Comparative Genomics Provides Insights into Adaptive Evolution in Tactile-Foraging Birds
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Identification of Orthologues and Phylogenetic Tree Construction
2.3. Gene Family Expansion and Contraction
2.4. Positive Selection Analysis
2.5. Identification of Convergent Amino Acid Substitutions
3. Results
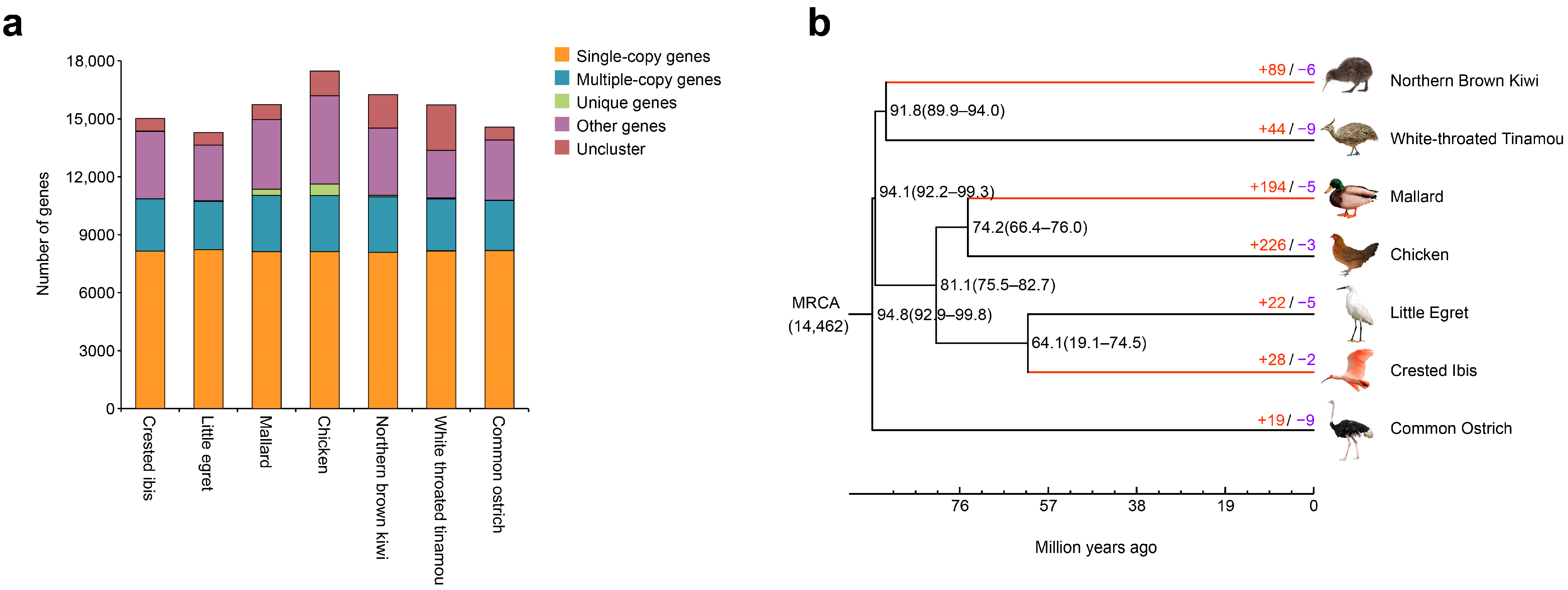
3.1. Identification of Orthologues and Phylogenetic Tree Construction
3.2. Gene Family Analysis
3.3. Positive Selection Analysis
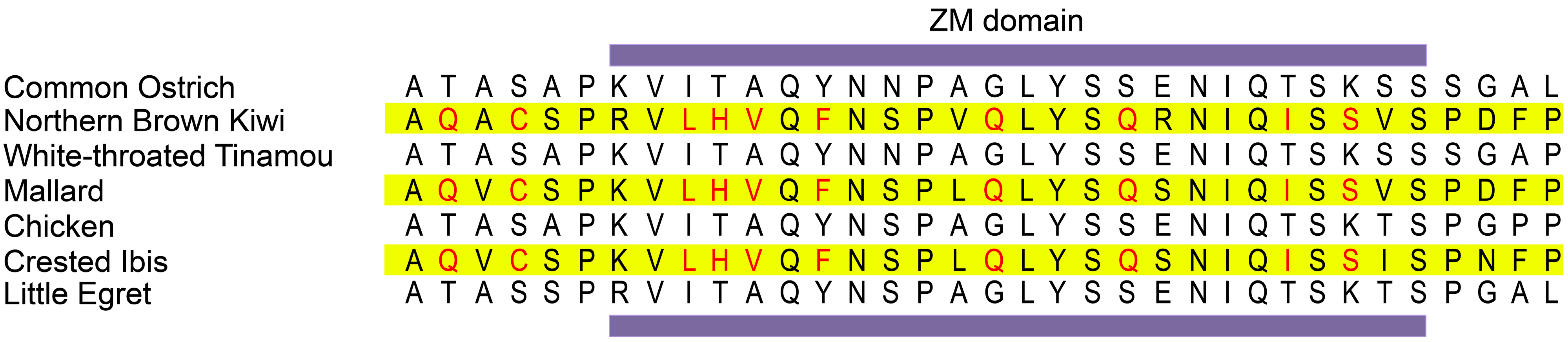
3.4. Identification of Convergent Amino Acid Substitutions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mcglone, F.; Wessberg, J.; Olausson, H. Discriminative and Affective Touch: Sensing and Feeling. Neuron 2014, 82, 737–755. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.B.; Whitmer, D.; Figueroa, R.; Williams, B.A.; Kleinfeld, D. Active spatial perception in the vibrissa scanning sensorimotor system. PLoS Biol. 2007, 5, e15. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Kang, H.; Wolfe, J.; Jadhav, S.P.; Feldman, D.E. Psychometric curve and behavioral strategies for whisker-based texture discrimination in rats. PLoS ONE 2011, 6, e20437. [Google Scholar] [CrossRef] [PubMed]
- Roohbakhsh, A.; Shamsizadeh, A.; Arababadi, M.K.; Ayoobi, F.; Fatemi, I.; Allahtavakoli, M.; Mohammad-Zadeh, M. Tactile learning in rodents: Neurobiology and neuropharmacology. Life Sci. 2016, 147, 1–8. [Google Scholar] [CrossRef]
- Krupa, D.J.; Matell, M.S.; Brisben, A.J.; Oliveira, L.M.; Nicolelis, M.A. Behavioral properties of the trigeminal somatosensory system in rats performing whisker-dependent tactile discriminations. J. Neurosci. 2001, 21, 5752–5763. [Google Scholar] [CrossRef]
- Catania, K.C.; Leitch, D.B.; Gauthier, D. Function of the Appendages in Tentacled Snakes (Erpeton tentaculatus). J. Exp. Biol. 2010, 213, 359–367. [Google Scholar] [CrossRef]
- Leitch, D.B.; Catania, K.C. Structure, Innervation and Response Properties of Integumentary Sensory Organs in Crocodilians. J. Exp. Biol. 2012, 215, 4217–4230. [Google Scholar] [CrossRef]
- Marasco, P.D.; Tsuruda, P.R.; Bautista, D.M.; Catania, K.C. Fine Structure of Eimer’s Organ in the Coast Mole (Scapanus orarius). Anat. Record. Anat. Rec. 2007, 290, 437–448. [Google Scholar] [CrossRef]
- Schneider, E.R.; Gracheva, E.O.; Bagriantsev, S.N. Evolutionary Specialization of Tactile Perception in Vertebrates. Physiology 2016, 31, 193–200. [Google Scholar] [CrossRef]
- du Toit, C.J.; Chinsamy, A.; Cunningham, S.J. Cretaceous origins of the vibrotactile bill-tip organ in birds. Proc. R. Soc. B Sci. 2020, 287, 20202322. [Google Scholar] [CrossRef]
- Piersma, T.; Aelst, R.V.; Kurk, K.; Berkhoudt, H.; Leo, R.M.M. A new pressure sensory mechanism for prey detection in birds: The use of principles of seabed dynamics? Proc. R. Soc. B Sci. 1998, 265, 1377–1383. [Google Scholar] [CrossRef]
- Cunningham, S.; Castro, I.; Alley, M. A New Prey-Detection Mechanism for Kiwi (Apteryx spp.) Suggests Convergent Evolution Between Paleognathous and Neognathous Birds. J. Anat. 2007, 211, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, S.J.; Castro, I.; Jensen, T.; Potter, M.A. Remote Touch Prey-Detection by Madagascar Crested Ibises Lophotibis cristata urschi. J. Avian Biol. 2010, 41, 350–353. [Google Scholar] [CrossRef]
