Lactic Acid Metabolism and Transporter Related Three Genes Predict the Prognosis of Patients with Clear Cell Renal Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Cohorts and Data Sources
2.2. Establishment and Verification of Prognostic Model
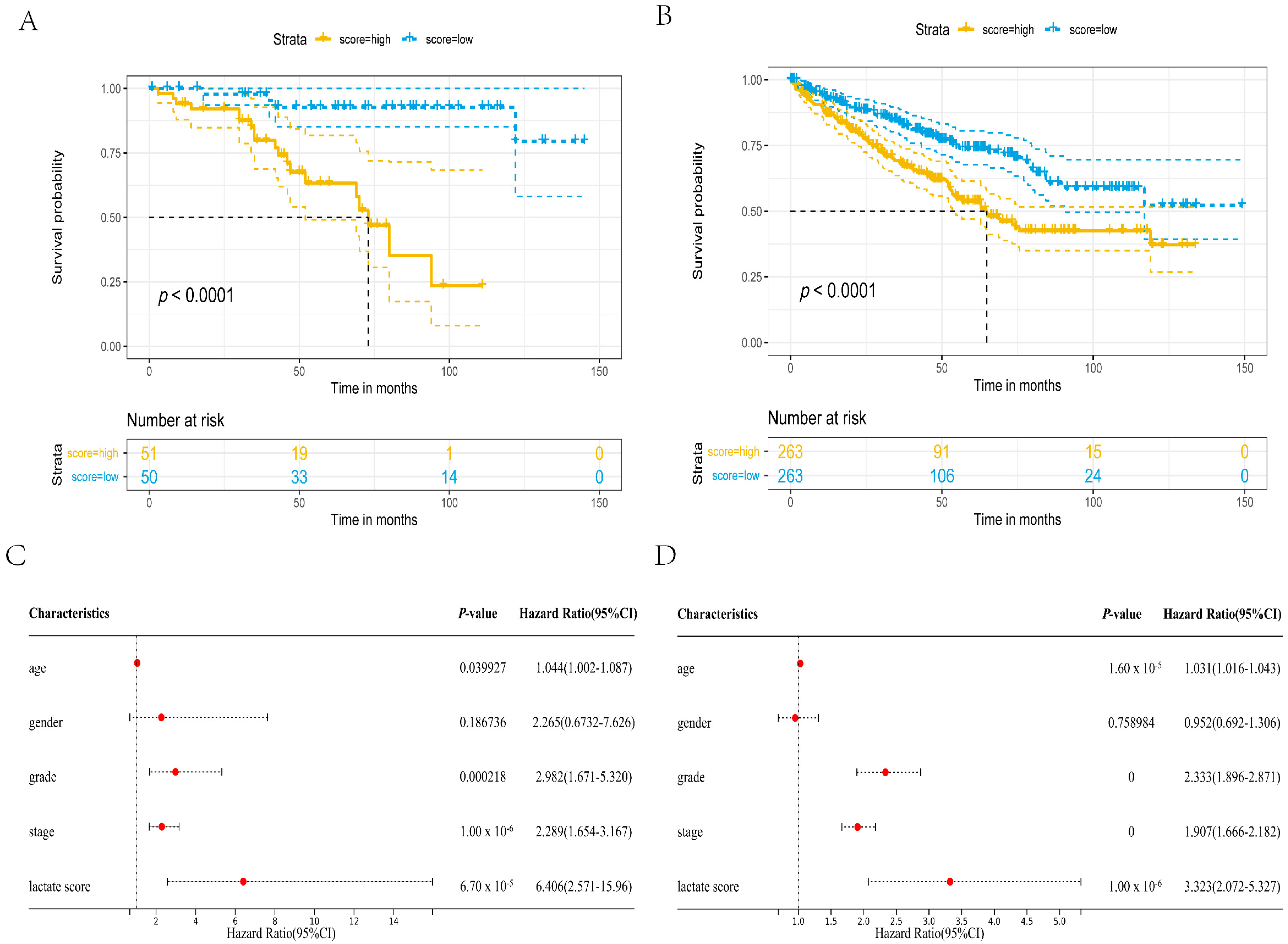
2.3. Univariate Cox Regression Analysis and Survival Analysis
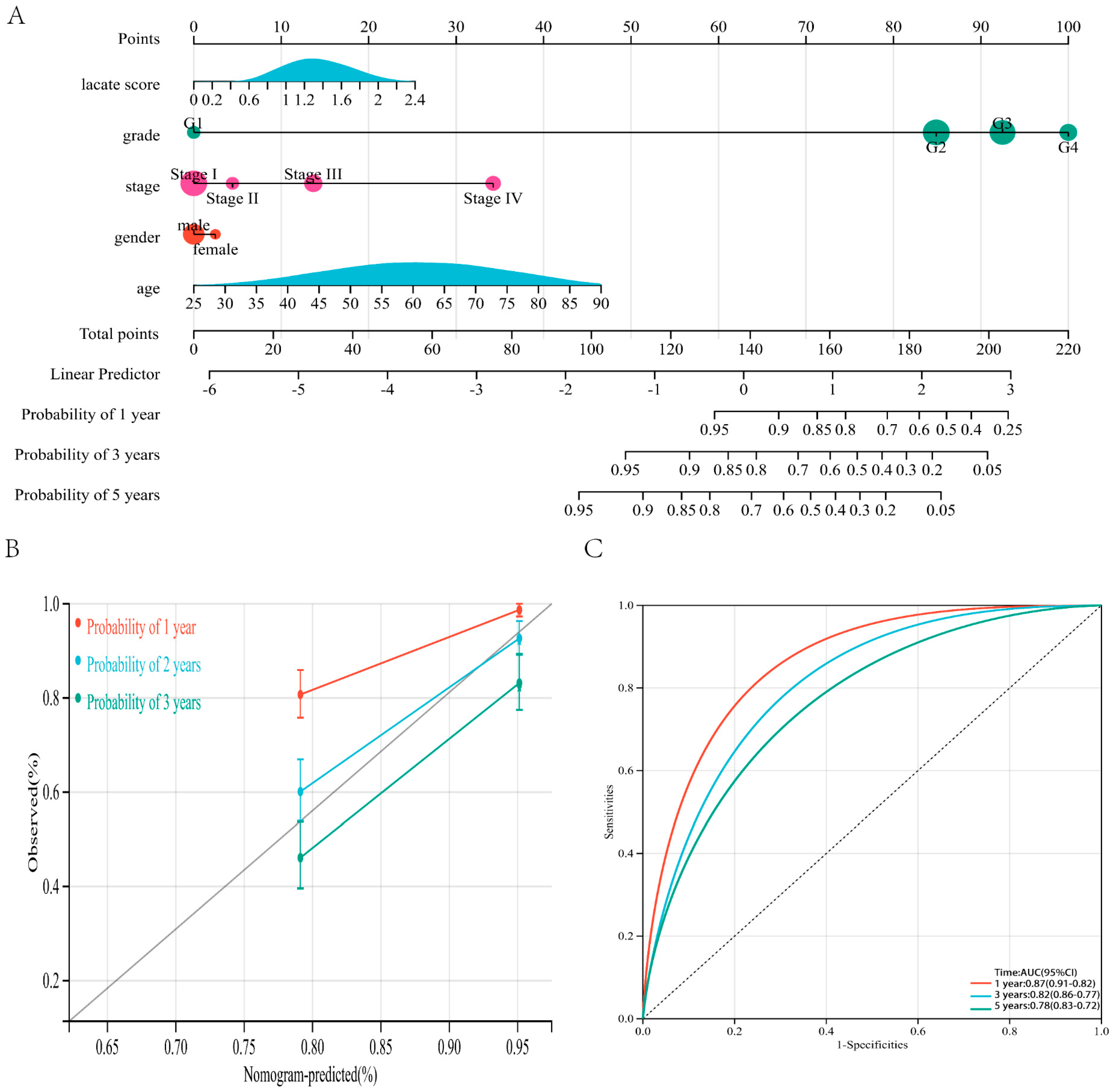
2.4. Constructing Nomogram Combining Risk Score and Clinicopathologic Factors
2.5. Immune Cell Infiltration in Tumor Microenvironment
2.6. Identification of Differentially Expressed Genes
2.7. Functional Enrichment Analysis
2.8. Statistical Analysis
3. Results
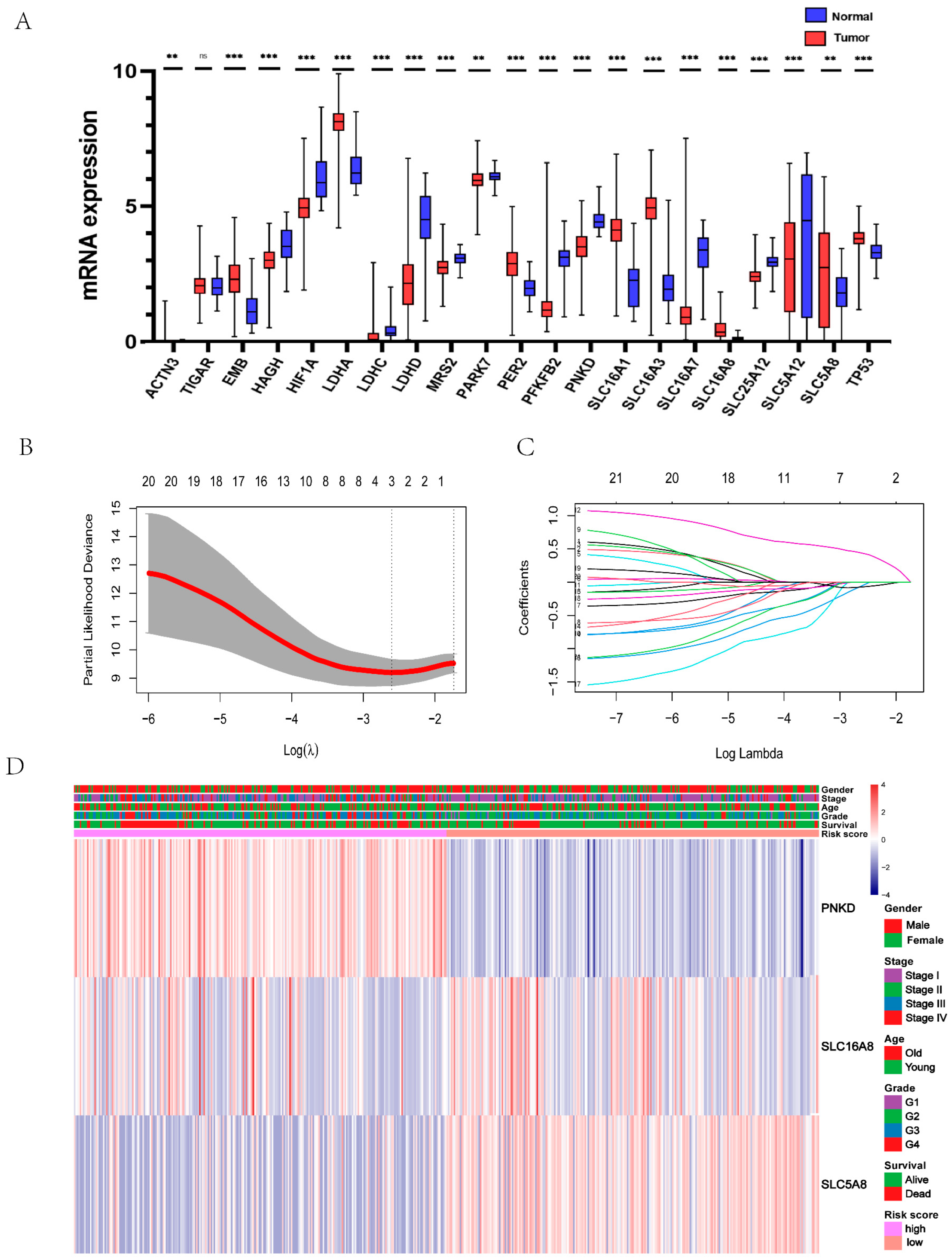
3.1. Identification of Lactic Acid Metabolism and Transporter Related Three Genes as Prognostic Model of ccRCC Patient
3.2. The Constructed Prognostic Model Can Be Used as a Prognostic Factor
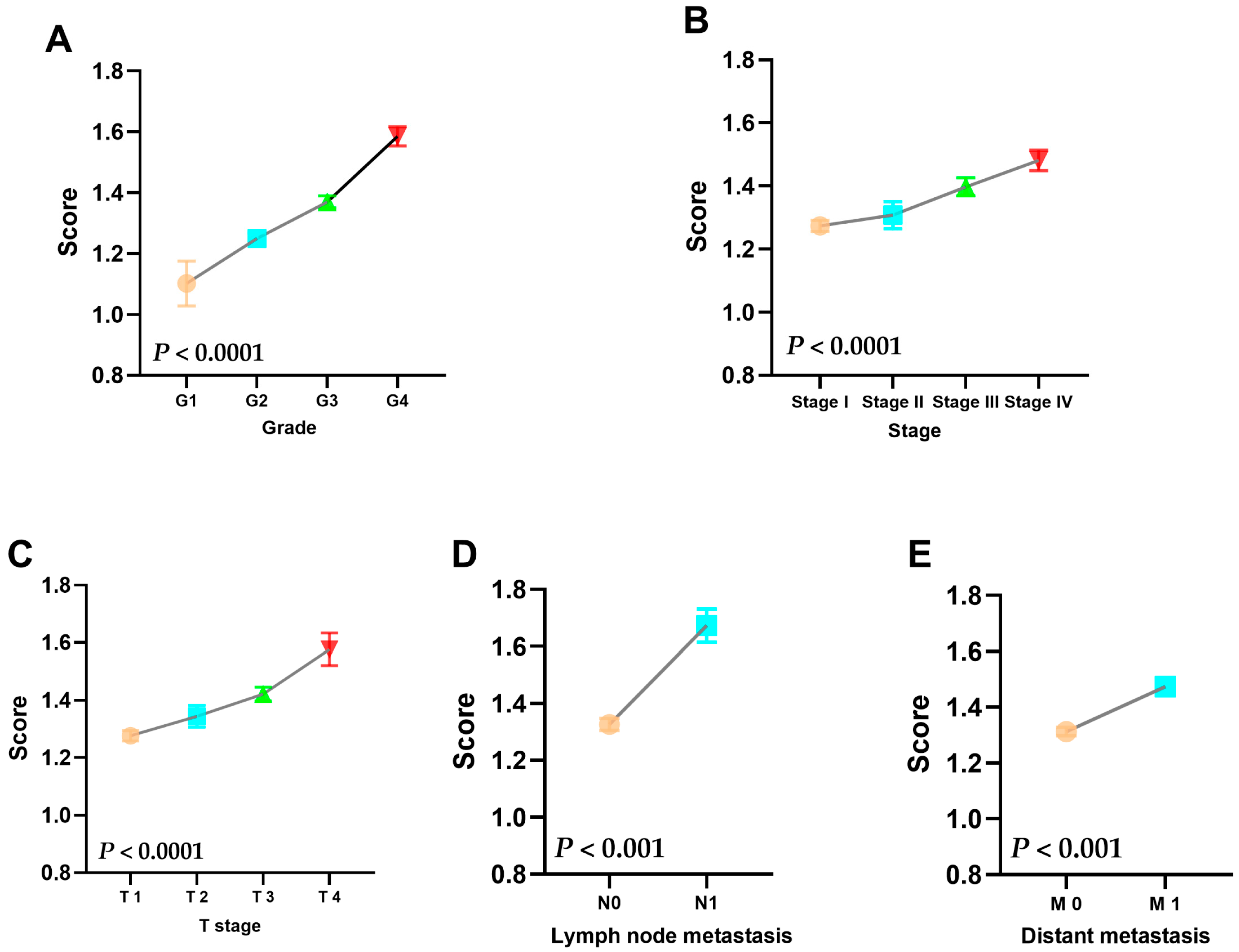
3.3. Risk Score Is Strongly Correlated with the Patient’s Clinicopathological Information
3.4. Nomogram Based on Prognostic Model and Clinicopathological Information can Predict the Prognosis of ccRCC Patients
3.5. Prognostic Model Is Associated with Immune Cell Infiltration
3.6. WGCNA Reveals That High Risk Score Is Related to Cell Cycle
