Potential Refinement of Recurrence Score by pSTAT3 Status
Abstract
:1. Introduction
2. Methods
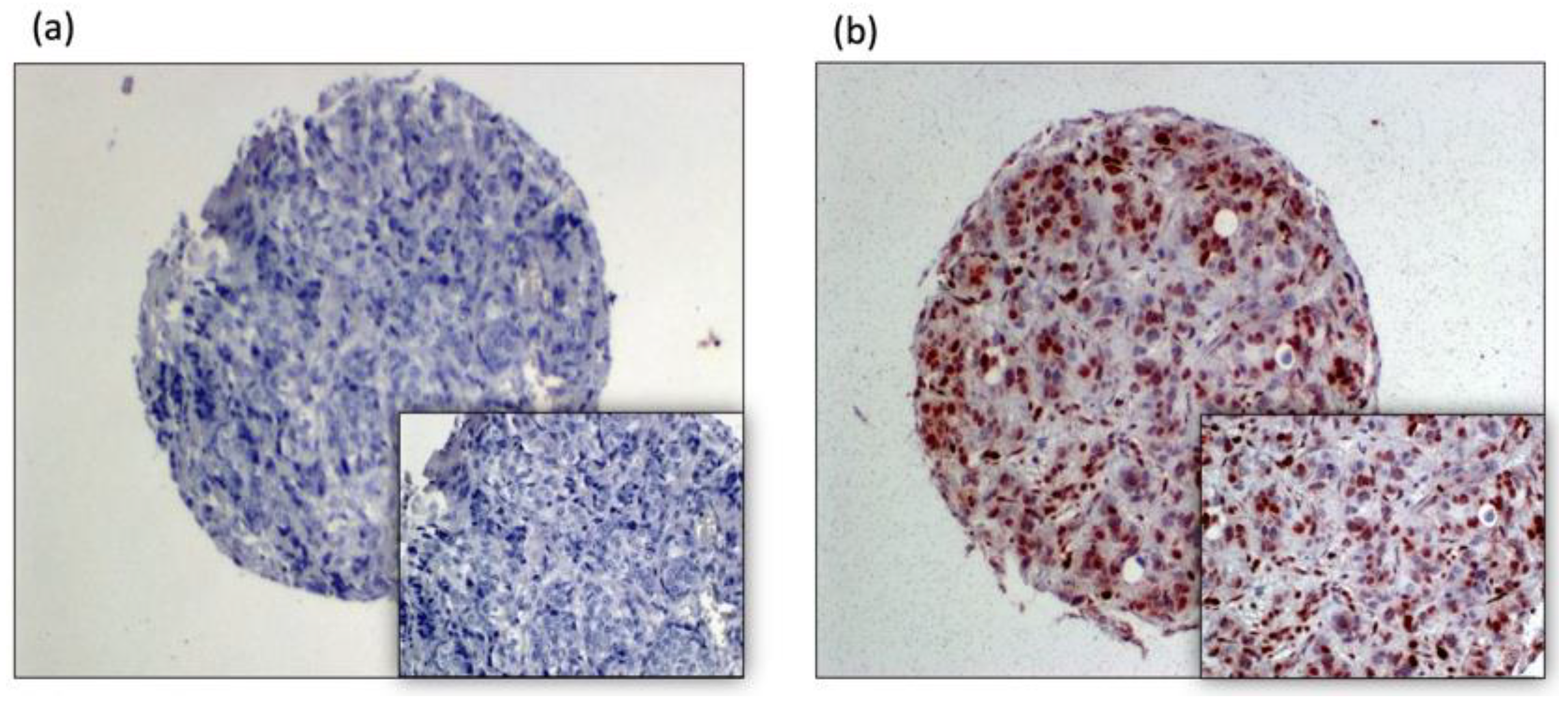
2.1. Tissue Analysis:
2.2. Clinical Data
2.3. Statistical Analysis
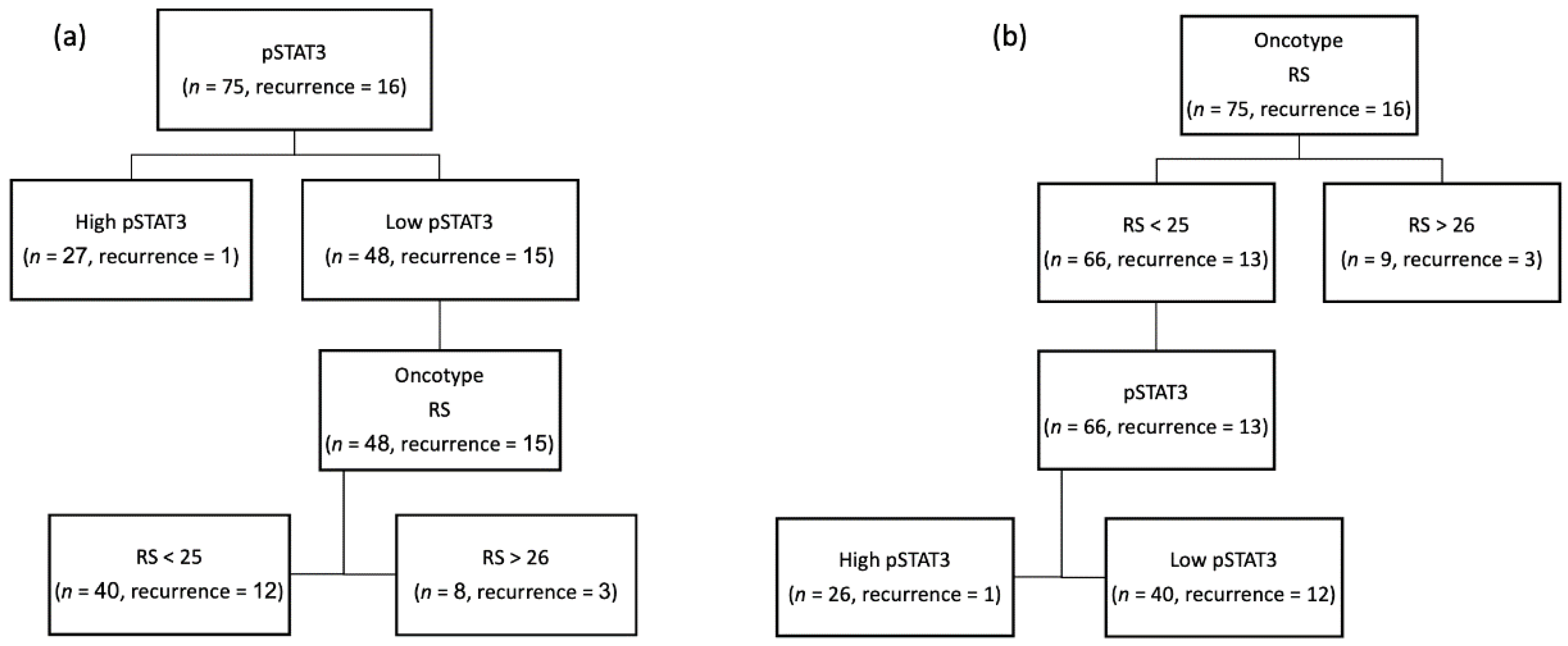
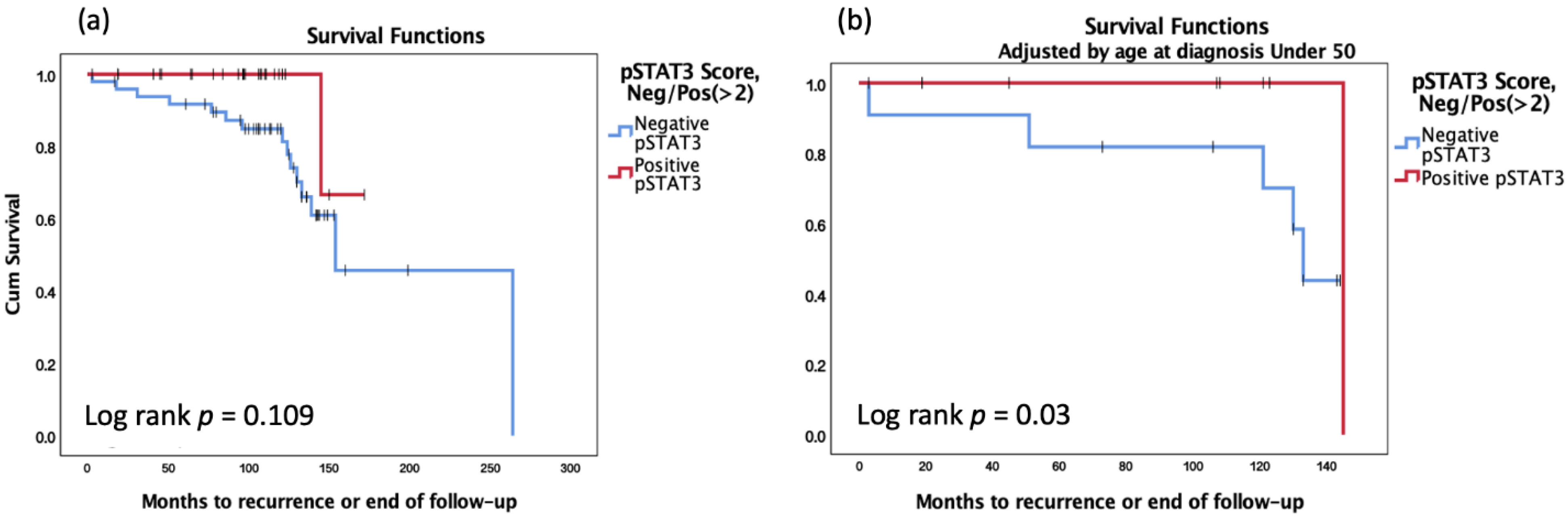
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, J.J.; Roth, J.A. The impact of the Oncotype Dx breast cancer assay in clinical practice: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2013, 141, 13–22. [Google Scholar] [CrossRef]
- Tiberi, D.; Masucci, L.; Shedid, D.; Roy, I.; Vu, T.; Patocskai, E.; Robidoux, A.; Wong, P. Limitations of Personalized Medicine and Gene Assays for Breast Cancer. Cureus 2017, 9, e1100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnenblick, A.; Uziely, B.; Nechushtan, H.; Kadouri, L.; Galun, E.; Axelrod, J.H.; Katz, D.; Daum, H.; Hamburger, T.; Maly, B.; et al. Tumor STAT3 tyrosine phosphorylation status, as a predictor of benefit from adjuvant chemotherapy for breast cancer. Breast Cancer Res. Treat. 2013, 138, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Sonnenblick, A.; Salgado, R.; Brohée, S.; Zahavi, T.; Peretz, T.; Eynden, G.V.D.; Rouas, G.; Salmon, A.; Francis, P.A.; Di Leo, A.; et al. p-STAT3 in luminal breast cancer: Integrated RNA-protein pooled analysis and results from the BIG 2-98 phase III trial. Int. J. Oncol. 2017, 52, 424–432. [Google Scholar] [CrossRef]
- Zhao, Y.; Schaafsma, E.; Cheng, C. Gene signature-based prediction of triple-negative breast cancer patient response to Neoadjuvant chemotherapy. Cancer Med. 2020, 9, 6281–6295. [Google Scholar] [CrossRef]
- Niu, G.; Wright, K.L.; Huang, M.; Song, L.; Haura, E.; Turkson, J.; Zhang, S.; Wang, T.; Sinibaldi, D.; Coppola, D.; et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 2002, 21, 2000–2008. [Google Scholar] [CrossRef] [Green Version]
