Knockdown of CDR1as Decreases Differentiation of Goat Skeletal Muscle Satellite Cells via Upregulating miR-27a-3p to Inhibit ANGPT1
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Vector Construction and Cell Transfection
2.3. RNA Extraction
2.4. miRNA Sequence
2.5. mRNA Sequencing and Data Processing
2.6. KEGG Pathway Analysis
2.7. Validation of RNA-Seq Data by qRT-PCR
2.8. Western Blot Assay
2.9. EdU Proliferation Assay
2.10. Immunofluorescence Analyses
2.11. Luciferase Assay Activity
2.12. Statistical Analyses
3. Results
3.1. Overview of mRNA and miRNA Sequencing Data Associated with CDR1as
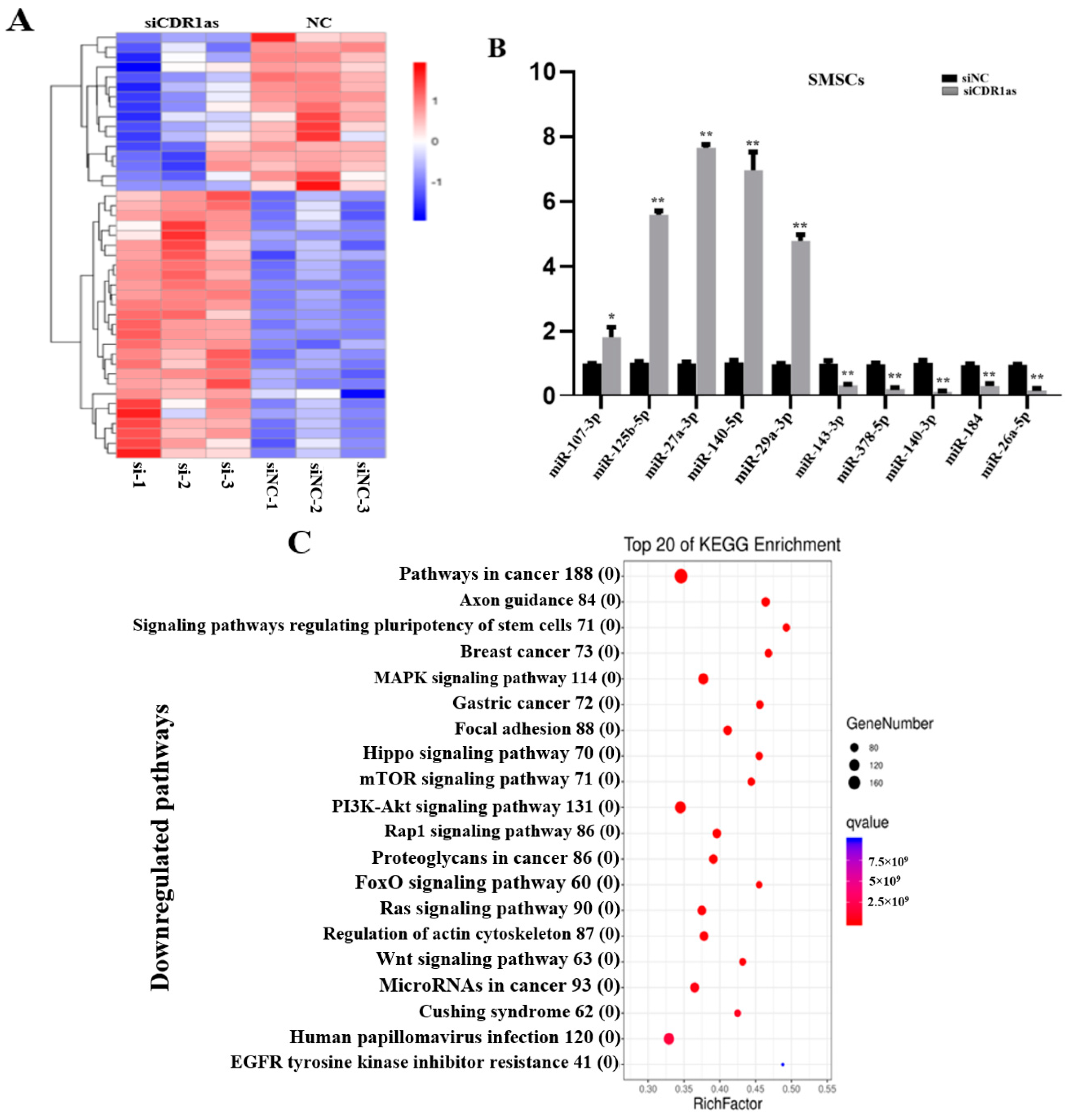
3.2. Differentially Expressed (DE) miRNAs in SMSCs Transfected with siCDR1as
3.3. DE mRNAs in SMSCs Transfected with siCDR1as
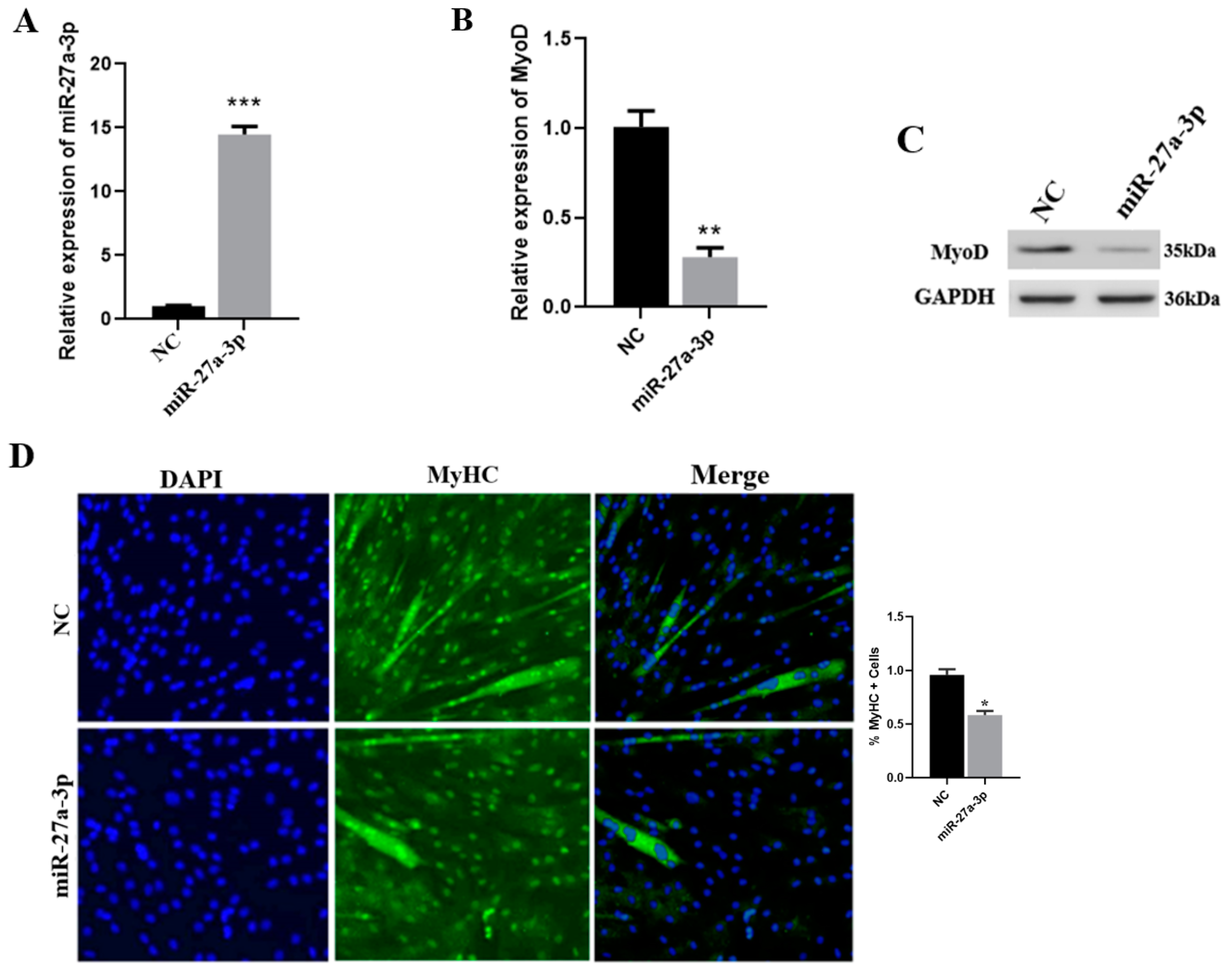
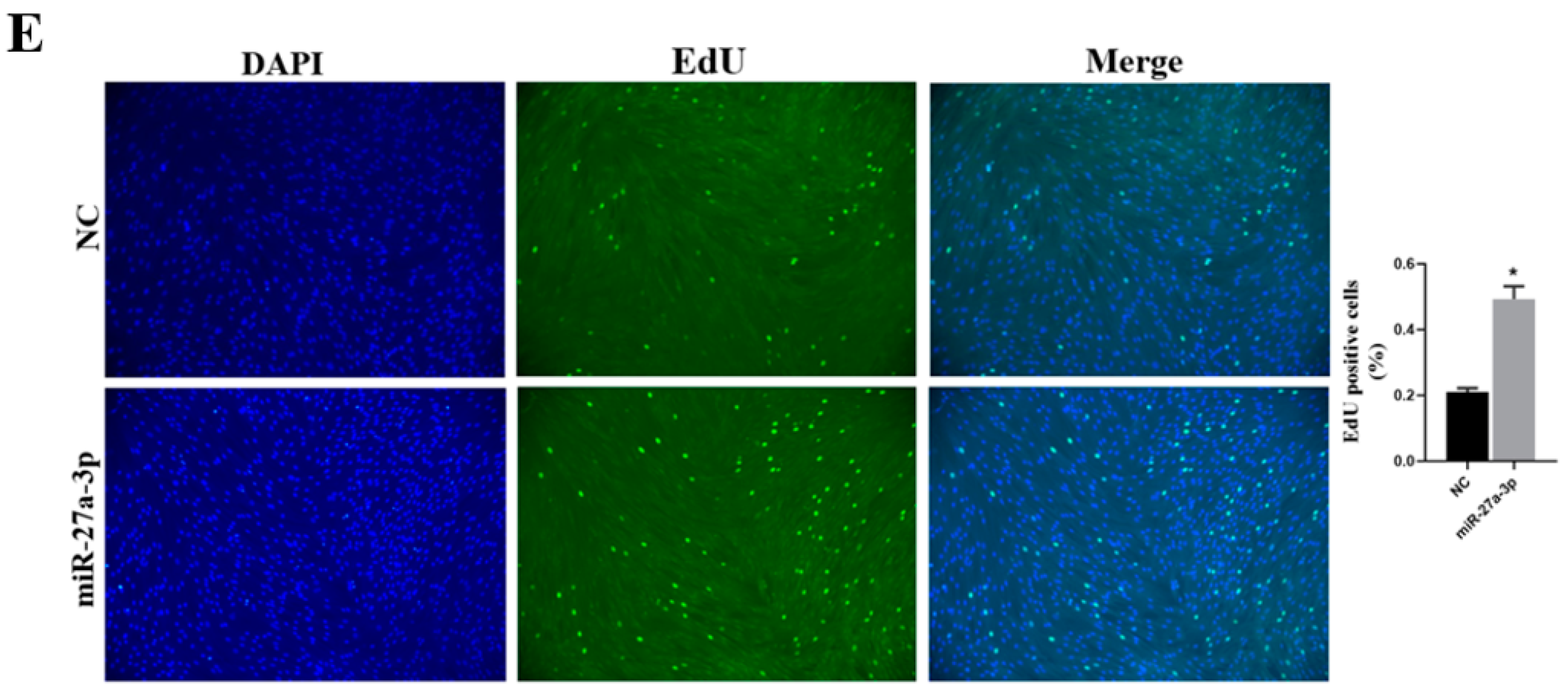
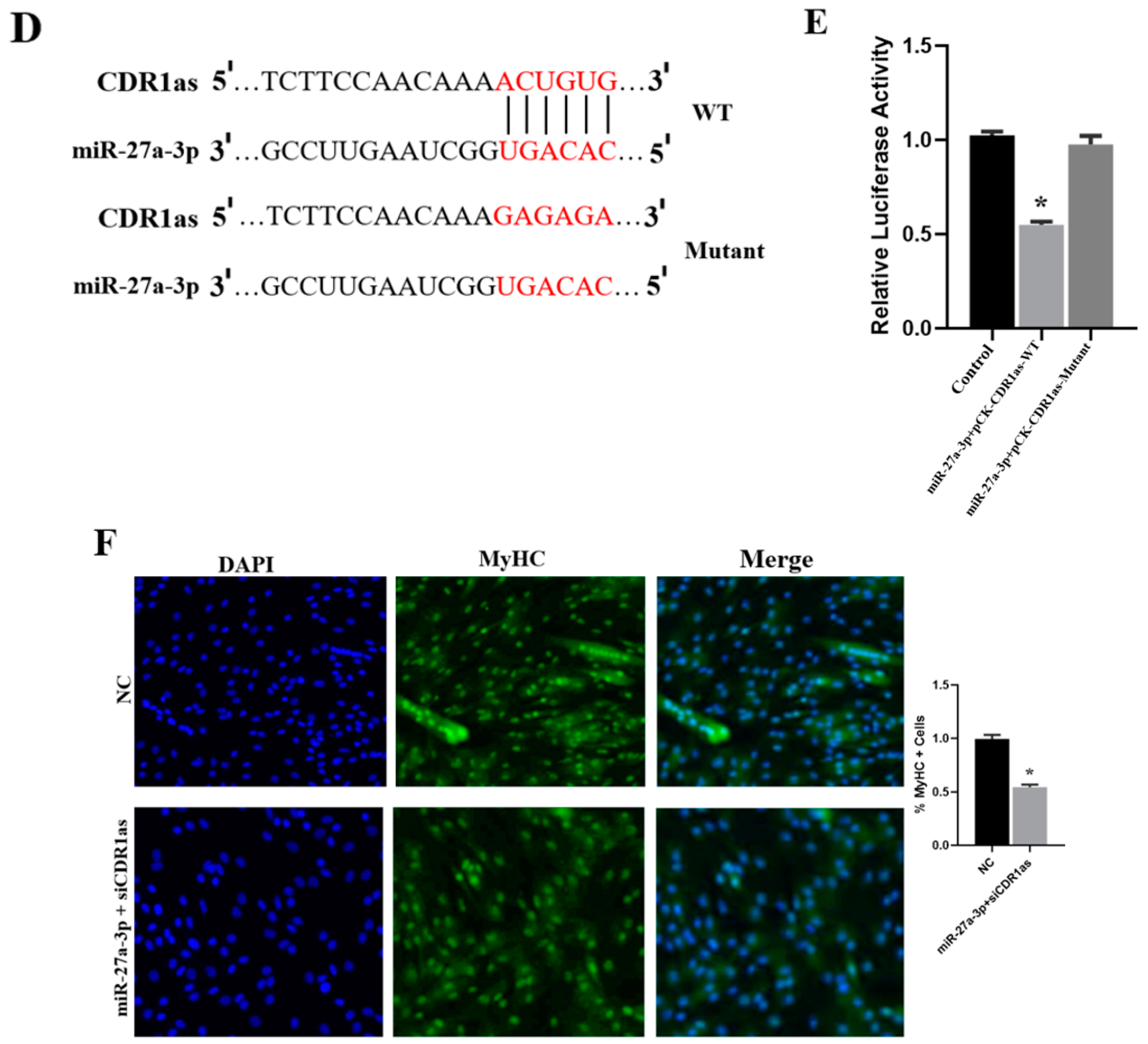
3.4. Effect of miR-27a-3p on SMSCs
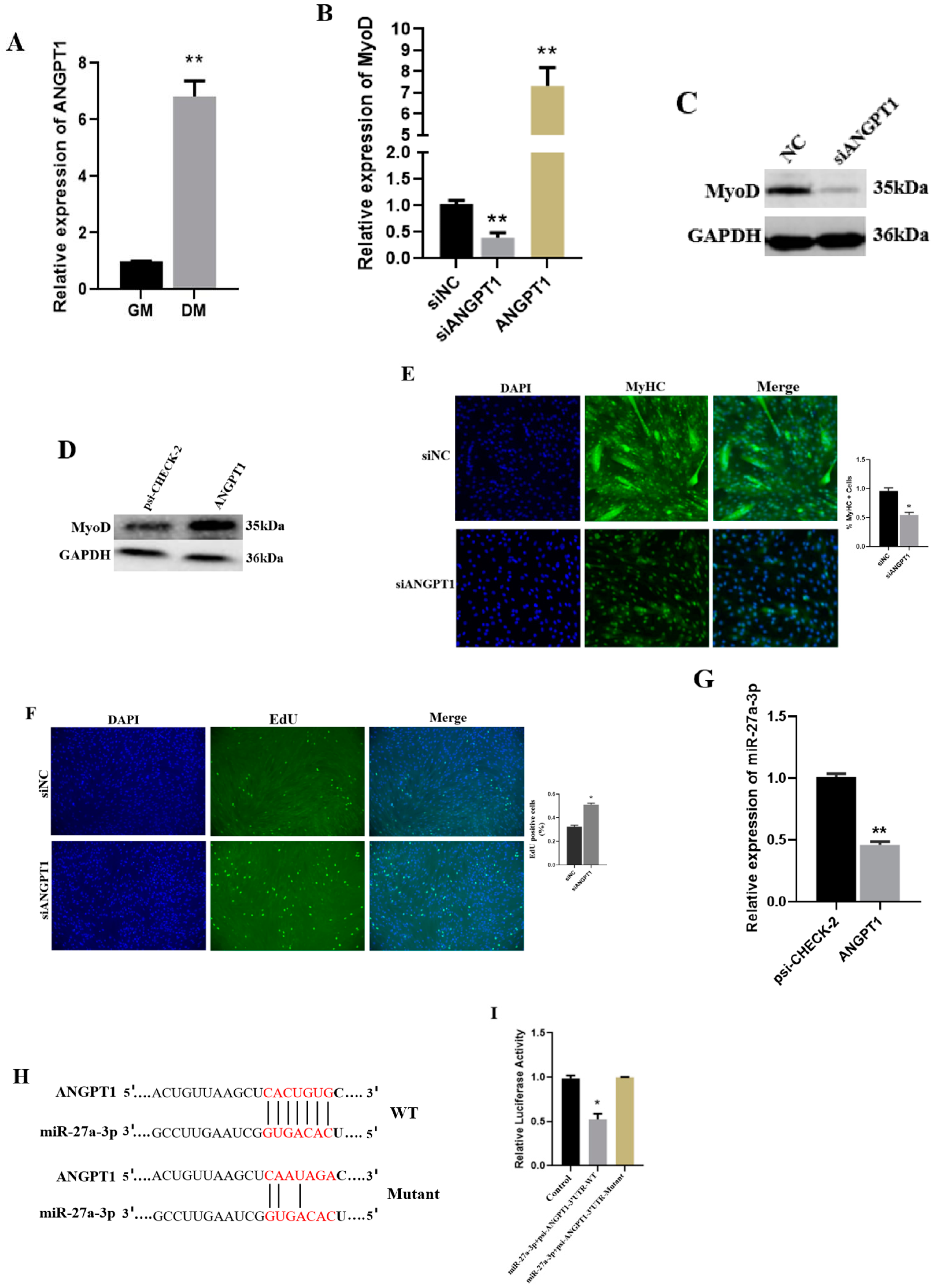
3.5. The Functional Role of ANGPT1 in SMSCs and Its Relationship with miR-27a-3p
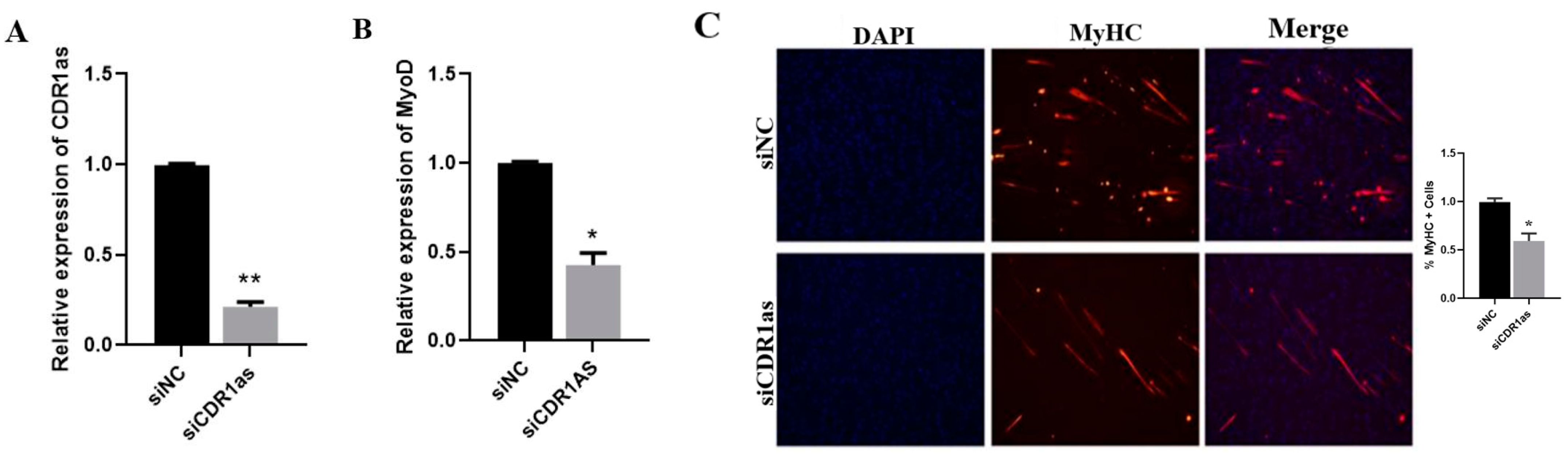
3.6. CDR1as Knockdown Inhibits Differentiation of SMSCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mohammadabadi, M. Tissue-specific mRNA expression profile of ESR2 gene in goat. Agric. Biotechnol. J. 2021, 12, 167–181. [Google Scholar]
- Buckingham, M.; Bajard, L.; Chang, T.; Daubas, P.; Hadchouel, J.; Meilhac, S.; Montarras, D.; Rocancourt, D.; Relaix, F. The formation of skeletal muscle: From somite to limb. J. Anat. 2003, 202, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Greenwood, P.L.; Cafe, L.M.; Zhou, G.; Zhang, W.; Dalrymple, B.P. Transcriptome analysis of cattle muscle identifies potential markers for skeletal muscle growth rate and major cell types. BMC Genom. 2015, 16, 177. [Google Scholar] [CrossRef] [PubMed]
- Giordani, L.; Parisi, A.; Le Grand, F. Chapter Six—Satellite Cell Self-Renewal. In Current Topics in Developmental Biology; Sassoon, D., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 126, pp. 177–203. [Google Scholar]