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, L.; Morandi, A.; Giannoni, E.; Chiarugi, P. Lactate: A Metabolic Driver in the Tumour Landscape. Trends Biochem. Sci. 2019, 44, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Nunes, A.; Afonso, J.; Granja, S.; Baltazar, F. Lactate and lactate transporters as key players in the maintenance of the warburg effect. Adv. Exp. Med. Biol. 2020, 1219, 51–74. [Google Scholar] [CrossRef] [PubMed]
- Halestrap, A.P. The SLC16 gene family—Structure, role and regulation in health and disease. Mol. Asp. Med. 2013, 34, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, Y.; Dong, D.; Wang, F.; Ma, X.; Guan, F.; Sun, L. MCT1 regulates aggressive and metabolic phenotypes in bladder cancer. J. Cancer 2018, 9, 2492–2501. [Google Scholar] [CrossRef]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2016, 38, 119–133. [Google Scholar] [CrossRef]
- Wettersten, H.I.; Aboud, O.A.; Lara, P.N.; Weiss, R.H. Metabolic reprogramming in clear cell renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 410–419. [Google Scholar] [CrossRef]
- Wei, G.; Sun, H.; Dong, K.; Hu, L.; Wang, Q.; Zhuang, Q.; Zhu, Y.; Zhang, X.; Shao, Y.; Tang, H.; et al. The thermogenic activity of adjacent adipocytes fuels the progression of ccRCC and compromises anti-tumor therapeutic efficacy. Cell Metab. 2021, 33, 2021–2039.e8. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshizato, T.; Shiraishi, Y.; Maekawa, S.; Okuno, Y.; Kamura, T.; Shimamura, T.; Sato-Otsubo, A.; Nagae, G.; Suzuki, H.; et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013, 45, 860–867. