HSF1-Activated Non-Coding Stress Response: Satellite lncRNAs and Beyond, an Emerging Story with a Complex Scenario
Abstract
1. Introduction
2. The Genomic Noncoding Sequences Transcribed under the Direct Control of HSF1
2.1. Satellite lncRNAs
HSF1 in the Control of Human SATIII Repeats’ Expression
2.2. Telomeric Repeat Containing RNA (TERRA) lncRNAs
HSF1 in the Control of Human TERRAs’ Expression
2.3. Short Interspersed Nuclear Element (SINE)-Containing lncRNAs
HSF1 in the Control of SINEs’ Expression
2.4. Enhancer RNAs (eRNAs)
HSF1 in the Control of eRNAs’ Expression
2.5. Nuclear-Enriched Abundant Transcript 1 (NEAT1)
HSF1 in the Control of Human NEAT1 Expression
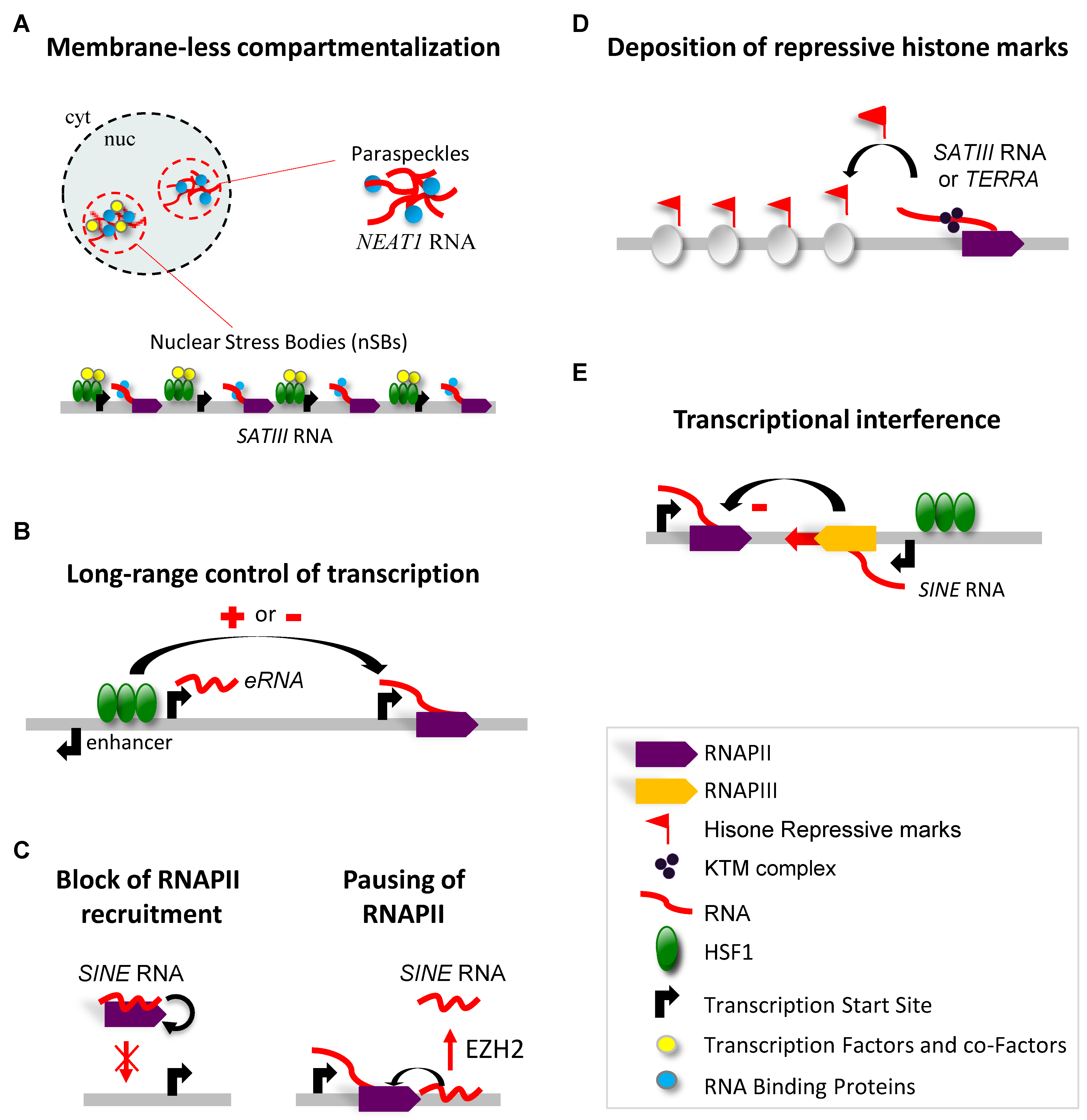
3. Molecular Functions of HSF1-Driven Production of lncRNAs in Response to Heat Stress
3.1. LncRNA-Dependent Nuclear Relocation of Factors Involved in the Control of Transcription
3.2. LncRNA-Dependent Nuclear Relocation of Factors Involved in the Control of Splicing
3.3. Interaction of lncRNAs with the Transcriptional Machinery
3.4. LncRNA-Dependent Recruitment of Repressive Complexes
3.5. LncRNA-Dependent Control of Genome Integrity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ciocca, D.R.; Arrigo, A.P.; Calderwood, S.K. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: An update. Arch. Toxicol. 2012, 87, 19–48. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S. HSF1, a versatile factor in tumorogenesis. Curr. Mol. Med. 2012, 12, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Nakai, A. Molecular basis of HSF regulation. Nat. Struct. Mol. Biol. 2016, 23, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Joutsen, J.; Sistonen, L. Tailoring of Proteostasis Networks with Heat Shock Factors. Cold Spring Harb. Perspect. Biol. 2019, 11, a034066. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef]
- Vihervaara, A.; Mahat, D.B.; Guertin, M.J.; Chu, T.; Danko, C.G.; Lis, J.T.; Sistonen, L. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Mahat, D.B.; Salamanca, H.H.; Duarte, F.M.; Danko, C.G.; Lis, J.T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-Wide Transcriptional Regulation. Mol. Cell 2016, 62, 63–78. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Jolly, C.; Metz, A.; Govin, J.; Vigneron, M.; Turner, B.M.; Khochbin, S.; Vourc’H, C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2003, 164, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, N.; Denegri, M.; Chiodi, I.; Corioni, M.; Valgardsdottir, R.; Cobianchi, F.; Riva, S.; Biamonti, G. Transcriptional Activation of a Constitutive Heterochromatic Domain of the Human Genome in Response to Heat Shock. Mol. Biol. Cell 2004, 15, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Koskas, S.; Decottignies, A.; Dufour, S.; Pezet, M.; Verdel, A.; Vourc’H, C.; Faure, V. Heat shock factor 1 promotes TERRA transcription and telomere protection upon heat stress. Nucleic Acids Res. 2017, 45, 6321–6333. [Google Scholar] [CrossRef] [PubMed]
- Horard, B.; Eymery, A.; Fourel, G.; Vassetzky, N.; Puechberty, J.; Roizes, G.; Lebrigand, K.; Barbry, P.; Laugraud, A.; Gautier, C.; et al. Global analysis of DNA methylation and transcription of human repetitive sequences. Epigenetics 2009, 4, 339–350. [Google Scholar] [CrossRef][Green Version]
- Deininger, P.L. Alu elements: Know the SINEs. Genome Biol. 2011, 12, 1–12. [Google Scholar] [CrossRef]
- Liu, W.-M.; Chu, W.-M.; Choudary, P.V.; Schmid, C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995, 23, 1758–1765. [Google Scholar] [CrossRef]
- Lellahi, S.M.; Rosenlund, I.A.; Hedberg, A.; Kiær, L.T.; Mikkola, I.; Knutsen, E.; Perander, M. The long noncoding RNA NEAT1 and nuclear paraspeckles are up-regulated by the transcription factor HSF1 in the heat shock response. J. Biol. Chem. 2018, 293, 18965–18976. [Google Scholar] [CrossRef]
- Seal, R.L.; Chen, L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A guide to naming human non-coding RNA genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef]
- Eymery, A.; Callanan, M.; Vourc’h, C. The Secret Message of Heterochromatin: New Insights into the Mechanisms and Function of Centromeric and Pericentric Repeat Sequence Transcription. Int. J. Dev. Biol. 2009, 53, 259–268. [Google Scholar] [CrossRef]
- Prosser, J.; Frommer, M.; Paul, C.; Vincent, P. Sequence relationships of three human satellite DNAs. J. Mol. Biol. 1986, 187, 145–155. [Google Scholar] [CrossRef]