- Li, L.; Chen, Y.; Nie, L.; Ding, X.; Zhang, X.; Zhao, W.; Xu, X.; Kyei, B.; Dai, D.; Zhan, S.; et al. MyoD-induced circular RNA CDR1as promotes myogenic differentiation of skeletal muscle satellite cells. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2019, 1862, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Z.; Valdez, M.R.; McAnally, J.; Richardson, J.; Olson, E.N. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development 2001, 128, 4623–4633. [Google Scholar] [CrossRef]
- Lee, E.H.; Woo, J.S.; Hwang, J.H.; Park, J.H.; Cho, C.H. Angiopoietin 1 enhances the proliferation and differentiation of skeletal myoblasts. J. Cell. Physiol. 2013, 228, 1038–1044. [Google Scholar] [CrossRef] [PubMed]
- Dallabrida, S.M.; Ismail, N.; Oberle, J.R.; Himes, B.E.; Rupnick, M.A. Angiopoietin-1 promotes cardiac and skeletal myocyte survival through integrins. Circ. Res. 2005, 96, e8–e24. [Google Scholar] [CrossRef]
- Jeansson, M.; Gawlik, A.; Anderson, G.; Li, C.; Kerjaschki, D.; Henkelman, M.; Quaggin, S.E. Angiopoietin-1 is essential in mouse vasculature during development and in response to injury. J. Clin. Investig. 2011, 121, 2278–2289. [Google Scholar] [CrossRef]
- Hagan, M.; Zhou, M.; Ashraf, M.; Kim, I.-M.; Su, H.; Weintraub, N.L.; Tang, Y. Long noncoding RNAs and their roles in skeletal muscle fate determination. Noncoding RNA Investig. 2017, 1, 24. [Google Scholar] [CrossRef]
- Apponi, L.H.; Corbett, A.H.; Pavlath, G.K. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol. Sci. 2011, 32, 652–658. [Google Scholar] [CrossRef]
- Mok, G.F.; Lozano-Velasco, E.; Münsterberg, A. microRNAs in skeletal muscle development. Semin. Cell Dev. Biol. 2017, 72, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Barazandeh, A.; Mohammadabadi, M.; Ghaderi-Zefrehei, M.; Rafeie, F.; Imumorin, I.G. Whole genome comparative analysis of CpG islands in