- Garcia, R.; Bowman, T.L.; Niu, G.; Yu, H.; Minton, S.; Muro-Cacho, C.A.; Cox, C.E.; Falcone, R.; Fairclough, R.; Parsons, S.; et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 2001, 20, 2499–2513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Lee, H.; Herrmann, A.; Buettner, R.; Jove, R. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Cancer 2014, 14, 736–746. [Google Scholar] [CrossRef]
- Knüpfer, H.; Preiß, R. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res. Treat. 2006, 102, 129–135. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Sonnenblick, A.; Shriki, A.; Galun, E.; Axelrod, J.H.; Daum, H.; Rottenberg, Y.; Hamburger, T.; Mali, B.; Peretz, T. Tissue microarray-based study of patients with lymph node-positive breast cancer shows tyrosine phosphorylation of signal transducer and activator of transcription 3 (tyrosine705-STAT3) is a marker of good prognosis. Clin. Transl. Oncol. 2012, 14, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.R.; Nelson, E.A.; Yeh, J.E.; Pinello, L.; Yuan, G.-C.; Frank, D.A. STAT5 Outcompetes STAT3 to Regulate the Expression of the Oncogenic Transcriptional Modulator BCL6. Mol. Cell. Biol. 2013, 33, 2879–2890. [Google Scholar] [CrossRef] [Green Version]
- Dolled-Filhart, M.; Camp, R.L.; Kowalski, D.P.; Smith, B.L.; Rimm, D.L. Tissue Microarray Analysis of Signal Transducers and Activators of Transcription 3 (Stat3) and Phospho-Stat3 (Tyr705) in Node-negative Breast Cancer Shows Nuclear Localization Is Associated with a Better Prognosis. Clin. Cancer Res. 2003, 9, 594–600. [Google Scholar] [PubMed]
- Walker, S.R.; Xiang, M.; Frank, D.A. Distinct roles of STAT3 and STAT5 in the pathogenesis and targeted therapy of breast cancer. Mol. Cell. Endocrinol. 2014, 382, 616–621. [Google Scholar] [CrossRef] [Green Version]