- Vissel, B.; Choo, K.H. Human α Satellite DNA—Consensus Sequence and Conserved Regions. Nucleic Acids Res. 1987, 15, 6751. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Rattner, J. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 1988, 16, 11645–11661. [Google Scholar] [CrossRef] [PubMed]
- Manuelidis, L. Consensus sequence of mouse satellite DNA indicates it is derived from tandem 116 basepair repeats. FEBS Lett. 1981, 129, 25–28. [Google Scholar] [CrossRef]
- Prosser, J.; Reisner, A.; Bradley, M.; Ho, K.; Vincent, P. Buoyant density and hybridization analysis of human DNA sequences, including three satellite DNAs. Biochim. Biophys. Acta (BBA) Nucleic Acids Protein Synth. 1981, 656, 93–102. [Google Scholar] [CrossRef]
- Frommer, M.; Prosser, J.; Vincent, P.C. Human Satellite I Sequences Include a Male Specific 2.47 Kb Tandemly Repeated Unit Containing One Alu Family Member Per Repeat. Nucleic Acids Res. 1984, 12, 2887–2900. [Google Scholar] [CrossRef]
- Lee, C.; Wevrick, R.; Fisher, R.B.; Ferguson-Smith, M.A.; Lin, C.C. Human centromeric DNAs. Qual. Life Res. 1997, 100, 291–304. [Google Scholar] [CrossRef]
- Altemose, N.; Miga, K.H.; Maggioni, M.; Willard, H.F. Genomic Characterization of Large Heterochromatic Gaps in the Human Genome Assembly. PLoS Comput. Biol. 2014, 10, e1003628. [Google Scholar] [CrossRef]
- Altemose, N. A Classical Revival: Human Satellite DNAs Enter the Genomics Era. Preprints 2022, 2022020009. [Google Scholar] [CrossRef]
- Hoyt, S.J.; Storer, J.M.; Hartley, G.A.; Grady, P.G.; Gershman, A.; de Lima, L.G.; Limouse, C.; Halabian, R.; Wojenski, L.; Rodriguez, M.; et al. From Telomere to Telomere: The Transcriptional and Epigenetic State of Human Repeat Elements. bioXiv 2021. [Google Scholar] [CrossRef]
- Saksouk, N.; Barth, T.K.; Ziegler-Birling, C.; Olova, N.; Nowak, A.; Rey, E.; Mateos-Langerak, J.; Urbach, S.; Reik, W.; Torres-Padilla, M.-E.; et al. Redundant Mechanisms to Form Silent Chromatin at Pericentromeric Regions Rely on BEND3 and DNA Methylation. Mol. Cell 2014, 56, 580–594. [Google Scholar] [CrossRef]
- Probst, A.V.; Almouzni, G. Heterochromatin Establishment in the Context of Genome-Wide Epigenetic Reprogramming. Trends Genet. 2011, 27, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Padeken, J.; Zeller, P.; Towbin, B.; Katic, I.; Kalck, V.; Methot, S.P.; Gasser, S.M. Syner-gistic Lethality between Brca1 and H3k9me2 Loss Reflects Satellite Derepression. Genes Dev. 2019, 33, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.X.; Fan, S.; Xiong, J.; Yuan, R.-Q.; Meng, Q.; Gao, M.; Goldberg, I.D.; Fuqua, S.A.; Pestell, R.G.; Rosen, E.M. Role of BRCA1 in heat shock response. Oncogene 2003, 22, 10–27. [Google Scholar] [CrossRef]
- Zhu, Q.; Pao, G.M.; Huynh, A.M.; Suh, H.; Tonnu, N.; Nederlof, P.M.; Gage, F.H.; Verma, I.M. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 2011, 477, 179–184. [Google Scholar] [CrossRef]
- Nurk, S.; Sergey, K.; Arang, R.; Mikko, R.; Andrey, V.B.; Alla, M.; Mitchell, R.V.; Nicolas, A.; Lev, U.; Ariel, G.; et al. The Complete Sequence of a Human Genome. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jolly, C.; Konecny, L.; Grady, D.L.; Kutskova, Y.A.; Cotto, J.J.; Morimoto, R.I.; Vourc’H, C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 2002, 156, 775–781. [Google Scholar] [CrossRef]
- Penin, J.; Dufour, S.; Faure, V.; Fritah, S.; Seigneurin-Berny, D.; Col, E.; Verdel, A.; Vourc’H, C. Chromosome Y pericentric heterochromatin is a primary target of HSF1 in male cells. Chromosoma 2021, 130, 53–60. [Google Scholar] [CrossRef]
- Eymery, A.; Souchier, C.; Vourc’h, C.; Jolly, C. Heat Shock Factor 1 Binds to and Transcribes Satellite Ii and Iii Sequences at Several Pericentromeric Regions in Heat-Shocked Cells. Exp. Cell Res. 2010, 316, 1845–1855. [Google Scholar] [CrossRef]
- Valgardsdottir, R.; Chiodi, I.; Giordano, M.; Rossi, A.; Bazzini, S.; Ghigna, C.; Riva, S.; Biamonti, G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res. 2007, 36, 423–434. [Google Scholar] [CrossRef]
- Lu, J.; Gilbert, D.M.; Li, Z.; Wang, L.; Hays, T.S.; Cai, Y. Proliferation-dependent and cell cycle–regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007, 179, 411–421. [Google Scholar] [CrossRef]
- Probst, A.V.; Okamoto, I.; Casanova, M.; el Marjou, F.; le Baccon, P.; Almouzni, G. A Strand-Specific Burst in Transcription of Pericentric Satellites Is Required for Chromocenter Formation and Early Mouse Development. Dev. Cell 2010, 19, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Rudert, F.; Bronner, S.; Garnier, J.M.; Dolle, P. Transcripts from Opposite Strands of γ Satellite DNA Are Differentially Expressed During Mouse Development. Mamm. Genome 1995, 6, 76–83. [Google Scholar] [PubMed]
