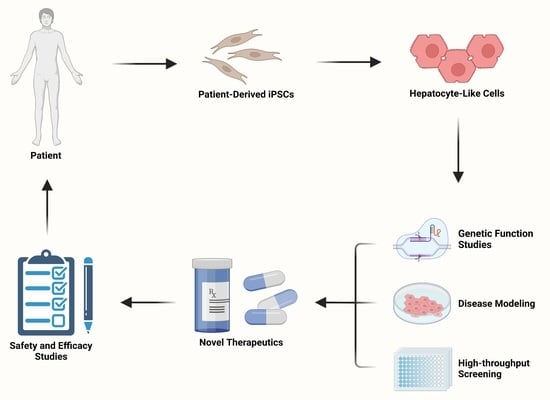
Advancements in Disease Modeling and Drug Discovery Using iPSC-Derived Hepatocyte-like Cells
Abstract
:1. Introduction
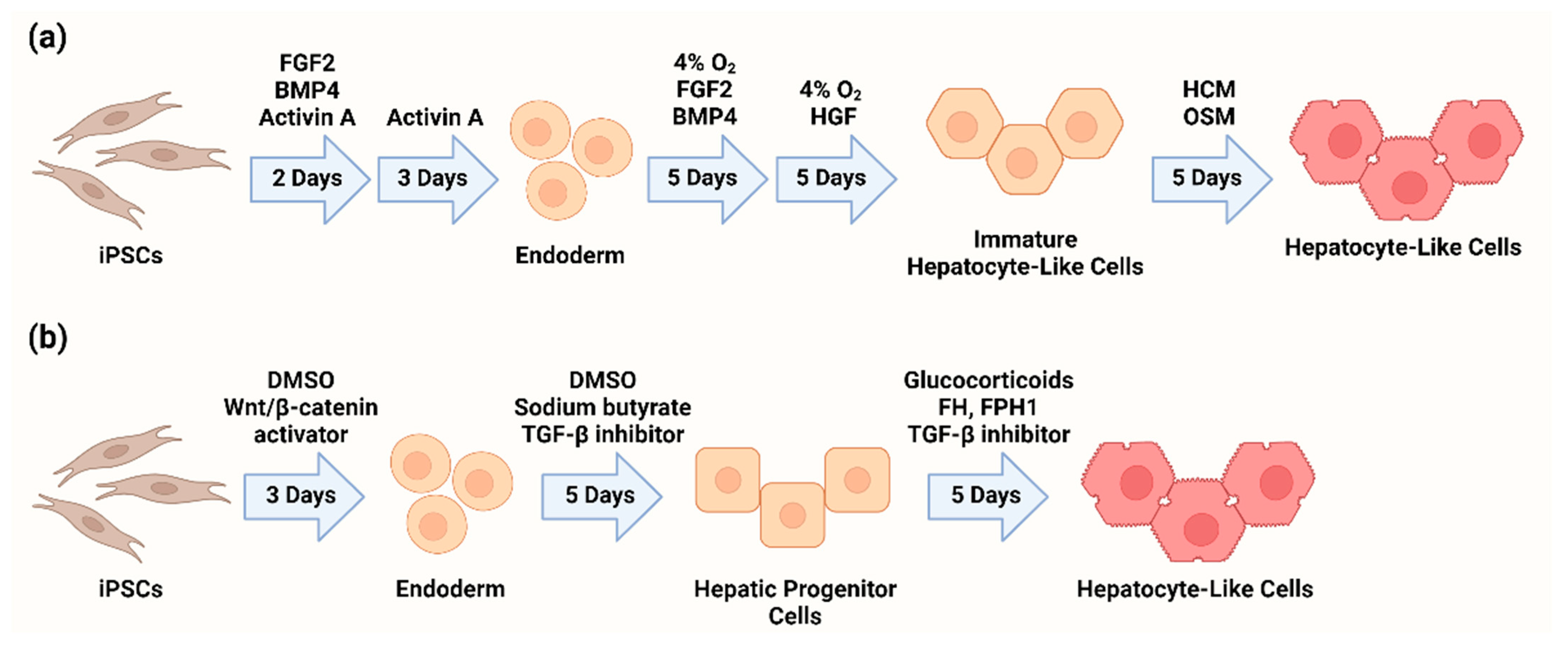
2. Differentiation of Hepatocyte-like Cells from iPSCs
3. Limitations and Recent Improvements in HLC Differentiation and Maturation
4. Genetic Function Studies
5. Disease Modeling Using iPSC-Derived Hepatocyte-like Cells
6. High-Throughput Screens for Drug Discovery and Development
7. Summary and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tapper, E.B.; Parikh, N.D. Mortality due to cirrhosis and liver cancer in the United States, 1999–2016: Observational study. BMJ 2018, 362, k2817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asrani, S.K.; Larson, J.J.; Yawn, B.; Therneau, T.M.; Kim, W.R. Underestimation of Liver-Related Mortality in the United States. Gastroenterology 2013, 145, 375–382.e2. [Google Scholar] [CrossRef] [Green Version]
- Hirode, G.; Saab, S.; Wong, R.J. Trends in the Burden of Chronic Liver Disease among Hospitalized US Adults. JAMA Netw. Open 2020, 3, e201997. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Li, L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell 2015, 6, 562–574. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, S.R.; Paul-Heng, M.; Krycer, J.R.; Fazakerley, D.J.; Sharland, A.F.; Hoy, X.A.J. Lipid and glucose metabolism in hepatocyte cell lines and primary mouse hepatocytes: A comprehensive resource for in vitro studies of hepatic metabolism. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E578–E589. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Sirota, M.; Fan-Minogue, H.; Hadley, D.; Butte, A.J. Relating hepatocellular carcinoma tumor samples and cell lines using gene expression data in translational research. BMC Med. Genom. 2015, 8 (Suppl. S2), S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiamehr, M.; Alexanova, A.; Viiri, L.E.; Heiskanen, L.; Vihervaara, T.; Kauhanen, D.; Ekroos, K.; Laaksonen, R.; Käkelä, R.; Aalto-Setälä, K. hiPSC-derived hepatocytes closely mimic the lipid profile of primary hepatocytes: A future personalised cell model for studying the lipid metabolism of the liver. J. Cell. Physiol. 2019, 234, 3744–3761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves-Bezerra, M.; Furey, N.; Johnson, C.G.; Bissig, K.D. Using CRISPR/Cas9 to model human liver disease. JHEP Rep. 2019, 1, 392. [Google Scholar] [CrossRef] [Green Version]
- Mann, J.P.; Semple, R.K.; Armstrong, M.J. How useful are monogenic rodent models for the study of human non-alcoholic fatty liver disease? Front. Endocrinol. 2016, 7, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogaards, J.J.P.; Bertrand, M.; Jackson, P.; Oudshoorn, M.J.; Weaver, R.J.; Van Bladeren, P.J.; Walther, B. Determining the best animal model for human cytochrome P450 activities: A comparison of mouse, rat, rabbit, dog, micropig, monkey and man. Xenobiotica 2000, 30, 1131–1152. [Google Scholar] [CrossRef]
- Azuma, H.; Paulk, N.; Ranade, A.; Dorrell, C.; Al-Dhalimy, M.; Ellis, E.; Strom, S.; Kay, M.A.; Finegold, M.; Grompe, M. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat. Biotechnol. 2007, 25, 903–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, M.; Kawai, K.; Mitsui, T.; Taniguchi, K.; Monnai, M.; Wakui, M.; Ito, M.; Suematsu, M.; Peltz, G.; Nakamura, M.; et al. The reconstituted ‘humanized liver’ in TK-NOG mice is mature and functional. Biochem. Biophys. Res. Commun. 2011, 405, 405–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayo, M.A.; Mallanna, S.K.; Di Furio, F.; Jing, R.; Tolliver, L.B.; Bures, M.; Urick, A.; Noto, F.K.; Pashos, E.E.; Greseth, M.D.; et al. A Drug Screen using Human iPSC-Derived Hepatocyte-like Cells Reveals Cardiac Glycosides as a Potential Treatment for Hypercholesterolemia. Cell Stem Cell 2017, 20, 478–489.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Wong, L.Y.; Tian, X.Y.; Wei, R.; Lai, W.H.; Au, K.W.; Luo, Z.; Ward, C.; Ho, W.I.; Ibañez, D.P.; et al. A familial hypercholesterolemia human liver chimeric mouse model using induced pluripotent stem cell-derived hepatocytes. J. Vis. Exp. 2018, 139, e57556. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Zhao, Y.; Liu, Y.; Ye, F.; Song, Z.; Qin, H.; Meng, S.; Chen, Y.; Zhou, R.; Song, X.; et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 2007, 45, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Cai, J.; Liu, Y.; Zhao, D.; Yong, J.; Duo, S.; Song, X.; Guo, Y.; Zhao, Y.; Qin, H.; et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009, 19, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.J.; Hay, D.C.; Park, I.H.; Fletcher, J.; Hannoun, Z.; Payne, C.M.; Dalgetty, D.; Black, J.R.; Ross, J.A.; Samuel, K.; et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology 2010, 51, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Liu, Y. A transcriptomic study suggesting human iPSC-derived hepatocytes potentially offer a better in vitro model of hepatotoxicity than most hepatoma cell lines. Cell Biol. Toxicol. 2017, 33, 407–421. [Google Scholar] [CrossRef]