camelid and other mammalian genomes. Mamm. Biol. 2019, 98, 73–79. [Google Scholar] [CrossRef]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Chen, X.L.; Huang, Z.Q.; Wen, W.X.; Xu, M.; Chen, D.W.; Yu, B.; He, J.; Luo, J.Q.; Yu, J.; et al. MicroRNA-27a promotes porcine myoblast proliferation by downregulating myostatin expression. Animal 2014, 8, 1867–1872. [Google Scholar] [CrossRef]
- Zou, C.; Li, J.; Luo, W.; Li, L.; Hu, A.; Fu, Y.; Hou, Y.; Li, C. Transcriptome analysis reveals long intergenic non-coding RNAs involved in skeletal muscle growth and development in pig. Sci. Rep. 2017, 7, 8704. [Google Scholar] [CrossRef]
- Guo, J.; Zhao, W.; Zhan, S.; Li, L.; Zhong, T.; Wang, L.; Dong, Y.; Zhang, H. Identification and Expression Profiling of miRNAome in Goat longissimus dorsi Muscle from Prenatal Stages to a Neonatal Stage. PLoS ONE 2016, 11, e0165764. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Q.; Hua, L.; Chen, J.; Zhang, J.; Bai, H.; Li, H.; Xu, B.; Shi, Z.; Cao, H.; et al. Comprehensive Analysis of Differentially Expressed mRNA, lncRNA and circRNA and Their ceRNA Networks in the Longissimus Dorsi Muscle of Two Different Pig Breeds. Int. J. Mol. Sci. 2019, 20, 1107. [Google Scholar] [CrossRef]
- Das, A.; Das, A.; Das, D.; Abdelmohsen, K.; Panda, A.C. Circular RNAs in myogenesis. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2019, 1863, 194372. [Google Scholar] [CrossRef]
- Hsiao, K.-Y.; Sun, H.S.; Tsai, S.-J. Circular RNA—New member of noncoding RNA with novel functions. Exp. Biol. Med. 2017, 242, 1136–1141. [Google Scholar] [CrossRef]
- Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016, 32, 309–316. [Google Scholar] [CrossRef]