- Jehan, Z.; Vallinayagam, S.; Tiwari, S.; Pradhan, S.; Singh, L.; Suresh, A.; Reddy, H.M.; Ahuja, Y.; Jesudasan, R.A. Novel noncoding RNA from human Y distal heterochromatic block (Yq12) generates testis-specific chimeric CDC2L2. Genome Res. 2007, 17, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Eymery, A.; Horard, B.; El Atifi-Borel, M.; Fourel, G.; Berger, F.; Vitte, A.-L.; Van den Broeck, A.; Brambilla, E.; Fournier, A.; Callanan, M.; et al. A transcriptomic analysis of human centromeric and pericentric sequences in normal and tumor cells. Nucleic Acids Res. 2009, 37, 6340–6354. [Google Scholar] [CrossRef]
- Ting, D.T.; Lipson, D.; Paul, S.; Brannigan, B.W.; Akhavanfard, S.; Coffman, E.J.; Contino, G.; Deshpande, V.; Iafrate, A.J.; Letovsky, S.; et al. Aberrant Overexpression of Satellite Repeats in Pancreatic and Other Epithelial Cancers. Science 2011, 331, 593–596. [Google Scholar] [CrossRef]
- Kanne, J.; Hussong, M.; Isensee, J.; Munoz-Lopez, A.; Wolffgramm, J.; Hess, F.; Grimm, C.; Bessonov, S.; Meder, L.; Wang, J.; et al. Pericentromeric Satellite Iii Transcripts Induce Etoposide Resistance. Cell Death Dis. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Blackburn, E.H. Telomere states and cell fates. Nature 2000, 408, 53–56. [Google Scholar] [CrossRef]
- Cech, T.R. Beginning to Understand the End of the Chromosome. Cell 2004, 116, 273–279. [Google Scholar] [CrossRef]
- Feuerhahn, S.; Iglesias, N.; Panza, A.; Porro, A.; Lingner, J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010, 584, 3812–3818. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat–Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Bettin, N.; Oss Pegorar, C.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Diman, A.; Decottignies, A. Genomic Origin and Nuclear Localization of Terra Telomeric Repeat-Containing RNA: From Darkness to Dawn. FEBS J. 2018, 285, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Farnung, B.O.; Wischnewski, H.; Khoriauli, L.; Vitelli, V.; Chawla, R.; Giulotto, E.; Azzalin, C.M. CpG-Island Promoters Drive Transcription of Human Telomeres. RNA 2009, 15, 2186–2194. [Google Scholar] [CrossRef]
- Porro, A.; Feuerhahn, S.; Lingner, J. TERRA-Reinforced Association of LSD1 with MRE11 Promotes Processing of Uncapped Telomeres. Cell Rep. 2014, 6, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blanco, R.; de Silanes, I.L.; Muñoz, P.; Gómez-López, G.; Flores, J.M.; Blasco, M.A. Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations. Proc. Natl. Acad. Sci. USA 2009, 106, 19393–19398. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Morcillo, G.; Barettino, D.; Carmona, M.J.; Carretero, M.T. Telomeric DNA sequences differentially activated by heat shock in two Chironomus subspecies. Chromosoma 1988, 96, 139–144. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef]
- Dewannieux, M.; Heidmann, T. LINEs, SINEs and processed pseudogenes: Parasitic strategies for genome modeling. Cytogenet. Genome Res. 2005, 110, 35–48. [Google Scholar] [CrossRef]
- Schmid, C.W. Does SINE evolution preclude Alu function? Nucleic Acids Res. 1998, 26, 4541–4550. [Google Scholar] [CrossRef]
- Paulson, K.; Schmid, C.W. Transcriptional inactivity of Alu repeats in HeLa cells. Nucleic Acids Res. 1986, 14, 6145–6158. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.M.; Carroll, M.L.; Nguyen, S.V.; Salem, A.-H.; Oldridge, M.; Wilkie, A.O.M.; Batzer, M.A.; Deininger, P.L. Potential Gene Conversion and Source Genes for Recently Integrated Alu Elements. Genome Res. 2000, 10, 1485–1495. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Vavrova-Anderson, J.; Oler, A.J.; Cowling, V.H.; Cairns, B.R.; White, R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.M.; Deininger, P.L.; Efstratiadis, A. Nonviral Retroposons: Genes, Pseudogenes, and Transposable Elements Generated by the Reverse Flow of Genetic In-formation. Annu. Rev. Biochem. 1986, 55, 631–661. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.R.; Sheffield, R.F.; Rudin, C.M. Human Alu Element Retrotransposition Induced by Genotoxic Stress. Nat. Genet. 2003, 35, 219–220. [Google Scholar] [CrossRef]