- Du, C.; Feng, Y.; Qiu, D.; Xu, Y.; Pang, M.; Cai, N.; Xiang, A.P.; Zhang, Q. Highly efficient and expedited hepatic differentiation from human pluripotent stem cells by pure small-molecule cocktails. Stem Cell Res. Ther. 2018, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Siller, R.; Greenhough, S.; Naumovska, E.; Sullivan, G.J. Small-Molecule-Driven Hepatocyte Differentiation of Human Pluripotent Stem Cells. Stem Cell Rep. 2015, 4, 939–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Li, R.; Cahan, P.; Zhao, Y.; Yourick, J.J.; Sprando, R.L. Hepatocyte-like cells derived from human induced pluripotent stem cells using small molecules: Implications of a transcriptomic study. Stem Cell Res. Ther. 2020, 11, 393. [Google Scholar] [CrossRef]
- Baxter, M.; Withey, S.; Harrison, S.; Segeritz, C.P.; Zhang, F.; Atkinson-Dell, R.; Rowe, C.; Gerrard, D.T.; Sison-Young, R.; Jenkins, R.; et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015, 62, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.; Kumar, M.; Tricot, T.; Elia, I.; Ordovas, L.; Jacobs, F.; One, J.; De Smedt, J.; Eelen, G.; Bird, M.; et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat. Commun. 2020, 11, 1393. [Google Scholar] [CrossRef] [Green Version]
- Bell, C.C.; Lauschke, V.M.; Vorrink, S.U.; Palmgren, H.; Duffin, R.; Andersson, T.B.; Ingelman-Sundberg, M. Transcriptional, Functional, and Mechanistic Comparisons of Stem Cell–Derived Hepatocytes, HepaRG Cells, and Three-Dimensional Human Hepatocyte Spheroids as Predictive In Vitro Systems for Drug-Induced Liver Injury. Drug Metab. Dispos. 2017, 45, 419–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulvestad, M.; Nordell, P.; Asplund, A.; Rehnstrom, M.; Jacobsson, S.; Holmgren, G.; Davidson, L.; Brolen, G.; Edsbagge, J.; Björquist, P.; et al. Drug metabolizing enzyme and transporter protein profiles of hepatocytes derived from human embryonic and induced pluripotent stem cells. Biochem. Pharmacol. 2013, 86, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Qosa, H.; Ribeiro, A.J.S.; Hartman, N.R.; Volpe, D.A. Characterization of a commercially available line of iPSC hepatocytes as models of hepatocyte function and toxicity for regulatory purposes. J. Pharmacol. Toxicol. Methods 2021, 110, 107083. [Google Scholar] [CrossRef]
- Kvist, A.J.; Kanebratt, K.P.; Walentinsson, A.; Palmgren, H.; O’Hara, M.; Björkbom, A.; Andersson, L.C.; Ahlqvist, M.; Andersson, T.B. Critical differences in drug metabolic properties of human hepatic cellular models, including primary human hepatocytes, stem cell derived hepatocytes, and hepatoma cell lines. Biochem. Pharmacol. 2018, 155, 124–140. [Google Scholar] [CrossRef]
- Yamashita, H.; Fukuda, K.; Hattori, F. Hepatocyte-like Cells Derived from Human Pluripotent Stem Cells Can Be Enriched by a Combination of Mitochondrial Content and Activated Leukocyte Cell Adhesion Molecule. JMA J. 2019, 2, 174. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.T.; Henderson, C.A.; Warren, C.R.; Friesen, M.; Becker, C.E.; Musunuru, K.; Cowan, C.A. Asialoglycoprotein receptor 1 is a specific cell-surface marker for isolating hepatocytes derived from human pluripotent stem cells. Development 2016, 143, 1475–1481. [Google Scholar] [CrossRef] [Green Version]
- Shan, J.; Schwartz, R.E.; Ross, N.T.; Logan, D.J.; Thomas, D.; Duncan, S.A.; North, T.E.; Goessling, W.; Carpenter, A.E.; Bhatia, S.N. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013, 9, 514–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.; Ai, Y.; Xiao, S.; Wang, B.; Wang, Y. Functional hit 1 (FH1)-based rapid and efficient generation of functional hepatocytes from human mesenchymal stem cells: A novel strategy for hepatic differentiation. Ann. Transl. Med. 2021, 9, 1087. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Schrooders, Y.; Hauser, D.; van Herwijnen, M.; Albrecht, W.; ter Braak, B.; Brecklinghaus, T.; Castell, J.V.; Elenschneider, L.; Escher, S.; et al. Comparing in vitro human liver models to in vivo human liver using RNA-Seq. Arch. Toxicol. 2021, 95, 573–589. [Google Scholar] [CrossRef]