- Sonnenblick, A.; Brohée, S.; Fumagalli, D.; Vincent, D.; Venet, D.; Ignatiadis, M.; Salgado, R.; Eynden, G.V.D.; Rothé, F.; Desmedt, C.; et al. Constitutive phosphorylated STAT3-associated gene signature is predictive for trastuzumab resistance in primary HER2-positive breast cancer. BMC Med. 2015, 13, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohrherr, J.; Uras, I.Z.; Moll, H.P.; Casanova, E. STAT3: Versatile Functions in Non-Small Cell Lung Cancer. Cancers 2020, 12, 1107. [Google Scholar] [CrossRef]
- Lieblein, J.C.; Ball, S.; Hutzen, B.; Sasser, A.K.; Lin, H.-J.; Huang, T.H.; Hall, B.M.; Lin, J. STAT3 can be activated through paracrine signaling in breast epithelial cells. BMC Cancer 2008, 8, 302–314. [Google Scholar] [CrossRef] [Green Version]
- Marotta, L.L.; Almendro, V.; Marusyk, A.; Shipitsin, M.; Schemme, J.; Walker, S.R.; Bloushtain-Qimron, N.; Kim, J.J.; Choudhury, S.A.; Maruyama, R.; et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J. Clin. Investig. 2011, 121, 2723–2735. [Google Scholar] [CrossRef]
- Dien, J.; Amin, H.M.; Chiu, N.; Wong, W.; Frantz, C.; Chiu, B.; Mackey, J.R.; Lai, R. Signal Transducers and Activators of Transcription-3 Up-Regulates Tissue Inhibitor of Metalloproteinase-1 Expression and Decreases Invasiveness of Breast Cancer. Am. J. Pathol. 2006, 169, 633–642. [Google Scholar] [CrossRef] [Green Version]
- Chapman, R.S.; Lourenco, P.C.; Tonner, E.; Flint, D.J.; Selbert, S.; Takeda, K.; Akira, S.; Clarke, A.; Watson, C.J. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 1999, 13, 2604–2616. [Google Scholar] [CrossRef] [Green Version]
- Sarosiek, K.A.; Malumbres, R.; Nechushtan, H.; Gentles, A.J.; Avisar, E.; Lossos, I.S. Novel IL-21 signaling pathway up-regulates c-Myc and induces apoptosis of diffuse large B-cell lymphomas. Blood 2010, 115, 570–580. [Google Scholar] [CrossRef]