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Sattler, U.G.A.; Mueller-Klieser, W. Lactate: A Metabolic Key Player in Cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Certo, M.; Tsai, C.-H.; Pucino, V.; Ho, P.-C.; Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2020, 21, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Zitvogel, L.; Sautes-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef]
- Watson, M.J.; Vignali, P.D.A.; Mullett, S.J.; Overacre-Delgoffe, A.E.; Peralta, R.M.; Grebinoski, S.; Menk, A.V.; Rittenhouse, N.L.; DePeaux, K.; Whetstone, R.D.; et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 2021, 591, 645–651. [Google Scholar] [CrossRef]
- Zisman, A.; Pantuck, A.J.; Dorey, F.; Said, J.W.; Shvarts, O.; Quintana, D.; Gitlitz, B.J.; Dekernion, J.B.; Figlin, R.A.; Belldegrun, A.S. Improved Prognostication of Renal Cell Carcinoma Using an Integrated Staging System. J. Clin. Oncol. 2001, 19, 1649–1657. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.-Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef]
- Gottfried, E.; Kunz-Schughart, L.; Ebner, S.; Mueller-Klieser, W.; Hoves, S.; Andreesen, R.; Mackensen, A.; Kreutz, M. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood 2006, 107, 2013–2021. [Google Scholar] [CrossRef]