- Poorna, M.R.; Sudhindran, S.; Thampi, M.V.; Mony, U. Differentiation of induced pluripotent stem cells to hepatocyte-like cells on cellulose nanofibril substrate. Colloids Surf. B Biointerfaces 2021, 198, 111466. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.T.; Longaker, M.T. Comparison of several attachment methods for human iPS, embryonic and adipose-derived stem cells for tissue engineering. J. Tissue Eng. Regen. Med. 2012, 6, s80. [Google Scholar] [CrossRef] [PubMed]
- Kehtari, M.; Zeynali, B.; Soleimani, M.; Kabiri, M.; Seyedjafari, E. Fabrication of a co-culture micro-bioreactor device for efficient hepatic differentiation of human induced pluripotent stem cells (hiPSCs). Artif. Cells Nanomed. Biotechnol. 2018, 46, 161–170. [Google Scholar] [CrossRef]
- Koui, Y.; Kido, T.; Ito, T.; Oyama, H.; Chen, S.W.; Katou, Y.; Shirahige, K.; Miyajima, A. An In Vitro Human Liver Model by iPSC-Derived Parenchymal and Non-parenchymal Cells. Stem Cell Rep. 2017, 9, 490–498. [Google Scholar] [CrossRef] [Green Version]
- Nagata, S.; Ozawa, F.; Nie, M.; Takeuchi, S. 3D culture of functional human iPSC-derived hepatocytes using a core-shell microfiber. PLoS ONE 2020, 15, e0234441. [Google Scholar] [CrossRef]
- Minami, T.; Ishii, T.; Yasuchika, K.; Fukumitsu, K.; Ogiso, S.; Miyauchi, Y.; Kojima, H.; Kawai, T.; Yamaoka, R.; Oshima, Y.; et al. Novel hybrid three-dimensional artificial liver using human induced pluripotent stem cells and a rat decellularized liver scaffold. Regen. Ther. 2019, 10, 127–133. [Google Scholar] [CrossRef]
- Wang, B.; Jakus, A.E.; Baptista, P.M.; Soker, S.; Soto-Gutierrez, A.; Abecassis, M.M.; Shah, R.N.; Wertheim, J.A. Functional Maturation of Induced Pluripotent Stem Cell Hepatocytes in Extracellular Matrix—A Comparative Analysis of Bioartificial Liver Microenvironments. Stem Cells Transl. Med. 2016, 5, 1257–1267. [Google Scholar] [CrossRef]
- Takebe, T.; Sekine, K.; Kimura, M.; Yoshizawa, E.; Ayano, S.; Koido, M.; Funayama, S.; Nakanishi, N.; Hisai, T.; Kobayashi, T.; et al. Massive and Reproducible Production of Liver Buds Entirely from Human Pluripotent Stem Cells. Cell Rep. 2017, 21, 2661–2670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bin Ramli, M.N.; Lim, Y.S.; Koe, C.T.; Demircioglu, D.; Tng, W.; Gonzales, K.A.U.; Tan, C.P.; Szczerbinska, I.; Liang, H.; Soe, E.L.; et al. Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 2020, 159, 1471–1486.e12. [Google Scholar] [CrossRef] [PubMed]
- Jalan-Sakrikar, N.; De Assuncao, T.M.; Navarro-Corcuera, A.; Hamdan, F.H.; Loarca, L.; Kirkeby, L.A.; Resch, Z.T.; O’Hara, S.P.; Juran, B.D.; Lazaridis, K.N.; et al. Induced Pluripotent Stem Cells from Subjects With Primary Sclerosing Cholangitis Develop a Senescence Phenotype Following Biliary Differentiation. Hepatol. Commun. 2022, 6, 345–360. [Google Scholar] [CrossRef]
- Sampaziotis, F.; de Brito, M.C.; Madrigal, P.; Bertero, A.; Saeb-Parsy, K.; Soares, F.A.C.; Schrumpf, E.; Melum, E.; Karlsen, T.H.; Bradley, J.A.; et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 2015, 33, 845–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbey, D.; Elwyn, S.; Hand, N.J.; Musunuru, K.; Rader, D.J. Self-Organizing Human Induced Pluripotent Stem Cell Hepatocyte 3D Organoids Inform the Biology of the Pleiotropic TRIB1 Gene. Hepatol. Commun. 2020, 4, 1316–1331. [Google Scholar] [CrossRef] [PubMed]
- Akbari, S.; Sevinç, G.G.; Ersoy, N.; Basak, O.; Kaplan, K.; Sevinç, K.; Ozel, E.; Sengun, B.; Enustun, E.; Ozcimen, B.; et al. Robust, Long-Term Culture of Endoderm-Derived Hepatic Organoids for Disease Modeling. Stem Cell Rep. 2019, 13, 627–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, S.; Schwartz, R.E.; March, S.; Galstian, A.; Gural, N.; Shan, J.; Prabhu, M.; Mota, M.M.; Bhatia, S.N. Human iPSC-Derived Hepatocyte-like Cells Support Plasmodium Liver-Stage Infection In Vitro. Stem Cell Rep. 2015, 4, 348–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, Y.Z.; Zheng, Y.W.; Miyakawa, K.; Murata, S.; Zhang, R.R.; Sekine, K.; Ueno, Y.; Takebe, T.; Wakita, T.; Ryo, A.; et al. Recapitulation of hepatitis B virus–host interactions in liver organoids from human induced pluripotent stem cells. EBioMedicine 2018, 35, 114–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.W.; Himeno, M.; Koui, Y.; Sugiyama, M.; Nishitsuji, H.; Mizokami, M.; Shimotohno, K.; Miyajima, A.; Kido, T. Modulation of hepatitis B virus infection by epidermal growth factor secreted from liver sinusoidal endothelial cells. Sci. Rep. 2020, 10, 14349. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Ye, Z.; Kafka, K.; Stewart, D.; Anders, R.; Schwarz, K.B.; Jang, Y.Y. Biliary Atresia Relevant Human Induced Pluripotent Stem Cells Recapitulate Key Disease Features in a Dish. J. Pediatr. Gastroenterol. Nutr. 2019, 68, 56–63. [Google Scholar] [CrossRef]