- Cunningham, S.J.; Corfield, J.R.; Iwaniuk, A.N.; Castro, I.; Alley, M.R.; Birkhead, T.R.; Parsons, S. The Anatomy of the Bill Tip of Kiwi and Associated Somatosensory Regions of the Brain: Comparisons with Shorebirds. PLoS ONE 2013, 8, e80036. [Google Scholar] [CrossRef]
- Demery, Z.P.; Chappell, J.; Martin, G.R. Vision, touch and object manipulation in Senegal parrots Poicephalus senegalus. Proc. R. Soc. B Sci. 2011, 278, 3687–3693. [Google Scholar] [CrossRef] [PubMed]
- Berkhoudt, H. The Morphology and Distribution of Cutaneous Mechanoreceptors (Herbst and Grandry Corpuscles) in Bill and Tongue of the Mallard (Anas platyrhynchos L.). Neth. J. Zool. 1980, 30, 1–34. [Google Scholar] [CrossRef]
- Avilova, K.V.; Fedorenko, A.G.; Lebedeva, N.V. The Mechanoreceptor Organs of the Lamellirostral Birds (Anseriformes, Aves). Biol. Bull. Russ. Acad. Sci. 2018, 45, 51–60. [Google Scholar] [CrossRef]
- Dubbeldam, J.L. The Sensory Trigeminal System in Birds: Input, Organization and Effects of Peripheral Damage. A Review. Arch. Physiol. Biochem. 1998, 106, 338–345. [Google Scholar] [CrossRef]
- Kishida, R.; Dubbeldam, J.L.; Goris, R.C. Primary Sensory Ganglion Cells Projecting to the Principal Trigeminal Nucleus in the Mallard, Anas platyrhynchos. J. Comp. Neurol. 1985, 240, 171–179. [Google Scholar] [CrossRef]
- Schneider, E.R.; Anderson, E.O.; Mastrotto, M.; Matson, J.D.; Schulz, V.P.; Gallagher, P.G.; Lamotte, R.H.; Gracheva, E.O.; Bagriantsev, S.N. Molecular Basis of Tactile Specialization in the Duck Bill. Proc. Natl. Acad. Sci. USA 2017, 114, 13036–13041. [Google Scholar] [CrossRef]
- Gutiérrez-Ibáñez, C.; Iwaniuk, A.N.; Wylie, D.R. The Independent Evolution of the Enlargement of the Principal Sensory Nucleus of the Trigeminal Nerve in Three Different Groups of Birds. Brain Behav. Evol. 2009, 74, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Crole, M.R.; Soley, J.T. Comparative Distribution and Arrangement of Herbst Corpuscles in the Oropharynx of the Ostrich (Struthio camelus) and Emu (Dromaius novaehollandiae). Anat. Rec. 2014, 297, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Crole, M.R.; Soley, J.T. Bony Pits in the Ostrich (Struthio camelus) and Emu (Dromaius novaehollandiae) Bill Tip. Anat. Rec. 2017, 300, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Gussekloo, S.W.S.; Bout, R.G. The kinematics of feeding and drinking in palaeognathous birds in relation to cranial morphology. J. Exp. Biol. 2005, 208, 3395–3407. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R.; Katzir, G. Visual-fields in ostriches. Nature 1995, 374, 19–20. [Google Scholar] [CrossRef]
- Martin, G.R.; Ashash, U.; Katzir, G. Ostrich ocular optics. Brain Behav. Evolut. 2001, 58, 115–120. [Google Scholar] [CrossRef]
- Wylie, D.R.; Gutiérrez-Ibáñez, C.; Iwaniuk, A.N. Integrating brain, behavior, and phylogeny to understand the evolution of sensory systems in birds. Front. Neurosci. 2015, 9, 281. [Google Scholar] [CrossRef]
- Martin, G.R.; Wilson, K.J.; Martin Wild, J.; Parsons, S.; Fabiana Kubke, M.; Corfield, J. Kiwi Forego Vision in the Guidance of Their Nocturnal Activities. PLoS ONE 2007, 2, e198. [Google Scholar] [CrossRef]