- Pandey, R.; Mandal, A.K.; Jha, V.; Mukerji, M. Heat Shock Factor Binding in Alu Repeats Expands Its Involvement in Stress through an Antisense Mechanism. Genome Biol. 2012, 12, 1–17. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A Class of Long Noncoding RNAs Synthesized at Enhancers: Figure 1. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3k27ac Separates Active from Poised Enhancers and Predicts Developmental State. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Wang, D.; Garcia-Bassets, I.; Benner, C.; Li, W.; Su, X.; Zhou, Y.; Qiu, J.; Liu, W.; Kaikkonen, M.; Ohgi, K.A.; et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011, 474, 390–394. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007, 8, 1–16. [Google Scholar] [CrossRef]
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A Novel Nuclear Domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef]
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. Men Epsilon/β Nuclear-Retained Non-Coding RNAs Are up-Regulated Upon Muscle Differentiation and Are Essential Components of Paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-End Processing of Long Noncoding RNA Initiates Construction of Nuclear Paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. Menepsilon/β Noncoding RNAs Are Essential for Structural Integrity of Nuclear Paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Li, R.; Harvey, A.R.; Hodgetts, S.I.; Fox, A.H. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 2017, 23, 872–881. [Google Scholar] [CrossRef]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural Analyses of Neat1 LncRNAs Suggest Long-Range RNA Interactions That May Contribute to Paraspeckle Architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef]
- Chen, L.L.; Carmichael, G.G. Altered Nuclear Retention of mRNAs Containing Inverted Repeats in Human Embryonic Stem Cells: Functional Role of a Nuclear Noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef]
- Biamonti, G.; Vourc’H, C. Nuclear Stress Bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000695. [Google Scholar] [CrossRef]
- Krol, J. Paraspeckles: Nuclear nests helping to raise mature miRNAs. Nat. Struct. Mol. Biol. 2017, 24, 783–784. [Google Scholar] [CrossRef]
- Biamonti, G. Nuclear stress bodies: A heterochromatin affair? Nat. Rev. Mol. Cell Biol. 2004, 5, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Biamonti, G.; Caceres, J.F. Cellular stress RNA splicing. Trends Biochem. Sci. 2009, 34, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Vourc’H, C.; Biamonti, G. Transcription of Satellite DNAs in Mammals. Long Non-Coding RNAs 2010, 51, 95–118. [Google Scholar] [CrossRef]
- Hussong, M.; Kaehler, C.; Kerick, M.; Grimm, C.; Franz, A.; Timmermann, B.; Welzel, F.; Isensee, J.; Hucho, T.; Krobitsch, S.; et al. The bromodomain protein BRD4 regulates splicing during heat shock. Nucleic Acids Res. 2017, 45, 382–394. [Google Scholar] [CrossRef]
- Fritah, S.; Col, E.; Boyault, C.; Govin, J.; Sadoul, K.; Chiocca, S.; Christians, E.; Khochbin, S.; Jolly, C.; Vourc’H, C. Heat-Shock Factor 1 Controls Genome-wide Acetylation in Heat-shocked Cells. Mol. Biol. Cell 2009, 20, 4976–4984. [Google Scholar] [CrossRef]
- Miozzo, F.; Saberan-Djoneidi, D.; Mezger, V. HSFs, Stress Sensors and Sculptors of Transcription Compartments and Epigenetic Landscapes. J. Mol. Biol. 2015, 427, 3793–3816. [Google Scholar] [CrossRef]
- Col, E.; Hoghoughi, N.; Dufour, S.; Penin, J.; Koskas, S.; Faure, V.; Ouzounova, M.; Hernandez-Vargash, H.; Reynoird, N.; Daujat, S.; et al. Bromodomain Factors of Bet Family Are New Essential Actors of Pericentric Heterochromatin Transcriptional Activation in Response to Heat Shock. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Goenka, A.; Sengupta, S.; Pandey, R.; Parihar, R.; Mohanta, G.C.; Mukerji, M.; Ganesh, S. Human satellite-III non-coding RNAs modulate heat shock-induced transcriptional repression. J. Cell Sci. 2016, 129, 3541–3552. [Google Scholar] [CrossRef]
- Marmorstein, R.; Zhou, M.-M. Writers and Readers of Histone Acetylation: Structure, Mechanism, and Inhibition. Cold Spring Harb. Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef]
- Bond, U. Heat shock but not other stress inducers leads to the disruption of a subset of snRNPs and inhibition of in vitro splicing in HeLa cells. EMBO J. 1988, 7, 3509–3518. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M.; Pepperkok, R.; Carvalho, M.T.; Lamond, A.I. Transcription-Dependent Colocalization of the U1, U2, U4/U6, and U5 Snrnps in Coiled Bodies. J. Cell. Biol. 1992, 117, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chiodi, I.; Corioni, M.; Giordano, M.; Valgardsdottir, R.; Ghigna, C.; Cobianchi, F.; Xu, R.M.; Riva, S.; Biamonti, G. RNA Recognition Motif 2 Directs the Recruitment of Sf2/Asf to Nuclear Stress Bodies. Nucleic Acids Res. 2004, 32, 4127–4141. [Google Scholar] [CrossRef] [PubMed]