- Warren, C.R.; O’Sullivan, J.F.; Friesen, M.; Becker, C.E.; Zhang, X.; Liu, P.; Wakabayashi, Y.; Morningstar, J.E.; Shi, X.; Choi, J.; et al. Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Cell Stem Cell 2017, 20, 547–557.e7. [Google Scholar] [CrossRef] [PubMed]
- Pashos, E.E.; Park, Y.S.; Wang, X.; Raghavan, A.; Yang, W.; Abbey, D.; Peters, D.T.; Arbelaez, J.; Hernandez, M.; Kuperwasser, N.; et al. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell 2017, 20, 558–570.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaserman, J.E.; Hurley, K.; Dodge, M.; Villacorta-Martin, C.; Vedaie, M.; Jean, J.C.; Liberti, D.C.; James, M.F.; Higgins, M.I.; Lee, N.J.; et al. A Highly Phenotyped Open Access Repository of Alpha-1 Antitrypsin Deficiency Pluripotent Stem Cells. Stem Cell Rep. 2020, 15, 242–255. [Google Scholar] [CrossRef]
- Omer, L.; Hudson, E.A.; Hudgins, L.C.; Boyd, N.L. Cohort Generation and Characterization of Patient-Specific Familial Hypercholesterolemia Induced Pluripotent Stem Cells. Stem Cells Dev. 2021, 30, 632–640. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Corbett, J.L.; Cai, J.; Beeson, G.C.; Beeson, C.C.; Chan, S.S.; Dimmock, D.P.; Lazcares, L.; Geurts, A.M.; Lemasters, J.J.; et al. A Screen Using iPSC-Derived Hepatocytes Reveals NAD + as a Potential Treatment for mtDNA Depletion Syndrome. Cell Rep. 2018, 25, 1469–1484.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, T.; Park, S.B.; Inuzuka, T.; Zhang, M.; Allen, J.N.; Chayama, K.; Liang, T.J. Genetically edited hepatic cells expressing the NTCP-S267F variant are resistant to hepatitis B virus infection. Mol. Ther. Methods Clin. Dev. 2021, 23, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Shen, J.; Wang, R.; Gu, H.; Zheng, J.; Xue, F.; Liu, X.; Liu, W.; Fu, R.; Zheng, L.; et al. Targeted genome engineering in human induced pluripotent stem cells from patients with hemophilia B using the CRISPR-Cas9 system. Stem Cell Res. Ther. 2018, 9, 92. [Google Scholar]
- Wei, R.; Yang, J.; Cheng, C.W.; Ho, W.I.; Li, N.; Hu, Y.; Hong, X.; Fu, J.; Yang, B.; Liu, Y.; et al. CRISPR-targeted genome editing of human induced pluripotent stem cell-derived hepatocytes for the treatment of Wilson’s disease. JHEP Rep. 2022, 4, 100389. [Google Scholar] [CrossRef]
- Fattahi, F.; Asgari, S.; Pournasr, B.; Seifinejad, A.; Totonchi, M.; Taei, A.; Aghdami, N.; Salekdeh, G.H.; Baharvand, H. Disease-Corrected Hepatocyte-Like Cells from Familial Hypercholesterolemia-Induced Pluripotent Stem Cells. Mol. Biotechnol. 2012, 54, 863–873. [Google Scholar] [CrossRef]
- Duncan, S.A.; Liu, J.-T.; Corbett, J.L.; Heslop, J.A. Enhanced genome editing in human iPSCs with CRISPR-CAS9 by co-targeting ATP1a1. PeerJ 2020, 8, e9060. [Google Scholar] [CrossRef]
- Tokgozoglu, L.; Kayikcioglu, M. Familial Hypercholesterolemia: Global Burden and Approaches. Curr. Cardiol. Rep. 2021, 23, 151. [Google Scholar] [CrossRef] [PubMed]
- Cayo, M.A.; Cai, J.; Delaforest, A.; Noto, F.K.; Nagaoka, M.; Clark, B.S.; Collery, R.F.; Si-Tayeb, K.; Duncan, S.A. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology 2012, 56, 2163–2171. [Google Scholar] [CrossRef] [Green Version]
- van der Ploeg, A.T.; Reuser, A.J. Pompe’s disease. Lancet 2008, 372, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Jonouchi, T.; Osafune, K.; Takita, J.; Sakurai, H. A Liver Model of Infantile-Onset Pompe Disease Using Patient-Specific Induced Pluripotent Stem Cells. Front. Cell Dev. Biol. 2019, 7, 316. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Deshmukh, A.; Prasad, N.; Jang, Y.Y. Alcohol increases liver progenitor populations and induces disease phenotypes in human IPSC-derived mature stage hepatic cells. Int. J. Biol. Sci. 2016, 12, 1052–1062. [Google Scholar] [CrossRef] [Green Version]
- Dao Thi, V.L.; Wu, X.; Rice, C.M. Stem cell–derived culture models of hepatitis e virus infection. Cold Spring Harb. Perspect. Med. 2019, 9, a031799. [Google Scholar] [CrossRef]
- Sakurai, F.; Kunito, T.; Takayama, K.; Hashimoto, R.; Tachibana, M.; Sakamoto, N.; Wakita, T.; Mizuguchi, H. Hepatitis C virus-induced innate immune responses in human iPS cell-derived hepatocyte-like cells. Virus Res. 2017, 242, 7–15. [Google Scholar] [CrossRef]
- Schöbel, A.; Rösch, K.; Herker, E. Functional innate immunity restricts Hepatitis C Virus infection in induced pluripotent stem cell-derived hepatocytes. Sci. Rep. 2018, 8, 3893. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Conlon, D.M.; Bi, X.; Slovik, K.J.; Shi, J.; Edelstein, H.I.; Millar, J.S.; Javaheri, A.; Cuchel, M.; Pashos, E.E.; et al. Lack of MTTP Activity in Pluripotent Stem Cell-Derived Hepatocytes and Cardiomyocytes Abolishes apoB Secretion and Increases Cell Stress. Cell Rep. 2017, 19, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017, 2, e94954. [Google Scholar] [CrossRef] [Green Version]