- Puig-Kröger, A.; Pello, O.M.; Muñiz-Pello, O.; Selgas, R.; Criado, G.; Bajo, M.A.; Sánchez-Tomero, J.A.; Alvarez, V.; del Peso, G.; Sánchez-Mateos, P.; et al. Peritoneal dialysis solutions inhibit the differentiation and maturation of human monocyte-derived dendritic cells: Effect of lactate and glucose-degradation products. J. Leukoc. Biol. 2003, 73, 482–492. [Google Scholar] [CrossRef]
- Balgi, A.D.; Diering, G.H.; Donohue, E.; Lam, K.; Fonseca, B.D.; Zimmerman, C.; Numata, M.; Roberge, M. Regulation of mTORC1 Signaling by pH. PLoS ONE 2011, 6, e21549. [Google Scholar] [CrossRef]
- Guo, C.; Huang, T.; Wang, Q.-H.; Li, H.; Khanal, A.; Kang, E.-H.; Zhang, W.; Niu, H.-T.; Dong, Z.; Cao, Y.-W. Monocarboxylate transporter 1 and monocarboxylate transporter 4 in cancer-endothelial co-culturing microenvironments promote proliferation, migration, and invasion of renal cancer cells. Cancer Cell Int. 2019, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Feng, Q.; Wang, Z.; Li, W.; Sun, Z.; Wilhelm, J.; Huang, G.; Vo, T.; Sumer, B.D.; Gao, J. Tumor-Targeted Inhibition of Monocarboxylate Transporter 1 Improves T-Cell Immunotherapy of Solid Tumors. Adv. Healthc. Mater. 2020, 10, e2000549. [Google Scholar] [CrossRef] [PubMed]
- Doherty, J.; Cleveland, J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Investig. 2013, 123, 3685–3692. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Choi, S.Y.; Niu, X.; Kang, N.; Xue, H.; Killam, J.; Wang, Y. Lactic Acid and an Acidic Tumor Microenvironment suppress Anticancer Immunity. Int. J. Mol. Sci. 2020, 21, 8363. [Google Scholar] [CrossRef]