- Yost, H.; Lindquist, S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 1986, 45, 185–193. [Google Scholar] [CrossRef]
- Matera, A.G.; Ward, D.C. Nucleoplasmic Organization of Small Nuclear Ribonucleoproteins in Cultured Human Cells. J. Cell Biol. 1993, 121, 715–727. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, M.; Manley, J.L. Manley. Phosphorylation Switches the General Splicing Re-pressor Srp38 to a Sequence-Specific Activator. Nat. Struct. Mol. Biol. 2008, 15, 1040–1048. [Google Scholar] [CrossRef]
- Shin, C.; Feng, Y.; Manley, J. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 2004, 427, 553–558. [Google Scholar] [CrossRef]
- Weighardt, F.; Cobianchi, F.; Cartegni, L.; Chiodi, I.; Villa, A.; Riva, S.; Biamonti, G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 1999, 112, 1465–1476. [Google Scholar] [CrossRef]
- Ninomiya, K.; Adachi, S.; Natsume, T.; Iwakiri, J.; Terai, G.; Asai, K.; Hirose, T. LncRNA-Dependent Nuclear Stress Bodies Promote Intron Retention through Sr Protein Phosphorylation. EMBO J. 2020, 39, e102729. [Google Scholar] [CrossRef]
- Metz, A.; Soret, J.; Vourc’H, C.; Tazi, J.; Jolly, C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J. Cell Sci. 2004, 117, 4551–4558. [Google Scholar] [CrossRef]
- Denegri, M.; Chiodi, I.; Corioni, M.; Cobianchi, F.; Riva, S.; Biamonti, G. Stress-Induced Nuclear Bodies Are Sites of Accumulation of Pre-mRNA Processing Factors. Mol. Biol. Cell 2001, 12, 3502–3514. [Google Scholar] [CrossRef]
- Ninomiya, K.; Iwakiri, J.; Aly, M.K.; Sakaguchi, Y.; Adachi, S.; Natsume, T.; Terai, G.; Asai, K.; Suzuki, T.; Hirose, T. M(6) a Modification of Hsatiii LncRNAs Regulates Temperature-Dependent Splicing. EMBO J. 2021, 40, e107976. [Google Scholar] [CrossRef] [PubMed]
- Valgardsdottir, R.; Chiodi, I.; Giordano, M.; Cobianchi, F.; Riva, S.; Biamonti, G. Struc-tural and Functional Characterization of Noncoding Repetitive RNAs Transcribed in Stressed Human Cells. Mol. Biol Cell 2005, 16, 2597–2604. [Google Scholar] [CrossRef] [PubMed]
- Shalgi, R.; Hurt, J.A.; Lindquist, S.; Burge, C.B. Widespread Inhibition of Posttranscriptional Splicing Shapes the Cellular Transcriptome Following Heat Shock. Cell Rep. 2014, 7, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Shao, C.; Wu, Q.J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.T.; Zhang, Y.; Wang, Y.; et al. Neat1 Scaffolds RNA-Binding Proteins and the Microprocessor to Globally Enhance pri-miRna Processing. Nat. Struct Mol. Biol. 2017, 24, 816–824. [Google Scholar] [CrossRef]
- Lakhotia, S.C.; Mallik, M.; Singh, A.K.; Ray, M. The Large Noncoding Hsr omega-N Transcripts Are Essential for Thermotolerance and Remobilization of Hnrnps, Hp1 and RNA Polymerase II during Recovery from Heat Shock in Drosophila. Chromosoma 2012, 121, 49–70. [Google Scholar] [CrossRef]
- Mallik, M.; Lakhotia, S.C. The Developmentally Active and Stress-Inducible Noncoding Hsr omega Gene Is a Novel Regulator of Apoptosis in Drosophila. Genetics 2009, 183, 831–852. [Google Scholar] [CrossRef]
- Lakhotia, S.C. Long non-coding RNAs coordinate cellular responses to stress. Wiley Interdiscip. Rev. RNA 2012, 3, 779–796. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Rajendra, T.K.; Lal, A.K.; Lakhotia, S.C. Omega Speckles—A Novel Class of Nuclear Speckles Containing Hnrnps Associated with Noncoding Hsr-Omega RNA in Drosophila. J. Cell Sci. 2000, 113, 3485–3497. [Google Scholar] [CrossRef]
- Jolly, C.; Lakhotia, S.C. Human sat III and Drosophila hsr omega transcripts: A common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2007, 35, 2812. [Google Scholar] [CrossRef][Green Version]
- Singh, A.K.; Lakhotia, S.C. Dynamics of hnRNPs and omega speckles in normal and heat shocked live cell nuclei of Drosophila melanogaster. Chromosoma 2015, 124, 367–383. [Google Scholar] [CrossRef]
- Kwak, H.; Fuda, N.J.; Core, L.J.; Lis, J.T. Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science 2013, 339, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2011, 3, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.-H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef]
- Tippens, N.D.; Vihervaara, A.; Lis, J.T. Enhancer Transcription: What, Where, When, and Why? Genes Dev. 2018, 32, 1–3. [Google Scholar] [CrossRef]
- A Allen, T.; Von Kaenel, S.; A Goodrich, J.; Kugel, J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004, 11, 816–821. [Google Scholar] [CrossRef]