- Brooks, B.M.; Pradhan, M.; Cheng, Y.S.; Gorshkov, K.; Farkhondeh, A.; Chen, C.Z.; Beers, J.; Liu, C.; Baumgaertel, K.; Rodems, S.; et al. Generation of an induced pluripotent stem cell line (TRNDi031-A) from a patient with Alagille syndrome type 1 carrying a heterozygous p. C312X (c. 936 T > A) mutation in JAGGED-1. Stem Cell Res. 2021, 54, 102447. [Google Scholar] [CrossRef]
- Nikasa, P.; Tricot, T.; Mahdieh, N.; Baharvand, H.; Totonchi, M.; Hejazi, M.S.; Verfaillie, C.M. Patient-Specific Induced Pluripotent Stem Cell-Derived Hepatocyte-Like Cells as a Model to Study Autosomal Recessive Hypercholesterolemia. Stem Cells Dev. 2021, 30, 714–724. [Google Scholar] [CrossRef]
- Hayashi, H.; Osaka, S.; Sakabe, K.; Fukami, A.; Kishimoto, E.; Aihara, E.; Sabu, Y.; Mizutani, A.; Kusuhara, H.; Naritaka, N.; et al. Modeling Human Bile Acid Transport and Synthesis in Stem Cell-Derived Hepatocytes with a Patient-Specific Mutation. Stem Cell Rep. 2021, 16, 309–323. [Google Scholar] [CrossRef]
- Imagawa, K.; Takayama, K.; Isoyama, S.; Tanikawa, K.; Shinkai, M.; Harada, K.; Tachibana, M.; Sakurai, F.; Noguchi, E.; Hirata, K.; et al. Generation of a bile salt export pump deficiency model using patient-specific induced pluripotent stem cell-derived hepatocyte-like cells. Sci. Rep. 2017, 7, 41806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Eldridge, L.; Chaudhari, P.; Zhang, L.; Anders, R.A.; Schwarz, K.B.; Ye, Z.; Jang, Y.Y. Derivation of a disease-specific human induced pluripotent stem cell line from a biliary atresia patient. Stem Cell Res. 2017, 24, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Yoshitoshi-Uebayashi, E.Y.; Toyoda, T.; Yasuda, K.; Kotaka, M.; Nomoto, K.; Okita, K.; Yasuchika, K.; Okamoto, S.; Takubo, N.; Nishikubo, T.; et al. Modelling urea-cycle disorder citrullinemia type 1 with disease-specific iPSCs. Biochem. Biophys. Res. Commun. 2017, 486, 613–619. [Google Scholar] [CrossRef] [Green Version]
- Luce, E.; Steichen, C.; Allouche, M.; Messina, A.; Heslan, J.; Lambert, T.; Weber, A.; Nguyen, T.H.; Christophe, O.; Dubart-Kupperschmitt, A. In vitro recovery of FIX clotting activity as a marker of highly functional hepatocytes in a hemophilia B iPSC model. Hepatology 2021, 75, 866–880. [Google Scholar] [CrossRef]
- Martorell, L.; Luce, E.; Vazquez, J.L.; Richaud-Patin, Y.; Jimenez-Delgado, S.; Corrales, I.; Borras, N.; Casacuberta-Serra, S.; Weber, A.; Parra, R.; et al. Advanced cell-based modeling of the royal disease: Characterization of the mutated F9 mRNA. J. Thromb. Haemost. 2017, 15, 2188–2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sa-Ngiamsuntorn, K.; Wongkajornsilp, A.; Phanthong, P.; Borwornpinyo, S.; Kitiyanant, N.; Chantratita, W.; Hongeng, S. A robust model of natural hepatitis C infection using hepatocyte-like cells derived from human induced pluripotent stem cells as a long-term host. Virol. J. 2016, 13, 59. [Google Scholar]
- Guo, J.; Duan, L.; He, X.; Li, S.; Wu, Y.; Xiang, G.; Bao, F.; Yang, L.; Shi, H.; Gao, M.; et al. A Combined Model of Human iPSC-Derived Liver Organoids and Hepatocytes Reveals Ferroptosis in DGUOK Mutant mtDNA Depletion Syndrome. Adv. Sci. 2021, 8, 2004680. [Google Scholar] [CrossRef]
- Graffmann, N.; Ncube, A.; Martins, S.; Fiszl, A.R.; Reuther, P.; Bohndorf, M.; Wruck, W.; Beller, M.; Czekelius, C.; Adjaye, J. A stem cell based in vitro model of NAFLD enables the analysis of patient specific individual metabolic adaptations in response to a high fat diet and AdipoRon interference. Biol. Open 2021, 10, bio054189. [Google Scholar] [CrossRef] [PubMed]
- Sinton, M.C.; Meseguer-Ripolles, J.; Lucendo-Villarin, B.; Wernig-Zorc, S.; Thomson, J.P.; Carter, R.N.; Lyall, M.J.; Walker, P.D.; Thakker, A.; Meehan, R.R.; et al. A human pluripotent stem cell model for the analysis of metabolic dysfunction in hepatic steatosis. iScience 2021, 24, 101931. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.E.; van den Boogert, M.A.W.; Rios-Ocampo, W.A.; Jansen, J.C.; Conlon, D.; Chong, P.L.E.; Levels, J.H.M.; Eilers, R.E.; Sachdev, V.V.; Zelcer, N.; et al. Defective Lipid Droplet–Lysosome Interaction Causes Fatty Liver Disease as Evidenced by Human Mutations in TMEM199 and CCDC115. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 583–597. [Google Scholar] [CrossRef]
- Gurevich, I.; Burton, S.A.; Munn, C.; Ohshima, M.; Goedland, M.E.; Czysz, K.; Rajesh, D. iPSC-derived hepatocytes generated from NASH donors provide a valuable platform for disease modeling and drug discovery. Biol. Open 2020, 9, bio055087. [Google Scholar] [CrossRef] [PubMed]