- Sun, N.; Nasello, C.; Deng, L.; Wang, N.; Zhang, Y.; Xu, Z.; Song, Z.; Kwan, K.; King, R.A.; Pang, Z.P.; et al. The PNKD gene is associated with Tourette Disorder or Tic disorder in a multiplex family. Mol. Psychiatry 2017, 23, 1487–1495. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-M.; Meng, Q.-H.; Kong, F.-F.; Wang, K.; Du, L.-J. SLC5A8 regulates the biological behaviors of cervical cancer cells through mediating the Wnt signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4679–4686. [Google Scholar]



| TCGA Cohort (526) | E-MTAB-1980 Cohort (101) | |
|---|---|---|
| Age (years) | ||
| ≥65 | 194 (36.9%) | 44 (43.6%) |
| <65 | 332 (63.1%) | 57 (56.4%) |
| Sex | ||
| Male | 343 (65.2%) | 77 (23.8%) |
| Female | 183 (34.8%) | 24 (23.8% |
| Grade | ||
| G1 | 13 (2.5%) | 13 (12.9%) |
| G2 | 224 (42.6%) | 59 (58.4%) |
| G3 | 204 (38.8%) | 22 (21.8%) |
| G4 | 74 (14.1%) | 5 (5.0%) |
| Unknown | 8 (1.5%) | 2 (2.0%) |
| Stage | ||
| I | 261 (49.6%) | 67 (66.3%) |
| II | 57 (10.8%) | 11 (10.9%) |
| III | 125 (23.8%) | 14 (13.9%) |
| IV | 83 (15.8%) | 13 (12.9%) |
| T stage | ||
| T1 | 267 (50.8%) | 68 (67.3%) |
| T2 | 69 (13.1%) | 11 (10.9%) |
| T3 | 179 (34.0%) | 21 (20.85) |
| T4 | 11 (2.1%) | 1 (1.0%) |
| N stage | ||
| N2 | \ | 4 (4%) |
| N1 | 16 (3.1%) | 3 (3.0%) |
| N0 | 238 (45.2%) | 94 (93.1%) |