- Li, P.; Yang, X.; Yuan, W.; Yang, C.; Zhang, X.; Han, J.; Wang, J.; Deng, X.; Yang, H.; Li, P.; et al. CircRNA-Cdr1as Exerts Anti-Oncogenic Functions in Bladder Cancer by Sponging MicroRNA-135a. Cell. Physiol. Biochem. 2018, 46, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.; Meng, L.; Sang, Y.; Liu, S.; Ding, P.; Ju, Y.; Liu, F.; Gu, L.; Lian, Y.; Li, J.; et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018, 426, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Kyei, B.; Li, L.; Yang, L.; Zhan, S.; Zhang, H. CDR1as/miRNAs-Related Regulatory Mechanisms in Muscle Development and Diseases. Gene 2020, 730, 144315. [Google Scholar] [CrossRef]
- Zhao, W.; Chen, L.; Zhong, T.; Wang, L.; Guo, J.; Dong, Y.; Feng, J.; Song, T.; Li, L.; Zhang, H. The differential proliferation and differentiation ability of skeletal muscle satellite cell in Boer and Nanjiang brown goats. Small Rumin. Res. 2018, 169, 99–107. [Google Scholar] [CrossRef]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J.; et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Mofarrahi, M.; McClung, J.M.; Kontos, C.D.; Davis, E.C.; Tappuni, B.; Moroz, N.; Pickett, A.E.; Huck, L.; Harel, S.; Danialou, G.; et al. Angiopoietin-1 enhances skeletal muscle regeneration in mice. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2015, 308, R576–R589. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Choi, S.; Liu, X.; Zhang, M.; Schageman, J.J.; Lee, S.Y.; Hart, R.; Lin, L.; Thurmond, F.A.; Williams, R.S. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 2003, 278, 8826–8836. [Google Scholar] [CrossRef] [PubMed]
- Raissadati, A.; Nykänen, A.I.; Tuuminen, R.; Syrjälä, S.O.; Krebs, R.; Arnaudova, R.; Rouvinen, E.; Wang, X.; Poller, W.; Lemström, K.B. Systemic overexpression of matricellular protein CCN1 exacerbates obliterative bronchiolitis in mouse tracheal allografts. Transpl. Int. 2015, 28, 1416–1425. [Google Scholar] [CrossRef]
- Sogos, V.; Balaci, L.; Ennas, M.G.; Dell’Era, P.; Presta, M.; Gremo, F. Developmentally regulated expression and localization of fibroblast growth factor receptors in the human muscle. Dev. Dyn. 1998, 211, 362–373. [Google Scholar] [CrossRef][Green Version]