- Iwaniuk, A.N.; Gutierrez-Ibanez, C.; Pakan, J.M.P.; Wylie, D.R. Allometric Scaling of the Tectofugal Pathway in Birds. Brain Behav. Evol. 2010, 75, 122–137. [Google Scholar] [CrossRef]
- Schneider, E.R.; Mastrotto, M.; Laursen, W.J.; Schulz, V.P.; Goodman, J.B.; Funk, O.H.; Gallagher, P.G.; Gracheva, E.O.; Bagriantsev, S.N. Neuronal Mechanism for Acute Mechanosensitivity in Tactile-Foraging Waterfowl. Proc. Natl. Acad. Sci. USA 2014, 111, 14941–14946. [Google Scholar] [CrossRef]
- Schneider, E.R.; Anderson, E.O.; Feketa, V.V.; Mastrotto, M.; Nikolaev, Y.A.; Gracheva, E.O.; Bagriantsev, S.N. A Cross-Species Analysis Reveals a General Role for Piezo2 in Mechanosensory Specialization of Trigeminal Ganglia from Tactile Specialist Birds. Cell Rep. 2019, 26, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.L.; Aoyama, M.; Sugita, S. Topography of ganglion cells in the retina of the duck (Anas platyrhynchos var. domesticus). Anim. Sci. J. 2007, 78, 286–292. [Google Scholar] [CrossRef]
- Budnik, V.; Mpodozis, J.; Varela, F.J.; Maturana, H.R. Regional Specialization of The Quail Retina—Ganglion-Cell Density and Oil Droplet Distribution. Neurosci. Lett. 1984, 51, 145–150. [Google Scholar] [CrossRef]
- Fernández-Juricic, E.; Erichsen, J.T.; Kacelnik, A. Visual perception and social foraging in birds. Trends Ecol. Evol. 2004, 19, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R.; Portugal, S.J. Differences in foraging ecology determine variation in visual fields in ibises and spoonbills (Threskiornithidae). Ibis 2011, 153, 662–671. [Google Scholar] [CrossRef]
- Ye, Y.; Jiang, Y.; Hu, C.; Liu, Y.; Qing, B.; Wang, C.; Fernández-Juricic, E.; Ding, C. What Makes a Tactile Forager Join Mixed-Species Flocks? A Case Study with the Endangered Crested Ibis (Nipponia nippon). Auk 2017, 134, 421–431. [Google Scholar] [CrossRef]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative Genomics Reveals Insights into Avian Genome Evolution and Adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Sanderson, M.J. r8s: Inferring Absolute Rates of Molecular Evolution and Divergence Times in the Absence of a Molecular Clock. Bioinformatics 2003, 19, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A Computational Tool for the Study of Gene Family Evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Wirthlin, M.; Lima, N.C.B.; Guedes, R.L.M.; Soares, A.E.R.; Almeida, L.G.P.; Cavaleiro, N.P.; Loss De Morais, G.; Chaves, A.V.; Howard, J.T.; Teixeira, M.M.; et al. Parrot Genomes and the Evolution of Heightened Longevity and Cognition. Curr. Biol. 2018, 28, 4001–4008. [Google Scholar] [CrossRef]
- Xu, S.; He, Z.; Guo, Z.; Zhang, Z.; Wyckoff, G.J.; Greenberg, A.; Wu, C.I.; Shi, S. Genome-Wide Convergence During Evolution of Mangroves from Woody Plants. Mol. Biol. Evol. 2017, 34, 1008–1015. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Peng, C.; Ren, J.; Deng, C.; Jiang, D.; Wang, J.; Qu, J.; Chang, J.; Yan, C.; Jiang, K.; Murphy, R.W.; et al. The genome of Shaw’s sea snake (Hydrophis curtus) reveals secondary adaptation to its marine environment. Mol. Biol. Evol. 2020, 37, 1744–1760. [Google Scholar] [CrossRef]