- Couto, J.P.; Daly, L.; Almeida, A.; Knauf, J.A.; Fagin, J.A.; Sobrinho-Simoes, M.; Lima, J.; Maximo, V.; Soares, P.; Lyden, D.; et al. STAT3 negatively regulates thyroid tumorigenesis. Proc. Natl. Acad. Sci. 2012, 109, E2361–E2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kim, J.C.K.; Lee, S.-E.; Quinley, C.; Kim, H.R.; Herdman, S.; Corr, M.; Raz, E. Signal Transducer and Activator of Transcription 3 (STAT3) Protein Suppresses Adenoma-to-carcinoma Transition in Apcmin/+ Mice via Regulation of Snail-1 (SNAI) Protein Stability. J. Biol. Chem. 2012, 287, 18182–18189. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Huang, J.; Li, W.; Chen, Y.; Liu, X.; Wang, J. Meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with breast cancer. Oncotarget 2018, 9, 13060–13067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segatto, I.; Baldassarre, G.; Belletti, B. STAT3 in Breast Cancer Onset and Progression: A Matter of Time and Context. Int. J. Mol. Sci. 2018, 19, 2818. [Google Scholar] [CrossRef] [Green Version]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| pSTAT3 Score, Negative (<2)/Positive (≥2) | ||||||
|---|---|---|---|---|---|---|
| Variable | n = 75 | |||||
| Sub-Variable | Negative | Positive | p Value | |||
| (n = 49) | n% | (n = 26) | n% | |||
| Age at diagnosis | Over 50 | 38 | 77.6% | 18 | 69.2% | 0.43 |
| Under 50 | 11 | 22.4% | 8 | 30.8% | ||
| Tumor type | Ductal | 35 | 71.4% | 17 | 65.4% | 0.966 |
| Lobular | 5 | 10.2% | 3 | 11.5% | ||
| Mucinous | 1 | 2.0% | 1 | 3.8% | ||
| Both Ductal and Lobular | 8 | 16.3% | 5 | 19.2% | ||
| Lymph Nodes | No | 38 | 77.6% | 20 | 76.9% | 0.951 |
| Yes | 11 | 22.4% | 6 | 23.1% | ||
| Chemotherapy | No | 33 | 67.3% | 20 | 76.9% | 0.357 |
| Yes | 16 | 32.7% | 6 | 23.1% | ||
| Hormone therapy | No | 6 | 12.2% | 2 | 7.7% | 0.706 |
| Yes | 43 | 87.8% | 24 | 92.3% | ||
| RS score | ≤26 | 41 | 83.7% | 25 | 96.2% | 0.15 |
| >26 | 8 | 10.3% | 1 | 3.8% | ||
| Recurrence | No | 34 | 69.4% | 25 | 96.2% | 0.007 |
| Yes | 15 | 30.6% | 1 | 3.8% | ||
| Age | Median (range) | 61.39 (29.7–81) | 57.91 (38.6–77.9) | 0.963 | ||
| Mean ± SD | 58.01 ± 12.07 | 57.88 ± 10.68 | ||||