- Ouyang, H.; Chen, X.; Li, W.; Li, Z.; Nie, Q.; Zhang, X. Circular RNA circSVIL Promotes Myoblast Proliferation and Differentiation by Sponging miR-203 in Chicken. Front. Genet. 2018, 9, 172. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Yang, J.; Dong, D.; Hao, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y. circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol. Ther.-Nucleic Acids 2018, 11, 272–283. [Google Scholar] [CrossRef]
- Su, Y.; Lv, X.; Yin, W.; Zhou, L.; Hu, Y.; Zhou, A.; Qi, F. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging 2019, 11, 8183–8203. [Google Scholar] [CrossRef]
- Rahmati, Y.; Asemani, Y.; Aghamiri, S.; Ezzatifar, F.; Najafi, S. CiRS-7/CDR1as; An oncogenic circular RNA as a potential cancer biomarker. Pathol. Res. Pract. 2021, 227, 153639. [Google Scholar] [CrossRef]
- Jiang, C.; Zeng, X.; Shan, R.; Wen, W.; Li, J.; Tan, J.; Li, L.; Wan, R. The Emerging Picture of the Roles of CircRNA-CDR1as in Cancer. Front. Cell Dev. Biol. 2020, 8, 590478. [Google Scholar] [CrossRef]
- Jama, A.; Huang, D.; Alshudukhi, A.A.; Chrast, R.; Ren, H. Lipin1 is required for skeletal muscle development by regulating MEF2c and MyoD expression. J. Physiol. 2019, 597, 889–901. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, Y.; Zhang, H.; Ma, G.; Wu, G.; Xiang, A.; Shi, X.E.; Yang, G.S.; Sun, S. MicroRNA-106a-5p Inhibited C2C12 Myogenesis via Targeting PIK3R1 and Modulating the PI3K/AKT Signaling. Genes 2018, 9, 333. [Google Scholar] [CrossRef] [PubMed]
- Romer, L.H.; Birukov, K.G.; Garcia, J.G.N. Focal adhesions: Paradigm for a signaling nexus. Circ. Res. 2006, 98, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y.; et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Jin, W.; Fu, S.; Li, D.; Zhang, Y.; Sun, G.; Jiang, R.; Han, R.; Li, Z.; et al. Analyses of MicroRNA and mRNA Expression Profiles Reveal the Crucial Interaction Networks and Pathways for Regulation of Chicken Breast Muscle Development. Front. Genet. 2019, 10, 197. [Google Scholar] [CrossRef]