- Iroegbu, J.D.; Ijomone, O.K.; Femi-Akinlosotu, O.M.; Ijomone, O.M. ERK/MAPK signalling in the developing brain: Perturbations and consequences. Neurosci. Biobehav. Rev. 2021, 131, 792–805. [Google Scholar] [CrossRef]
- Licausi, F.; Hartman, N. Role of mTOR Complexes in Neurogenesis. Int. J. Mol. Sci. 2018, 19, 1544. [Google Scholar] [CrossRef]
- Hiew, L.; Poon, C.; You, H.; Lim, L. TGF-β/Smad Signalling in Neurogenesis: Implications for Neuropsychiatric Diseases. Cells 2021, 10, 1382. [Google Scholar] [CrossRef]
- Corfield, J.R.; Eisthen, H.L.; Iwaniuk, A.N.; Parsons, S. Anatomical Specializations for Enhanced Olfactory Sensitivity in Kiwi, Apteryx mantelli. Brain Behav. Evol. 2014, 84, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Le Duc, D.; Renaud, G.; Krishnan, A.; Almén, M.S.; Huynen, L.; Prohaska, S.J.; Ongyerth, M.; Bitarello, B.D.; Schiöth, H.B.; Hofreiter, M.; et al. Kiwi Genome Provides Insights into Evolution of a Nocturnal Lifestyle. Genome Biol. 2015, 16, 147. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Yang, Z.; Maldonado, E.; Li, C.; Zhang, G.; Gilbert, M.T.P.; Jarvis, E.D.; O Brien, S.J.; Johnson, W.E.; Antunes, A. Olfactory Receptor Subgenomes Linked with Broad Ecological Adaptations in Sauropsida. Mol. Biol. Evol. 2015, 32, 2832–2843. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, V.; Grunewald, O.; Muller, J.; Zeitz, C.; Obermaier, C.D.; Devos, A.; Pelletier, V.; Bocquet, B.; Andrieu, C.; Bacquet, J.; et al. Novel TTLL5 Variants Associated with Cone-Rod Dystrophy and Early-Onset Severe Retinal Dystrophy. Int. J. Mol. Sci. 2021, 22, 6410. [Google Scholar] [CrossRef]
- Yusuf, I.H.; Garrett, A.M.; Maclaren, R.E.; Charbel Issa, P. Retinal cadherins and the retinal cadherinopathies: Current concepts and future directions. Prog. Retin. Eye Res. 2022, 101038. [Google Scholar] [CrossRef]
- Tanackovic, G.; Ransijn, A.; Ayuso, C.; Harper, S.; Berson, E.L.; Rivolta, C. A Missense Mutation in PRPF6 Causes Impairment of pre-mRNA Splicing and Autosomal-Dominant Retinitis Pigmentosa. Am. J. Hum. Genet. 2011, 88, 643–649. [Google Scholar] [CrossRef]
- Xu, M.; Yang, L.; Wang, F.; Li, H.; Wang, X.; Wang, W.; Ge, Z.; Wang, K.; Zhao, L.; Li, H.; et al. Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum. Genet. 2015, 134, 1069–1078. [Google Scholar] [CrossRef]
- Laurie, N.A.; Donovan, S.L.; Shih, C.; Zhang, J.; Mills, N.; Fuller, C.; Teunisse, A.; Lam, S.; Ramos, Y.; Mohan, A.; et al. Inactivation of the p53 pathway in retinoblastoma. Nature 2006, 444, 61–66. [Google Scholar] [CrossRef]
- Mcevoy, J.; Ulyanov, A.; Brennan, R.; Wu, G.; Pounds, S.; Zhang, J.; Dyer, M.A. Analysis of MDM2 and MDM4 single nucleotide polymorphisms, mRNA splicing and protein expression in retinoblastoma. PLoS ONE 2012, 7, e42739. [Google Scholar] [CrossRef]
- Lim, H.T.; Kim, D.H.; Kim, H. PAX6 aniridia syndrome. Curr. Opin. Ophthalmol. 2017, 28, 436–447. [Google Scholar] [CrossRef]
