, G.; Thakur, K.G. Molecular and Biochemical Characterization of YeeF/YezG, a Polymorphic Toxin-Immunity Protein Pair from Bacillus subtilis. Front. Microbiol. 2020, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- d’Adda di Fagagna, F.; Weller, G.R.; Doherty, A.J.; Jackson, S.P. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 2003, 4, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bose, B.; Auchtung, J.M.; Lee, C.A.; Grossman, A.D. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol. Microbiol. 2008, 70, 570–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minnullina, L.; Pudova, D.; Shagimardanova, E.; Shigapova, L.; Sharipova, M.; Mardanova, A. Comparative genome analysis of uropathogenic morganella morganii strains. Front. Cell. Infect. Microbiol. 2019, 9, 167. [Google Scholar] [CrossRef]
- Dhaese, P.; Dobbelaere, M.R.; Van Montagu, M. The temperate B. subtilis phage ϕ105 genome contains at least two distinct regions encoding superinfection immunity. Mol. Genet. Genom. 1985, 200, 490–492. [Google Scholar] [CrossRef]
- Birdsell, D.C.; Hathaway, G.M.; Rutberg, L. Characterization of Temperate Bacillus Bacteriophage φ105. J. Virol. 1969, 4, 264–270. [Google Scholar] [CrossRef] [Green Version]
- Rutberg, L.; Hoch, J.A.; Spizizen, J. Mechanism of Transfection with Deoxyribonucleic Acid from the Temperate Bacillus Bacteriophage d105. J. Virol. 1969, 4, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Garro, A.J.; Law, M.F. Relationship between lysogeny, spontaneous induction, and transformation efficiencies in Bacillus subtilis. J. Bacteriol. 1974, 120, 1256–1259. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.E.; Walsh, T.R. Toxin–antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 2017, 41, 343. [Google Scholar] [CrossRef] [PubMed]
- Brantl, S.; Müller, P. Toxin–Antitoxin Systems in Bacillus subtilis. Toxins 2019, 11, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Wang, H.; Wang, B.; Pan, L. Efficient production of extracellular pullulanase in Bacillus subtilis ATCC6051 using the host strain construction and promoter optimization expression system. Microb. Cell Fact. 2018, 17, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional Enzymes in Life and Disease. J. Biol. Chem. 2008, 283, 30433. [Google Scholar] [CrossRef] [Green Version]
- Sabirova, A.R.; Rudakova, N.L.; Balaban, N.P.; Ilyinskaya, O.N.; Demidyuk, I.V.; Kostrov, S.V.; Rudenskaya, G.N.; Sharipova, M.R. A novel secreted metzincin metalloproteinase from Bacillus intermedius. FEBS Lett. 2010, 584, 4419–4425. [Google Scholar] [CrossRef] [Green Version]
- Sharipova, M.; Balaban, N.; Kayumov, A.; Kirillova, Y.; Mardanova, A.; Gabdrakhmanova, L.; Leshchinskaya, I.; Rudenskaya, G.; Akimkina, T.; Safina, D.; et al. The expression of the serine proteinase gene of Bacillus intermedius in Bacillus subtilis. Microbiol. Res. 2008, 163, 39–50. [Google Scholar] [CrossRef]
- Sharipova, M.R.; Shagimardanova, E.I.; Chastukhina, I.B.; Shamsutdinov, T.R.; Balaban, N.P.; Mardanova, A.M.; Rudenskaya, G.N.; Demidyuk, I.V.; Kostrov, S.V. The expression of Bacillus intermedius glutamyl endopeptidase gene in Bacillus subtilis recombinant strains. Mol. Biol. Rep. 2007, 34, 79–87. [Google Scholar] [CrossRef]
- Sharipova, M.R.; Mardanova, A.M.; Rudakova, N.L.; Pudova, D.S. Bistability and Formation of the Biofilm Matrix as Adaptive Mechanisms during the Stationary Phase of Bacillus subtilis. Microbiology 2021, 90, 20–36. [Google Scholar] [CrossRef]
- Fujita, M.; Losick, R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005, 19, 2236–2244. [Google Scholar] [CrossRef] [Green Version]
- Salzberg, L.I.; Helmann, J.D. An antibiotic-inducible cell wall-associated protein that protects Bacillus subtilis from autolysis. J. Bacteriol. 2007, 189, 4671–4680. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Hashimoto, M.; Higashitsuji, Y.; Harada, H.; Hariyama, N.; Takahashi, L.; Iwashita, T.; Ooiwa, S.; Sekiguchi, J. Post-translational control of vegetative cell separation enzymes through a direct interaction with specific inhibitor IseA in Bacillus subtilis. Mol. Microbiol. 2008, 70, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, N.; Parashar, V.; Baker, M.D.; Dubnau, D.A.; Neiditch, M.B. An Atypical Phr Peptide Regulates the Developmental Switch Protein RapH. J. Bacteriol. 2011, 193, 6197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, A.; Olmos, J. Bacillus subtilis transcriptional regulators interaction. Biotechnol. Lett. 2004, 26, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, B.; Rao, L.; Wang, X.; Zhao, H.; Li, M.; Yu, F. Molecular Characteristics of Rifampin-Sensitive and -Resistant Isolates and Characteristics of rpoB Gene Mutations in Methicillin-Resistant Staphylococcus aureus. Infect. Drug Resist. 2021, 4, 4591–4600. [Google Scholar] [CrossRef]
- Wolf, D.; Rippa, V.; Mobarec, J.C.; Sauer, P.; Adlung, L.; Kolb, P.; Bischofs, I.B. The quorum-sensing regulator ComA from Bacillus subtilis activates transcription using topologically distinct DNA motifs. Nucleic Acids Res. 2016, 44, 2160–2172. [Google Scholar] [CrossRef] [PubMed]
- Toymentseva, A.A.; Mascher, T.; Sharipova, M.R. Regulatory Characteristics of Bacillus pumilus Protease Promoters. Curr. Microbiol. 2017, 74, 550–559. [Google Scholar] [CrossRef]


| Strains | Year Isolated | Location Isolated | Description | GenBank Accession Number | Total Length (bp) | No. of ORFs | Coverage |
|---|---|---|---|---|---|---|---|
| B. pumilus 7P | 1974 | Russia | Soil isolate | CP058911.1 | 3,609,117 | 3662 | 42.0× |
| B. pumilus 3-19 | 1994 | Russia | Derivative of 7P | CP054310.1 | 3,609,444 | 3679 | 50.0× |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pudova, D.S.; Toymentseva, A.A.; Gogoleva, N.E.; Shagimardanova, E.I.; Mardanova, A.M.; Sharipova, M.R. Comparative Genome Analysis of Two Bacillus pumilus Strains Producing High Level of Extracellular Hydrolases. Genes 2022, 13, 409. https://doi.org/10.3390/genes13030409
Pudova DS, Toymentseva AA, Gogoleva NE, Shagimardanova EI, Mardanova AM, Sharipova MR. Comparative Genome Analysis of Two Bacillus pumilus Strains Producing High Level of Extracellular Hydrolases. Genes. 2022; 13(3):409. https://doi.org/10.3390/genes13030409
Chicago/Turabian StylePudova, Daria S., Anna A. Toymentseva, Natalia E. Gogoleva, Elena I. Shagimardanova, Ayslu M. Mardanova, and Margarita R. Sharipova. 2022. "Comparative Genome Analysis of Two Bacillus pumilus Strains Producing High Level of Extracellular Hydrolases" Genes 13, no. 3: 409. https://doi.org/10.3390/genes13030409