- Jiang, B.-H.; Zheng, J.Z.; Vogt, P.K. An essential role of phosphatidylinositol 3-kinase in myogenic differentiation. Proc. Natl. Acad. Sci. USA 1998, 95, 14179. [Google Scholar] [CrossRef]
- Jiang, B.-H.; Aoki, M.; Zheng, J.Z.; Li, J.; Vogt, P.K. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc. Natl. Acad. Sci. USA 1999, 96, 2077. [Google Scholar] [CrossRef]
- Kooistra, M.R.H.; Dubé, N.; Bos, J.L. Rap1: A key regulator in cell-cell junction formation. J. Cell Sci. 2007, 120, 17. [Google Scholar] [CrossRef]
- Boettner, B.; Van Aelst, L. Control of cell adhesion dynamics by Rap1 signaling. Curr. Opin. Cell Biol. 2009, 21, 684–693. [Google Scholar] [CrossRef]
- Lynch, G.S.; Ryall, J.G. Role of β-Adrenoceptor Signaling in Skeletal Muscle: Implications for Muscle Wasting and Disease. Physiol. Rev. 2008, 88, 729–767. [Google Scholar] [CrossRef]
- Aboalola, D.; Han, V.K.M. Insulin-Like Growth Factor Binding Protein-6 Promotes the Differentiation of Placental Mesenchymal Stem Cells into Skeletal Muscle Independent of Insulin-Like Growth Factor Receptor-1 and Insulin Receptor. Stem Cells Int. 2019, 2019, 9245938. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, I.; Tripathi, G.; Carter, E.J.; Cobb, L.J.; Salih, D.A.M.; Lovett, F.A.; Holding, C.; Pell, J.M. Akt2, a novel functional link between p38 mitogen-activated protein kinase and phosphatidylinositol 3-kinase pathways in myogenesis. Mol. Cell Biol. 2004, 24, 3607–3622. [Google Scholar] [CrossRef] [PubMed]
- Brack, A.S.; Conboy, I.M.; Conboy, M.J.; Shen, J.; Rando, T.A. A Temporal Switch from Notch to Wnt Signaling in Muscle Stem Cells Is Necessary for Normal Adult Myogenesis. Cell Stem Cell 2008, 2, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Bragato, C.; Brioschi, L.; Spreafico, M.; Esposito, S.; Pezzotta, A.; Pizzetti, F.; Moreno-Fortuny, A.; Bellipanni, G.; Giordano, A.; et al. HDAC8 regulates canonical Wnt pathway to promote differentiation in skeletal muscles. J. Cell. Physiol. 2019, 234, 6067–6076. [Google Scholar] [CrossRef] [PubMed]