- Völkner, C.; Pantoom, S.; Liedtke, M.; Lukas, J.; Hermann, A.; Frech, M.J. Assessment of FDA-Approved Drugs as a Therapeutic Approach for Niemann-Pick Disease Type C1 Using Patient-Specific iPSC-Based Model Systems. Cells 2022, 11, 319. [Google Scholar] [CrossRef]
- Soga, M.; Ishitsuka, Y.; Hamasaki, M.; Yoneda, K.; Furuya, H.; Matsuo, M.; Ihn, H.; Fusaki, N.; Nakamura, K.; Nakagata, N.; et al. HPGCD outperforms HPBCD as a potential treatment for niemann-pick disease type C during disease modeling with iPS cells. Stem Cells 2015, 33, 1075–1088. [Google Scholar] [CrossRef]
- Laemmle, A.; Poms, M.; Hsu, B.; Borsuk, M.; Rüfenacht, V.; Robinson, J.; Sadowski, M.C.; Nuoffer, J.; Häberle, J.; Willenbring, H. Aquaporin 9 induction in human iPSC-derived hepatocytes facilitates modeling of ornithine transcarbamylase deficiency. Hepatology 2021, 00, 1–14. [Google Scholar] [CrossRef]
- Pournasr, B.; Duncan, S.A. Generation of isogenic Propionyl-CoA carboxylase beta subunit (PCCB) deficient induced pluripotent stem cell lines. Stem Cell Res. 2020, 48. [Google Scholar] [CrossRef]
- Giadone, R.M.; Liberti, D.C.; Matte, T.M.; Rosarda, J.D.; Torres-Arancivia, C.; Ghosh, S.; Diedrich, J.K.; Pankow, S.; Skvir, N.; Jean, J.C.; et al. Expression of Amyloidogenic Transthyretin Drives Hepatic Proteostasis Remodeling in an Induced Pluripotent Stem Cell Model of Systemic Amyloid Disease. Stem Cell Rep. 2020, 15, 515–528. [Google Scholar] [CrossRef]
- Giadone, R.M.; Rosarda, J.D.; Akepati, P.R.; Thomas, A.C.; Boldbaatar, B.; James, M.F.; Wilson, A.A.; Sanchorawala, V.; Connors, L.H.; Berk, J.L.; et al. A library of ATTR amyloidosis patient-specific induced pluripotent stem cells for disease modelling and in vitro testing of novel therapeutics. Amyloid 2018, 25, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Petters, J.; Cimmaruta, C.; Iwanov, K.; Chang, M.L.; Völkner, C.; Knuebel, G.; Escobar, H.M.; Frech, M.J.; Hermann, A.; Rolfs, A.; et al. Generation of induced pluripotent stem cell lines AKOSi002-A and AKOSi003-A from symptomatic female adults with Wilson disease. Stem Cell Res. 2020, 43, 101708. [Google Scholar] [CrossRef] [PubMed]
- Parisi, S.; Polishchuk, E.V.; Allocca, S.; Ciano, M.; Musto, A.; Gallo, M.; Perone, L.; Ranucci, G.; Iorio, R.; Polishchuk, R.S.; et al. Characterization of the most frequent ATP7B mutation causing Wilson disease in hepatocytes from patient induced pluripotent stem cells. Sci. Rep. 2018, 8, 6247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.M.; Yik, W.Y.; Zhang, P.; Lu, W.; Huang, N.; Kim, B.R.; Shibata, D.; Zitting, M.; Chow, R.H.; Moser, A.B.; et al. Induced pluripotent stem cell models of Zellweger spectrum disorder show impaired peroxisome assembly and cell type-specific lipid abnormalities. Stem Cell Res. Ther. 2015, 6, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parafati, M.; Bae, S.H.; Kirby, R.J.; Fitzek, M.; Iyer, P.; Engkvist, O.; Smith, D.M.; Malany, S. Pluripotent Stem Cell-Derived Hepatocytes Phenotypic Screening Reveals Small Molecules Targeting the CDK2/4-C/EBPα/DGAT2 Pathway Preventing ER-Stress Induced Lipid Accumulation. Int. J. Mol. Sci. 2020, 21, 9557. [Google Scholar] [CrossRef]
- Jing, R.; Duncan, C.B.; Duncan, S.A. A small-molecule screen reveals that HSP90β promotes the conversion of induced pluripotent stem cell-derived endoderm to a hepatic fate and regulates HNF4A turnover. Development 2017, 144, 1764–1774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koui, Y.; Himeno, M.; Mori, Y.; Nakano, Y.; Saijou, E.; Tanimizu, N.; Kamiya, Y.; Anzai, H.; Maeda, N.; Wang, L.; et al. Development of human iPSC-derived quiescent hepatic stellate cell-like cells for drug discovery and in vitro disease modeling. Stem Cell Rep. 2021, 16, 3050–3063. [Google Scholar] [CrossRef]
- Kim, J.W.; Im, I.; Kim, H.; Jeon, J.S.; Kang, E.H.; Jo, S.; Chun, H.S.; Yoon, S.; Kim, J.H.; Kim, S.K.; et al. Live-cell screening platform using human-induced pluripotent stem cells expressing fluorescence-tagged cytochrome P450 1A1. FASEB J. 2020, 34, 9141–9155. [Google Scholar] [CrossRef] [PubMed]
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.; Vernetti, L.A.; Gough, A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology 2021, 450, 152667. [Google Scholar] [CrossRef]
- Shinozawa, T.; Kimura, M.; Cai, Y.; Saiki, N.; Yoneyama, Y.; Ouchi, R.; Koike, H.; Maezawa, M.; Zhang, R.R.; Dunn, A.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell–Derived Organoids. Gastroenterology 2021, 160, 831–846.e10. [Google Scholar] [CrossRef]