- Imani, S.; Ijaz, I.; Shasaltaneh, M.D.; Fu, S.; Cheng, J.; Fu, J. Molecular genetics characterization and homology modeling of the CHM gene mutation: A study on its association with choroideremia. Mutat. Res. Rev. Mutat. Res. 2018, 775, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Dejgaard, S.Y.; Presley, J.F. Rab18: New insights into the function of an essential protein. Cell. Mol. Life Sci. 2019, 76, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Bem, D.; Yoshimura, S.; Nunes-Bastos, R.; Bond, F.F.; Kurian, M.A.; Rahman, F.; Handley, M.T.W.; Hadzhiev, Y.; Masood, I.; Straatman-Iwanowska, A.A.; et al. Loss-of-Function Mutations in RAB18 Cause Warburg Micro Syndrome. Am. J. Hum. Genet. 2011, 88, 499–507. [Google Scholar] [CrossRef]
- Mclaughlin, P.J.; Sassani, J.W.; Klocek, M.S.; Zagon, I.S. Diabetic keratopathy and treatment by modulation of the opioid growth factor (OGF)–OGF receptor (OGFr) axis with naltrexone: A review. Brain Res. Bull. 2010, 81, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Matos-Rodrigues, G.E.; Tan, P.B.; Rocha-Martins, M.; Charlier, C.F.; Gomes, A.L.; Cabral-Miranda, F.; Grigaravicius, P.; Hofmann, T.G.; Frappart, P.; Martins, R.A.P. Progenitor death drives retinal dysplasia and neuronal degeneration in a mouse model of Atrip-Seckel syndrome. Dis. Model. Mech. 2020, 13, dmm045807. [Google Scholar] [CrossRef] [PubMed]
- Van Camp, G.; Snoeckx, R.L.; Hilgert, N.; van den Ende, J.; Fukuoka, H.; Wagatsuma, M.; Suzuki, H.; Smets, R.M.; Vanhoenacker, F.; Declau, F.; et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am. J. Hum. Genet. 2006, 79, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Huang, L.; Chen, Y.; Liu, S.; Chen, C.; Xiong, K.; Song, L.; Zhou, Y.; Yang, X.; Zhong, M. A novel contiguous deletion involving NDP, MAOB and EFHC2 gene in a patient with familial Norrie disease: Bilateral blindness and leucocoria without other deficits. J. Genet. 2017, 96, 1015–1020. [Google Scholar] [CrossRef]
- Tomas-Roca, L.; Tsaalbi-Shtylik, A.; Jansen, J.G.; Singh, M.K.; Epstein, J.A.; Altunoglu, U.; Verzijl, H.; Soria, L.; van Beusekom, E.; Roscioli, T.; et al. De novo mutations in PLXND1 and REV3L cause Möbius syndrome. Nat. Commun. 2015, 6, 7199. [Google Scholar] [CrossRef]
- Kloeckener-Gruissem, B.; Vandekerckhove, K.; Nürnberg, G.; Neidhardt, J.; Zeitz, C.; Nürnberg, P.; Schipper, I.; Berger, W. Mutation of Solute Carrier SLC16A12 Associates with a Syndrome Combining Juvenile Cataract with Microcornea and Renal Glucosuria. Am. J. Hum. Genet. 2008, 82, 772–779. [Google Scholar] [CrossRef]
- Wright, Z.C.; Singh, R.K.; Alpino, R.; Goldberg, A.F.X.; Sokolov, M.; Ramamurthy, V. ARL3 regulates trafficking of prenylated phototransduction proteins to the rod outer segment. Hum. Mol. Genet. 2016, 25, 2031–2044. [Google Scholar] [CrossRef]
- El Shamieh, S.; Neuillé, M.; Terray, A.; Orhan, E.; Condroyer, C.; Démontant, V.; Michiels, C.; Antonio, A.; Boyard, F.; Lancelot, M.; et al. Whole-Exome Sequencing Identifies KIZ as a Ciliary Gene Associated with Autosomal-Recessive Rod-Cone Dystrophy. Am. J. Hum. Genet. 2014, 94, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhou, G.; Chen, X.; Chen, H.; Wu, K.; Xiang, L.; Lei, X.; Zhang, X.; Wu, R.; Jin, Z. Targeted RP9 ablation and mutagenesis in mouse photoreceptor cells by CRISPR-Cas9. Sci. Rep. 2017, 7, 43062. [Google Scholar] [CrossRef] [PubMed]