- Watt, K.I.; Goodman, C.A.; Hornberger, T.A.; Gregorevic, P. The Hippo Signaling Pathway in the Regulation of Skeletal Muscle Mass and Function. Exerc. Sport Sci. Rev. 2018, 46, 92–96. [Google Scholar] [CrossRef]
- Sun, C.; De Mello, V.; Mohamed, A.; Ortuste Quiroga, H.P.; Garcia-Munoz, A.; Al Bloshi, A.; Tremblay, A.M.; von Kriegsheim, A.; Collie-Duguid, E.; Vargesson, N.; et al. Common and Distinctive Functions of the Hippo Effectors Taz and Yap in Skeletal Muscle Stem Cell Function. Stem Cells 2017, 35, 1958–1972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-W.; Cai, H.-F.; Wei, X.-F.; Sun, J.-J.; Lan, X.-Y.; Lei, C.-Z.; Lin, F.-P.; Qi, X.-L.; Plath, M.; Chen, H. miR-30-5p Regulates Muscle Differentiation and Alternative Splicing of Muscle-Related Genes by Targeting MBNL. Int. J. Mol. Sci. 2016, 17, 182. [Google Scholar] [CrossRef]
- Cui, H.; Banerjee, S.; Xie, N.; Ge, J.; Liu, R.M.; Matalon, S.; Thannickal, V.J.; Liu, G. MicroRNA-27a-3p Is a Negative Regulator of Lung Fibrosis by Targeting Myofibroblast Differentiation. Am. J. Respir. Cell Mol. Biol. 2016, 54, 843–852. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, X.; Huang, Z.; Chen, D.; Yu, B.; He, J.; Zheng, P.; Yu, J.; Luo, J.; Luo, Y.; et al. Effects of MicroRNA-27a on Myogenin Expression and Akt/FoxO1 Signal Pathway during Porcine Myoblast Differentiation. Anim. Biotechnol. 2018, 29, 183–189. [Google Scholar] [CrossRef]
- Chen, X.; Huang, Z.; Chen, D.; Yang, T.; Liu, G. MicroRNA-27a is induced by leucine and contributes to leucine-induced proliferation promotion in C2C12 cells. Int. J. Mol. Sci. 2013, 14, 14076–14084. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, X.; Yu, B.; He, J.; Chen, D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem. Biophys. Res. Commun. 2012, 423, 265–269. [Google Scholar] [CrossRef] [PubMed]

| Sample | Raw Reads | Clean Reads | Clean Reads (%) |