- Lin, N.; Zhou, X.; Geng, X.; Drewell, C.; Hübner, J.; Li, Z.; Zhang, Y.; Xue, M.; Marx, U.; Li, B. Repeated dose multi-drug testing using a microfluidic chip-based coculture of human liver and kidney proximal tubules equivalents. Sci. Rep. 2020, 10, 8879. [Google Scholar] [CrossRef] [PubMed]
- Ware, B.R.; Berger, D.R.; Khetani, S.R. Prediction of drug-induced liver injury in micropatterned co-cultures containing iPSC-derived human hepatocytes. Toxicol. Sci. 2015, 145, 252–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.M.; Kim, Y.; Shim, J.S.; Park, J.T.; Wang, R.H.; Leach, S.D.; Liu, J.O.; Deng, C.; Ye, Z.; Jang, Y.Y. Efficient drug screening and gene correction for treating liver disease using patient-specific stem cells. Hepatology 2013, 57, 2458–2468. [Google Scholar] [CrossRef] [Green Version]

| Disease | Configuration | Model Characteristics | Reference |
|---|---|---|---|
| Abetalipoproteinemia | 2D culture | Decreased ApoB secretion, intracellular lipid accumulation, increased cell death | [70] |
| Alagille Syndrome | 3D organoids containing hepatocytes and cholangiocytes | Impaired bile duct formation and regenerative capacity | [71] |
| 2D culture | Patient-derived, HLC phenotype uncharacterized | [72] | |
| Alcohol-induced Liver Injury | 2D culture | Reduced proliferation, oxidative mitochondrial injury, increased steatosis, and hepatocellular carcinoma markers | [66] |
| α-1 Antitrypsin Deficiency | 2D culture | AAT retention, enrichment of fibrosis- and cirrhosis-associated pathways | [54] |
| Autosomal Recessive Hypercholesterolemia | 2D culture | Reduced LDL uptake | [73] |
| Bile Salt Export Pump (BSEP) Deficiency | 2D culture | Impaired biliary excretion, altered localization of BSEP protein | [74,75] |
| Biliary Atresia | 2D culture | Decreased biliary marker expression, increased expression of fibrosis markers | [51,76] |
| Citrullinemia Type I | 3D organoids | Accumulation of ammonia, decreased ureagenesis | [47] |
| 2D culture | Decreased ureagenesis | [77] | |
| Familial Hypercholesterolemia | 2D culture | Inability to uptake LDL | [60] |
| Hemophilia B | 3D organoids | Production of inactive coagulation factor IX (F9) | [78] |
| 2D culture | Reduced expression and activity of F9 | [58] | |
| 2D culture | Aberrant splicing of F9 mRNA leading to reduced F9 expression | [79] | |
| Hepatitis B | 3D organoids | Higher susceptibility to HBV infection than 2D culture, increased duration of infectious virus production | [49] |
| 2D coculture of HLCs and liver non-parenchymal cells | Improved efficiency of infection relative to 2D monoculture due to epidermal growth factor (EGF) modulation of endocytosis | [50] | |
| Hepatitis C | 2D culture | Permissive to infection with HCV, upregulation of type I and III interferons in response to infection | [68] |
| 2D culture | Higher susceptibility to and propagation of HCV compared to Huh7 cells | [80] | |
| 2D culture | Supportive of full HCV life cycle, increased expression of interferon-stimulated genes | [69] | |
| Hepatitis E | 2D culture | Permissive host for hepatitis E virus natural isolates | [67] |
| Malaria | 2D culture | Permissive host for infection with Plasmodium species; chemical maturation allows for bioactivation of primaquine | [48] |
| mtDNA Depletion Syndrome | 2D culture | Decreased mtDNA copy number, disruption of mitochondrial ultrastructure, reduced mitochondrial respiration/intracellular ATP, increased reactive oxygen species levels | [56] |
| 2D culture and 3D organoids | Decreased mtDNA, reduced mitochondrial respiration, increased reactive oxygen species, increased sensitivity to iron overload | [81] | |
| Nonalcoholic Fatty Liver Disease | 2D culture | Patient-specific lipid droplet formation upon administration of oleic acid, decreased lipid metabolism-associated gene expression in higher levels of steatosis | [82] |
| 2D culture | Decreased electron transport chain activity, altered transcription of mitochondrial respiration pathways, increased pyruvate carboxylase activity, and fumarate accumulation in response to steatosis induction | [83] | |
| 2D culture | Defects in V-ATPase assembly leading to increased ApoB secretion | [84] | |
| 3D organoids with hepatocytes, macrophages, mesenchymal stem cells, and endothelial cells | Spontaneous lipid accumulation in absence of fatty acid supplementation | [85] | |
| Niemann–Pick Disease Type C (NPC) | 2D culture | Increased lysosomal accumulation of cholesterol, increased trafficking of NPC1 to lysosomes | [86] |
| 2D culture | Increased lysosomal cholesterol accumulation, increased cell size, upregulated autophagy and impaired autophagic flux | [87] | |
| Ornithine Transcarbamylase Deficiency | 2D culture | Decreased urea secretion | [88] |