- Gill, J.S.; Georgiou, M.; Kalitzeos, A.; Moore, A.T.; Michaelides, M. Progressive cone and cone-rod dystrophies: Clinical features, molecular genetics and prospects for therapy. Brit. J. Ophthalmol. 2019, 103, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Boone, P.M.; Yuan, B.; Gu, S.; Ma, Z.; Gambin, T.; Gonzaga-Jauregui, C.; Jain, M.; Murdock, T.J.; White, J.J.; Jhangiani, S.N.; et al. Hutterite-type cataract maps to chromosome 6p21.32-p21.31, cosegregates with a homozygous mutation in LEMD2, and is associated with sudden cardiac death. Mol. Genet. Genom. Med. 2016, 4, 77–94. [Google Scholar] [CrossRef]
- Ansar, M.; Chung, H.; Taylor, R.L.; Nazir, A.; Imtiaz, S.; Sarwar, M.T.; Manousopoulou, A.; Makrythanasis, P.; Saeed, S.; Falconnet, E.; et al. Bi-allelic Loss-of-Function Variants in DNMBP Cause Infantile Cataracts. Am. J. Hum. Genet. 2018, 103, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Longley, M.J.; Clark, S.; Man, C.Y.W.; Hudson, G.; Durham, S.E.; Taylor, R.W.; Nightingale, S.; Turnbull, D.M.; Copeland, W.C.; Chinnery, P.F. Mutant POLG2 Disrupts DNA Polymerase γ Subunits and Causes Progressive External Ophthalmoplegia. Am. J. Hum. Genet. 2006, 78, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, E.; Delvallée, C.; Mary, L.; Stoetzel, C.; Geoffroy, V.; Marks-Delesalle, C.; Holder-Espinasse, M.; Ghoumid, J.; Dollfus, H.; Muller, J. Identification and Characterization of Known Biallelic Mutations in the IFT27 (BBS19) Gene in a Novel Family with Bardet-Biedl Syndrome. Front. Genet. 2019, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Bin, J.; Madhavan, J.; Ferrini, W.; Mok, C.A.; Billingsley, G.; Hã On, E. BBS7 and TTC8 (BBS8) mutations play a minor role in the mutational load of Bardet-Biedl syndrome in a multiethnic population. Hum. Mutat. 2009, 30, E737–E746. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Morimoto, T.; Hotta, K.; Fujikado, T.; Nishida, K. A novel compound heterozygous mutation in TTC8 identified in a Japanese patient. Hum. Genome Var. 2019, 6, 14. [Google Scholar] [CrossRef]
- Ece Solmaz, A.; Onay, H.; Atik, T.; Aykut, A.; Cerrah Gunes, M.; Ozalp Yuregir, O.; Bas, V.N.; Hazan, F.; Kirbiyik, O.; Ozkinay, F. Targeted multi-gene panel testing for the diagnosis of Bardet Biedl syndrome: Identification of nine novel mutations across BBS1, BBS2, BBS4, BBS7, BBS9, BBS10 genes. Eur. J. Med. Genet. 2015, 58, 689–694. [Google Scholar] [CrossRef]
- Ludlam, W.G.; Aoba, T.; Cuéllar, J.; Bueno-Carrasco, M.T.; Makaju, A.; Moody, J.D.; Franklin, S.; Valpuesta, J.M.; Willardson, B.M. Molecular architecture of the Bardet–Biedl syndrome protein 2-7-9 subcomplex. J. Biol. Chem. 2019, 294, 16385–16399. [Google Scholar] [CrossRef] [PubMed]
- Radha Rama Devi, A.; Naushad, S.M.; Lingappa, L. Clinical and Molecular Diagnosis of Joubert Syndrome and Related Disorders. Pediatr. Neurol. 2020, 106, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Srour, M.; Hamdan, F.F.; Schwartzentruber, J.A.; Patry, L.; Ospina, L.H.; Shevell, M.I.; Désilets, V.; Dobrzeniecka, S.; Mathonnet, G.; Lemyre, E.; et al. Mutations in TMEM231 cause Joubert syndrome in French Canadians. J. Med. Genet. 2012, 49, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Baltus, A.E.; Mathew, R.S.; Murphy, E.A.; Evrony, G.D.; Gonzalez, D.M.; Wang, E.P.; Marshall-Walker, C.A.; Barry, B.J.; Murn, J.; et al. Microcephaly Gene Links Trithorax and REST/NRSF to Control Neural Stem Cell Proliferation and Differentiation. Cell 2012, 151, 1097–1112. [Google Scholar] [CrossRef] [PubMed]