- Espinoza, C.A.; Allen, T.A.; Hieb, A.R.; Kugel, J.F.; Goodrich, J.A. B2 RNA Binds Directly to RNA Polymerase II to Repress Transcript Synthesis. Nat. Struct. Mol. Biol. 2004, 11, 822–829. [Google Scholar] [CrossRef]
- Mariner, P.D.; Walters, R.D.; Espinoza, C.A.; Drullinger, L.F.; Wagner, S.D.; Kugel, J.F.; Goodrich, J.A. Human Alu RNA Is a Modular Transacting Repressor of mRNA Transcription during Heat Shock. Mol. Cell 2008, 29, 499–509. [Google Scholar] [CrossRef]
- Yakovchuk, P.; Goodrich, J.A.; Kugel, J.F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. USA 2009, 106, 5569–5574. [Google Scholar] [CrossRef]
- Zovoilis, A.; Cifuentes-Rojas, C.; Chu, H.-P.; Hernandez, A.J.; Lee, J.T. Destabilization of B2 RNA by EZH2 Activates the Stress Response. Cell 2016, 167, 1788–1802.e13. [Google Scholar] [CrossRef]
- Maison, C.; Bailly, D.; Peters, A.H.; Quivy, J.P.; Roche, D.; Taddei, A.; Lachner, M.; Jenuwein, T.; Almouzni, G. Higher-Order Structure in Pericentric Heterochromatin In-volves a Distinct Pattern of Histone Modification and an RNA Component. Nat. Genet. 2002, 30, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Muchardt, C.; Guillemé, M.; Seeler, J.-S.; Trouche, D.; Dejean, A.; Yaniv, M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 2002, 3, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.V.; Santos, F.; Reik, W.; Almouzni, G.; Dean, W. Structural Differences in Centromeric Heterochromatin Are Spatially Reconciled on Fertilisation in the Mouse Zygote. Chromosoma 2007, 116, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Kanellopoulou, C.; Muljo, S.A.; Kung, A.L.; Ganesan, S.; Drapkin, R.; Jenuwein, T.; Livingston, D.M.; Rajewsky, K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005, 19, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Gutbrod, M.J.; Roche, B.; Steinberg, J.I.; Lakhani, A.A.; Chang, K.; Schorn, A.J.; Martienssen, R.A. Dicer promotes genome stability via the bromodomain transcriptional co-activator BRD4. Nat. Commun. 2022, 13, 1–14. [Google Scholar] [CrossRef]
- Fukagawa, T.; Nogami, M.; Yoshikawa, M.; Ikeno, M.; Okazaki, T.; Takami, Y.; Nakayama, T.; Oshimura, M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004, 6, 784–791. [Google Scholar] [CrossRef]
- Giordano, M.; Infantino, L.; Biggiogera, M.; Montecucco, A.; Biamonti, G. Heat Shock Affects Mitotic Segregation of Human Chromosomes Bound to Stress-Induced Satellite III RNAs. Int. J. Mol. Sci. 2020, 21, 2812. [Google Scholar] [CrossRef]
- Pezer, Z.; Ugarkovic, D. Satellite DNA-associated siRNAs as mediators of heat shock response in insects. RNA Biol. 2012, 9, 587–595. [Google Scholar] [CrossRef]
- da Rocha, S.T.; Heard, E. Novel Players in X Inactivation: Insights into Xist-Mediated Gene Silencing and Chromosome Conformation. Nat. Struct. Mol. Biol. 2017, 24, 197–204. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Lingner, J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015, 25, 29–36. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. A ‘Higher Order’ of Telomere Regulation: Telomere Heterochromatin and Telomeric RNAs. EMBO J. 2009, 28, 2323–2336. [Google Scholar] [CrossRef] [PubMed]
- Tardat, M.; Déjardin, J. Telomere chromatin establishment and its maintenance during mammalian development. Chromosoma 2018, 127, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.J.; López-Silanes, I.; Megias, D.; Fraga, M.F.; Castells-García, Á.; Blasco, M.A. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Biscotti, M.A.; Canapa, A.; Forconi, M.; Olmo, E.; Barucca, M. Transcription of Tan-demly Repetitive DNA: Functional Roles. Chromosom. Res. 2015, 23, 463–477. [Google Scholar] [CrossRef]
- Catania, S.; Pidoux, A.L.; Allshire, R.C. Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin. PLoS Genet. 2015, 11, e1004986. [Google Scholar] [CrossRef]
- Ferri, F.; Bouzinba-Segard, H.; Velasco, G.; Hubé, F.; Francastel, C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009, 37, 5071–5080. [Google Scholar] [CrossRef]
- Grenfell, A.W.; Strzelecka, M.; Heald, R. Transcription brings the complex(ity) to the centromere. Cell Cycle 2017, 16, 235–236. [Google Scholar] [CrossRef]
- Quénet, D.; Dalal, Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 2014, 3, e03254. [Google Scholar] [CrossRef]
- Rosic, S.; Erhardt, S. No longer a nuisance: Long non-coding RNAs join CENP-A in epigenetic centromere regulation. Cell. Mol. Life Sci. 2016, 73, 1387–1398. [Google Scholar] [CrossRef]
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; et al. Centromere RNA Is a Key Component for the Assembly of Nucleoproteins at the Nucleolus and Cen-tromere. Genome Res. 2007, 17, 1146–1160. [Google Scholar] [CrossRef]