| Pompe Disease | 2D culture | Accumulation of glycogen in lysosomes | [65] |
| Primary Sclerosing Cholangitis | 3D organoids containing cholangiocytes | Altered organoid morphology, increased cellular senescence and inflammatory cytokine secretion | [44] |
| Propionic Acidemia | 2D culture | Knockout of propionyl CoA carboxylase | [89] |
| Transthyretin (TTR) Amyloidosis | 2D culture | Secretion of abnormal TTR, increased expression of transferrin and unfolded protein response signaling pathways | [90,91] |
| Wilson Disease | 2D culture | Patient-derived, HLC phenotype uncharacterized | [92] |
| 2D culture | Increased trafficking of ATP7B to the Golgi complex, increased rate of ATP7B degradation | [93] | |
| Zellweger Spectrum Disorder | 2D culture | Defective peroxisome assembly | [94] |
| Disease/Purpose | Assay Description | Results | Reference |
|---|---|---|---|
| α-1 Antitrypsin Deficiency | Screened over 3000 compounds from the Johns Hopkins Drug library using immunofluorescence to determine effect on AAT levels | Five hits confirmed to cause consistent reduction in AAT across multiple iPSC lines | [103] |
| Liver Fibrosis | Screened over 1400 compounds using a red fluorescent protein reporter line to assay inhibition of stellate cell activation | Two compounds suitable for oral administration identified as potential treatments for liver fibrosis | [97] |
| Familial Hypercholesterolemia | Screened over 2300 small molecules from the SPECTRUM collection drug library using an ELISA-based assay to detect ApoB secretion | Identified cardiac glycosides as potential treatment for lowering ApoB secretion | [13] |
| mtDNA Depletion Syndrome | Screened over 2300 small molecules from the SPECTRUM collection drug library using a luciferase ATP assay to identify drugs that could restore ATP levels | Identified NAD as being able to improve ATP production and mitochondrial function | [56] |
| Niemann–Pick Disease Type C | Used a series of 2-hydroxypropyl-cyclodextrins to determine impact on cholesterol accumulation and hepatic function | Identified HPGCD as potential treatment for NPC | [87] |
| Non-alcoholic Fatty Liver Disease | Screened 13,000 compounds from AstraZeneca chemogenic library using BODIPY staining to quantify intracellular neutral lipid droplets | 21 confirmed hits identified CDK2-4-C/EBPα/DGAT2 pathway as therapeutic target for lowering lipid accumulation | [95] |
| Identifying regulatory pathways for hepatic differentiation | Screened over 1100 small molecules using immunofluorescence to quantify HNF4α levels | Identification of HSP90β as a regulator of hepatic progenitor formation | [96] |
| Toxicity Screening | Generated mCherry-tagged CYP1A1 HLCs and screened 241 chemicals to identify aryl hydrocarbon receptor modulators | Five novel hits determined to up- or down-regulate expression of CYP1A1 in HLCs | [98] |
| Developed a 3D coculture model with macrophages and endothelial cells; screened 159 known toxic compounds for effects on hepatic function | Identified albumin expression assay as most-sensitive method for calculating TC50 values with this system | [99] | |
| Screened 238 marketed drugs using liver organoids in a multiplexed readout assay | Validated high predictive values for effects on viability, cholestatic, and/or mitochondrial toxicity | [100] | |
| Developed noncontact coculture model of liver spheroids and renal proximal tubule cells to assay liver and kidney toxicity simultaneously | Demonstrated toxicity profiles could be discriminated with known toxic CYP inhibitor compound CsA | [101] | |
| Screened 47 compounds for effects on albumin, urea, and ATP levels using micropatterned coculture of HLCs and murine embryonic fibroblasts | Micropatterned coculture model showed similar sensitivity for toxic drug identification to primary hepatocyte model | [102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blaszkiewicz, J.; Duncan, S.A. Advancements in Disease Modeling and Drug Discovery Using iPSC-Derived Hepatocyte-like Cells. Genes 2022, 13, 573. https://doi.org/10.3390/genes13040573
Blaszkiewicz J, Duncan SA. Advancements in Disease Modeling and Drug Discovery Using iPSC-Derived Hepatocyte-like Cells. Genes. 2022; 13(4):573. https://doi.org/10.3390/genes13040573
Chicago/Turabian StyleBlaszkiewicz, Josef, and Stephen A. Duncan. 2022. "Advancements in Disease Modeling and Drug Discovery Using iPSC-Derived Hepatocyte-like Cells" Genes 13, no. 4: 573. https://doi.org/10.3390/genes13040573
APA StyleBlaszkiewicz, J., & Duncan, S. A. (2022). Advancements in Disease Modeling and Drug Discovery Using iPSC-Derived Hepatocyte-like Cells. Genes, 13(4), 573. https://doi.org/10.3390/genes13040573