- Faheem, M.; Naseer, M.I.; Rasool, M.; Chaudhary, A.G.; Kumosani, T.A.; Ilyas, A.M.; Pushparaj, P.; Ahmed, F.; Algahtani, H.A.; Al-Qahtani, M.H.; et al. Molecular genetics of human primary microcephaly: An overview. BMC Med. Genom. 2015, 8 (Suppl. 1), S4. [Google Scholar] [CrossRef] [PubMed]
- Link, N.; Chung, H.; Jolly, A.; Withers, M.; Tepe, B.; Arenkiel, B.R.; Shah, P.S.; Krogan, N.J.; Aydin, H.; Geckinli, B.B.; et al. Mutations in ANKLE2, a ZIKA Virus Target, Disrupt an Asymmetric Cell Division Pathway in Drosophila Neuroblasts to Cause Microcephaly. Dev. Cell 2019, 51, 713–729. [Google Scholar] [CrossRef]
- Pavlakis, E.; Chiotaki, R.; Chalepakis, G. The role of Fras1/Frem proteins in the structure and function of basement membrane. Int. J. Biochem. Cell Biol. 2011, 43, 487–495. [Google Scholar] [CrossRef]
- Hoefele, J.; Wilhelm, C.; Schiesser, M.; Mack, R.; Heinrich, U.; Weber, L.T.; Biskup, S.; Daumer-Haas, C.; Klein, H.; Rost, I. Expanding the mutation spectrum for Fraser syndrome: Identification of a novel heterozygous deletion in FRAS1. Gene 2013, 520, 194–197. [Google Scholar] [CrossRef]
- Ronchi, D.; Di Fonzo, A.; Lin, W.; Bordoni, A.; Liu, C.; Fassone, E.; Pagliarani, S.; Rizzuti, M.; Zheng, L.; Filosto, M.; et al. Mutations in DNA2 Link Progressive Myopathy to Mitochondrial DNA Instability. Am. J. Hum. Genet. 2013, 92, 293–300. [Google Scholar] [CrossRef]
- Katsanis, N.; Ansley, S.J.; Badano, J.L.; Eichers, E.R.; Lewis, R.A.; Hoskins, B.E.; Scambler, P.J.; Davidson, W.S.; Beales, P.L.; Lupski, J.R. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 2001, 293, 2256–2259. [Google Scholar] [CrossRef]
- Ohno, K.; Kato, H.; Funahashi, S.; Hasegawa, T.; Sato, K. Characterization of CLP36/Elfin/PDLIM1 in the nervous system. J. Neurochem. 2009, 111, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Ohno, K.; Funahashi, S.; Miyazaki, K.; Nagano, A.; Sato, K. CLP36 interacts with palladin in dorsal root ganglion neurons. Neurosci. Lett. 2010, 476, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Ríos, H.; Paganelli, A.R.; Fosser, N.S. The role of PDLIM1, a PDZ-LIM domain protein, at the ribbon synapses in the chicken retina. J. Comp. Neurol. 2020, 528, 1820–1832. [Google Scholar] [CrossRef] [PubMed]
- Gerhold, K.A.; Pellegrino, M.; Tsunozaki, M.; Morita, T.; Leitch, D.B.; Tsuruda, P.R.; Brem, R.B.; Catania, K.C.; Bautista, D.M.; Nitabach, M.N. The star-nosed mole reveals clues to the molecular basis of mammalian touch. PLoS ONE 2013, 8, e55001. [Google Scholar] [CrossRef]
- Emerling, C.A.; Springer, M.S. Eyes underground: Regression of visual protein networks in subterranean mammals. Mol. Phylogenet. Evol. 2014, 78, 260–270. [Google Scholar] [CrossRef]
- Catania, K.C.; Kaas, J.H. Organization of the somatosensory cortex of the star-nosed mole. J. Comp. Neurol. (1911) 1995, 351, 549–567. [Google Scholar] [CrossRef]
- Rauschecker, J.P.; Tian, B.; Korte, M.; Egert, U. Crossmodal changes in the somatosensory vibrissa/barrel system of visually deprived animals. Proc. Natl. Acad. Sci. USA 1992, 89, 5063–5067. [Google Scholar] [CrossRef]