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human Centromeres Produce Chromosome-Specific and Array-Specific α Satellite Transcripts That Are Complexed with Cenp-A and Cenp-C. Dev. Cell 2017, 42, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Bouzinba-Segard, H.; Guais, A.; Francastel, C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA 2006, 103, 8709–8714. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.H.; O’Carroll, D.; Scherthan, H.; Mechtler, K.; Sauer, S.; Schöfer, C.; Weipoltshammer, K.; Pagani, M.; Lachner, M.; Kohlmaier, A.; et al. Loss of the Suv39h Histone Methyltransferases Impairs Mammalian Heterochromatin and Genome Stability. Cell 2001, 107, 323–337. [Google Scholar] [CrossRef]
- Rosic, S.; Kohler, F.; Erhardt, S. Repetitive Centromeric Satellite RNA Is Essential for Kinetochore Formation and Cell Division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Hoong, N.; Aslanian, A.; Hara, T.; Benner, C.; Heinz, S.; Miga, K.H.; Ke, E.; Verma, S.; Soroczynski, J.; et al. Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer. Mol. Cell 2018, 70, 842–853. [Google Scholar] [CrossRef]
- de Lange, T. Shelterin-Mediated Telomere Protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef]
- Deng, Z.; Wang, Z.; Stong, N.; Plasschaert, R.; Moczan, A.; Chen, H.-S.; Hu, S.; Wikramasinghe, P.; Davuluri, R.V.; Bartolomei, M.S.; et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012, 31, 4165–4178. [Google Scholar] [CrossRef]
- Feretzaki, M.; Nunes, P.R.; Lingner, J. Expression and differential regulation of human TERRA at several chromosome ends. RNA 2019, 25, 1470–1480. [Google Scholar] [CrossRef]
- Mendez-Bermudez, A.; Lototska, L.; Bauwens, S.; Giraud-Panis, M.J.; Croce, O.; Jamet, K.; Irizar, A.; Mowinckel, M.; Koundrioukoff, S.; Nottet, N.; et al. Ge-nome-Wide Control of Heterochromatin Replication by the Telomere Capping Protein Trf2. Mol. Cell 2018, 70, 449–461. [Google Scholar] [CrossRef]
- Petrova, N.V.; Velichko, A.K.; Kantidze, O.L.; Razin, S.V. Heat Shock-Induced Dis-sociation of Trf2 from Telomeres Does Not Initiate a Telomere-Dependent DNA Damage Response. Cell Biol. Int. 2014, 38, 675–681. [Google Scholar] [CrossRef]
- Velichko, A.K.; Petrova, N.V.; Kantidze, O.L.; Razin, S.V. Dual effect of heat shock on DNA replication and genome integrity. Mol. Biol. Cell 2012, 23, 3450–3460. [Google Scholar] [CrossRef] [PubMed]
- Masuta, Y.; Kawabe, A.; Nozawa, K.; Naito, K.; Kato, A.; Ito, H. Characterization of a heat-activated retrotransposon in Vigna angularis. Breed. Sci. 2018, 68, 168–176. [Google Scholar] [CrossRef]
- Vasilyeva, L.A.; Bubenshchikova, E.V.; Ratner, V.A. Heavy Heat Shock Induced Retrotransposon Transposition in Drosophila. Genet. Res. 1999, 74, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Mittelsten Scheid, O. How a Retrotransposon Exploits the Plant’s Heat Stress Response for Its Activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef] [PubMed]
- Casacuberta, E.; González, J. The impact of transposable elements in environmental adaptation. Mol. Ecol. 2013, 22, 1503–1517. [Google Scholar] [CrossRef] [PubMed]
- Chenais, B.; Caruso, A.; Hiard, S.; Casse, N. The Impact of Transposable Elements on Eukaryotic Genomes: From Genome Size Increase to Genetic Adaptation to Stressful Environments. Gene 2012, 509, 7–15. [Google Scholar] [CrossRef]
- Bersani, F.; Lee, E.; Kharchenko, P.V.; Xu, A.W.; Liu, M.; Xega, K.; MacKenzie, O.C.; Brannigan, B.W.; Wittner, B.S.; Jung, H.; et al. Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 15148–15153. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome Editing. The New Frontier of Genome Engineering with Crispr-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Helm, M.; Motorin, Y. Detecting RNA modifications in the epitranscriptome: Predict and validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Ji, L.; Zhang, Y.; Lv, P.; Cao, X.; Wang, Q.; Yan, Z.; Dong, S.; Du, D.; Zhang, F.; et al. The Nuclear Matrix Protein SAFB Cooperates with Major Satellite RNAs to Stabilize Heterochromatin Architecture Partially through Phase Separation. Mol. Cell 2020, 77, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Gaglia, G.; Rashid, R.; Yapp, C.; Joshi, G.N.; Li, C.G.; Lindquist, S.L.; Sarosiek, K.A.; Whitesell, L.; Sorger, P.K.; Santagata, S. Hsf1 Phase Transition Mediates Stress Adaptation and Cell Fate Decisions. Nat. Cell Biol. 2020, 22, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol. Cell 2018, 70, 1038–1053.e7. [Google Scholar] [CrossRef]