| Multivariate Analysis of the Clinical and Pathological Data for DFS | ||||
|---|---|---|---|---|
| Adjusted HR | 95.0% CI for HR | p Value | ||
| Lower | Upper | |||
| RS score, Neg/Pos (>26) | 0.548 | 0.089 | 3.394 | 0.518 |
| Lymph Nodes | 2.709 | 0.405 | 18.119 | 0.304 |
| Type: Ductal/Lobular/Mucinous/Ductal & Lobular | 0.024 | |||
| Type: Ductal vs. Lobular | 14.367 | 1.261 | 163.673 | 0.032 |
| Type: Ductal vs. Mucinous | 39.831 | 2.747 | 577.569 | 0.007 |
| Type: Ductal vs. Ductal and Lobular | 3.839 | 0.893 | 16.502 | 0.071 |
| Chemotherapy | 0.556 | 0.118 | 2.623 | 0.458 |
| Hormone therapy | 1.946 | 0.302 | 12.532 | 0.483 |
| Tumor Size (cm) | 0.981 | 0.438 | 2.198 | 0.963 |
| pSTAT3 Score, Negative/Positive (≥2) | 5.462 | 0.584 | 51.121 | 0.137 |
| Age at diagnosis under 50 | 5.778 | 1.092 | 30.560 | 0.039 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grinshpun, A.; Cohen, Y.; Zick, A.; Kadouri, L.; Hamburger, T.; Nisman, B.; Allweis, T.M.; Oprea, G.; Peretz, T.; Uziely, B.; et al. Potential Refinement of Recurrence Score by pSTAT3 Status. Genes 2022, 13, 438. https://doi.org/10.3390/genes13030438
Grinshpun A, Cohen Y, Zick A, Kadouri L, Hamburger T, Nisman B, Allweis TM, Oprea G, Peretz T, Uziely B, et al. Potential Refinement of Recurrence Score by pSTAT3 Status. Genes. 2022; 13(3):438. https://doi.org/10.3390/genes13030438
Chicago/Turabian StyleGrinshpun, Albert, Yogev Cohen, Aviad Zick, Luna Kadouri, Tamar Hamburger, Benjamin Nisman, Tanir M. Allweis, Gabriela Oprea, Tamar Peretz, Beatrice Uziely, and et al. 2022. "Potential Refinement of Recurrence Score by pSTAT3 Status" Genes 13, no. 3: 438. https://doi.org/10.3390/genes13030438
APA StyleGrinshpun, A., Cohen, Y., Zick, A., Kadouri, L., Hamburger, T., Nisman, B., Allweis, T. M., Oprea, G., Peretz, T., Uziely, B., & Sonnenblick, A. (2022). Potential Refinement of Recurrence Score by pSTAT3 Status. Genes, 13(3), 438. https://doi.org/10.3390/genes13030438