|---|---|---|---|
| siCDR1as-1 | 15,249,256 | 14,876,516 | 97.56% |
| siCDR1as-2 | 15,109,243 | 14,776,717 | 97.80% |
| siCDR1as-3 | 16,302,005 | 16,044,248 | 98.42% |
| siNC-1 | 17,583,292 | 17,101,617 | 97.26% |
| siNC-2 | 15,392,007 | 15,172,466 | 98.57% |
| siNC-3 | 16,631,937 | 16,245,764 | 97.68% |
| Sample | Raw Reads | Clean Reads | Clean Reads (%) |
|---|---|---|---|
| siCDR1as-1 | 51,865,796 | 51,084,670 | 98.49% |
| siCDR1as-2 | 64,928,768 | 63,448,702 | 97.72% |
| siCDR1as-3 | 62,668,742 | 61,124,018 | 97.53% |
| siNC-1 | 71,050,762 | 69,247,162 | 97.46% |
| siNC-2 | 60,328,494 | 58,720,344 | 97.33% |
| siNC-3 | 73,310,062 | 70,808,056 | 96.59% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyei, B.; Odame, E.; Li, L.; Yang, L.; Zhan, S.; Li, J.; Chen, Y.; Dai, D.; Cao, J.; Guo, J.; et al. Knockdown of CDR1as Decreases Differentiation of Goat Skeletal Muscle Satellite Cells via Upregulating miR-27a-3p to Inhibit ANGPT1. Genes 2022, 13, 663. https://doi.org/10.3390/genes13040663
Kyei B, Odame E, Li L, Yang L, Zhan S, Li J, Chen Y, Dai D, Cao J, Guo J, et al. Knockdown of CDR1as Decreases Differentiation of Goat Skeletal Muscle Satellite Cells via Upregulating miR-27a-3p to Inhibit ANGPT1. Genes. 2022; 13(4):663. https://doi.org/10.3390/genes13040663
Chicago/Turabian StyleKyei, Bismark, Emmanuel Odame, Li Li, Liu Yang, Siyuan Zhan, Juntao Li, Yuan Chen, Dinghui Dai, Jiaxue Cao, Jiazhong Guo, and et al. 2022. "Knockdown of CDR1as Decreases Differentiation of Goat Skeletal Muscle Satellite Cells via Upregulating miR-27a-3p to Inhibit ANGPT1" Genes 13, no. 4: 663. https://doi.org/10.3390/genes13040663
APA StyleKyei, B., Odame, E., Li, L., Yang, L., Zhan, S., Li, J., Chen, Y., Dai, D., Cao, J., Guo, J., Zhong, T., Wang, L., & Zhang, H. (2022). Knockdown of CDR1as Decreases Differentiation of Goat Skeletal Muscle Satellite Cells via Upregulating miR-27a-3p to Inhibit ANGPT1. Genes, 13(4), 663. https://doi.org/10.3390/genes13040663