- Das, S.; Bhattacharyya, N.P. Heat Shock Factor 1 Regulates Hsa-miR-432 Expression in Human Cervical Cancer Cell Line. Biochem. Biophys. Res. Commun. 2014, 453, 461–466. [Google Scholar] [CrossRef]
- Place, R.F.; Noonan, E.J. Non-coding RNAs turn up the heat: An emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones 2014, 19, 159–172. [Google Scholar] [CrossRef]
- Das, S.; Bhattacharyya, N. Heat Shock Factor 1-Regulated miRNAs Can Target Huntingtin and Suppress Aggregates of Mutant Huntingtin. MicroRNA 2016, 4, 185–193. [Google Scholar] [CrossRef]
- Dai, C.; Whitesell, L.; Rogers, A.B.; Lindquist, S. Heat Shock Factor 1 Is a Powerful Multifaceted Modifier of Carcinogenesis. Cell 2007, 130, 1005–1018. [Google Scholar] [CrossRef]

| ncRNA Production Activated by HSF1. | Length | Internal Repetitive Elements | Multiple Copies | HSE within Promoter Region | Genome Localization | Molecular Function in HS Cells |
|---|---|---|---|---|---|---|
| SATIII | from 2 kb to 5 kb and more | Yes (Tandem repeats, 5 b long) | Yes | Yes (in silico) | Multiple sites at heterochromatin pericentric regions | Titration of transcription factors |
| Reorientation of splicing decision | ||||||
| Maintenance of centromeric heterochromatin | ||||||
| Maintenance of repressive histone marks | ||||||
| Alu | ~280 b | No | Yes | Yes (ChIP) | Multiple and dispersed sites | RNAPII inhibition |
| Impact of gene expression through antisens RNA | ||||||
| Impact on transcriptional elongation | ||||||
| Recruitment of repressive transcriptional complexes | ||||||
| Alteration genome integrity through retrotransposition | ||||||
| eRNA | From 50 b to 2 kb | No | No | Yes (ChIP) | Multiple and dispersed sites | Control of gene expression |
| NEAT1 | ~3 kb (NEAT1-1) ~20 kb (NEAT1-2) | No | No | Yes (ChIP) | Unique locus | miRNA biogenesis |
| HSP genes down regulation following HS | ||||||
| TERRA | From 100 b to less than 100 kb | Yes (Tandem repeats, 6 b unit) | Yes | Yes (ChIP) | Multiple sites at telomeres | Genome protection through telomere protection |
| Telomeric heterochromatin reformation by recruiting repressive chromatin marks |
| Human | ||
|---|---|---|
| centromeric repetitive motif ([20]) | ||
| Minor SAT (120 bp) | all chromosomes | GGAAAATGATAAAAACCACACTGTAGAACATATTAGATGAGTGAGTTACACTGAAAAACACATTCGTTGGAAACGGGATTTGTAGAACAGTGTATATCAATGAGTTACAATGAGAAACAT |
| pericentric repetitive motif ([21]) | ||
| Major SAT (234 bp) | all chromosomes | CCTGGAATATGGCGAGAAAACTGAAAATCACGGAAAATGAGAAATACACACTTTAGGACGTGAAATATGGCGAGGAAAACTGAAAAAGGTGGAAAATTTAGAAATGTCCACTGTAGGACGTGGAATATGGCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACGAAATCACTAAAAAACGTGAAAAATGAGAAATGCACACTGAAGGA |
| Mouse | ||
| centromeric repetitive motif ([19]) | ||
| Alphoid (171 bp) | all chromosomes | CTTCTGTCTAGTTTTTATATGAAGATATTCCCGTTTCCAACCAAGGCCTCAAAGCGGTCCAAATATCCACAAGCTGATTCTACAAAAAGAGTGTTTCAAAACTGCTCTATGAAAAGGAAGGTTCAACTCTGTGAGTTGAATGTATACATCACAAAGAAGTTTCTGAGAATG |
| pericentric satellite repetitive motif ([22,23,24,25,26,27]) | ||
| SATI | chrs 3, 4, 13, 14, 15, 21, 22, Y | Alternance of fragments A (17 bp) = ACATAAAATATG/CAAAGT and B (25 bp) B1: ACAT/CCCAAATATAG/TATTA/TTATA/TCTGT and B2: ACCCAAAGT/GCCATAT/GCATTA/CTATACT |
| SATII | chrs 1, 2, 7, 10, 15, 16, 17, 22 | (CATTC)n degenerated |
| SATIII | chrs 1, 3, 5, 7, 9, 10, 17, Y, acrocentric | (CATTC)n |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vourc’h, C.; Dufour, S.; Timcheva, K.; Seigneurin-Berny, D.; Verdel, A. HSF1-Activated Non-Coding Stress Response: Satellite lncRNAs and Beyond, an Emerging Story with a Complex Scenario. Genes 2022, 13, 597. https://doi.org/10.3390/genes13040597
Vourc’h C, Dufour S, Timcheva K, Seigneurin-Berny D, Verdel A. HSF1-Activated Non-Coding Stress Response: Satellite lncRNAs and Beyond, an Emerging Story with a Complex Scenario. Genes. 2022; 13(4):597. https://doi.org/10.3390/genes13040597
Chicago/Turabian StyleVourc’h, Claire, Solenne Dufour, Kalina Timcheva, Daphné Seigneurin-Berny, and André Verdel. 2022. "HSF1-Activated Non-Coding Stress Response: Satellite lncRNAs and Beyond, an Emerging Story with a Complex Scenario" Genes 13, no. 4: 597. https://doi.org/10.3390/genes13040597
APA StyleVourc’h, C., Dufour, S., Timcheva, K., Seigneurin-Berny, D., & Verdel, A. (2022). HSF1-Activated Non-Coding Stress Response: Satellite lncRNAs and Beyond, an Emerging Story with a Complex Scenario. Genes, 13(4), 597. https://doi.org/10.3390/genes13040597