| Unknown | 272 (51.7%) | / |
| M stage | ||
| M1 | 78 (14.8%) | 12 (11.9%) |
| M0 | 418 (79.5%) | 89 (88.1%) |
| Unknown | 30 (5.7%) | / |
| Survival | ||
| Dead | 171 (32.5%) | 23 (22.8%) |
| Living | 355 (67.5%) | 78 (77.25) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Zhang, J.; Wang, T.; Yuan, Z.; Tang, H.; Zhang, D.; Chen, S.; Wang, X. Lactic Acid Metabolism and Transporter Related Three Genes Predict the Prognosis of Patients with Clear Cell Renal Cell Carcinoma. Genes 2022, 13, 620. https://doi.org/10.3390/genes13040620
Guo T, Zhang J, Wang T, Yuan Z, Tang H, Zhang D, Chen S, Wang X. Lactic Acid Metabolism and Transporter Related Three Genes Predict the Prognosis of Patients with Clear Cell Renal Cell Carcinoma. Genes. 2022; 13(4):620. https://doi.org/10.3390/genes13040620
Chicago/Turabian StyleGuo, Tuanjie, Jian Zhang, Tao Wang, Zhihao Yuan, Heting Tang, Dongliang Zhang, Siteng Chen, and Xiang Wang. 2022. "Lactic Acid Metabolism and Transporter Related Three Genes Predict the Prognosis of Patients with Clear Cell Renal Cell Carcinoma" Genes 13, no. 4: 620. https://doi.org/10.3390/genes13040620
APA StyleGuo, T., Zhang, J., Wang, T., Yuan, Z., Tang, H., Zhang, D., Chen, S., & Wang, X. (2022). Lactic Acid Metabolism and Transporter Related Three Genes Predict the Prognosis of Patients with Clear Cell Renal Cell Carcinoma. Genes, 13(4), 620. https://doi.org/10.3390/genes13040620