- Iwaniuk, A.; Clayton, D.; Wylie, D. Echolocation, vocal learning, auditory localization and the relative size of the avian auditory midbrain nucleus (MLd). Behav. Brain Res. 2006, 167, 305–317. [Google Scholar] [CrossRef]
- Corfield, J.R.; Parsons, S.; Harimoto, Y.; Acosta, M.L. Retinal Anatomy of the New Zealand Kiwi: Structural Traits Consistent With Their Nocturnal Behavior. Anat. Rec. 2015, 298, 771–779. [Google Scholar] [CrossRef]
- Gutierrez-Ibanez, C.; Iwaniuk, A.N.; Lisney, T.J.; Faunes, M.; Marin, G.J.; Wylie, D.R. Functional implications of species differences in the size and morphology of the isthmo optic nucleus (ION) in birds. PLoS ONE 2012, 7, e37816. [Google Scholar] [CrossRef]
- Corfield, J.R.; Long, B.; Krilow, J.M.; Wylie, D.R.; Iwaniuk, A.N. A unique cellular scaling rule in the avian auditory system. Brain Struct. Funct. 2016, 221, 2675–2693. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.E.; Leitch, D.B.; Wong, P.; Kaas, J.H.; Herculano-Houzel, S. Faster scaling of visual neurons in cortical areas relative to subcortical structures in non-human primate brains. Brain Struct. Funct. 2013, 218, 805–816. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Striedter, G.F.; Charvet, C.J. Developmental origins of species differences in telencephalon and tectum size: Morphometric comparisons between a parakeet (Melopsittacus undulatus) and a quail (Colinus virgianus). J. Comp. Neurol. 2008, 507, 1663–1675. [Google Scholar] [CrossRef] [PubMed]
- Charvet, C.J.; Striedter, G.F. Bigger brains cycle faster before neurogenesis begins: A comparison of brain development between chickens and bobwhite quail. Proc. R. Soc. B Sci. 2010, 277, 3469–3475. [Google Scholar] [CrossRef]
- Rster, W.H. Histological and electrophysiological investigations on the vibration-sensitive receptors (Herbst corpuscles) in the wing of the pigeon (Columba livia). J. Comp. Physiol. A 1990, 166, 663–673. [Google Scholar] [CrossRef]
- Shen, J.X.; Xu, Z.M. Response characteristics of Herbst corpuscles in the interosseous region of the pigeon’s hind limb. J. Comp. Physiol. A 1994, 175, 667–674. [Google Scholar] [CrossRef]
- Zelena, J.; Halata, Z.; Szeder, V.; Grim, M. Crural Herbst corpuscles in chicken and quail: Numbers and structure. Anat. Embryol. 1997, 196, 323–333. [Google Scholar] [CrossRef]
- Narayanan, P.; Sondermann, J.; Rouwette, T.; Karaca, S.; Urlaub, H.; Mitkovski, M.; Gomez-Varela, D.; Schmidt, M. Native Piezo2 Interactomics Identifies Pericentrin as a Novel Regulator of Piezo2 in Somatosensory Neurons. J. Proteome Res. 2016, 15, 2676–2687. [Google Scholar] [CrossRef]
- Nonomura, K.; Woo, S.; Chang, R.B.; Gillich, A.; Qiu, Z.; Francisco, A.G.; Ranade, S.S.; Liberles, S.D.; Patapoutian, A. Piezo2 senses airway stretch and mediates lung inflation-induced apnoea. Nature 2017, 541, 176–181. [Google Scholar] [CrossRef]
- Shin, S.M.; Moehring, F.; Itson-Zoske, B.; Fan, F.; Stucky, C.L.; Hogan, Q.H.; Yu, H. Piezo2 mechanosensitive ion channel is located to sensory neurons and nonneuronal cells in rat peripheral sensory pathway: Implications in pain. Pain 2021, 162, 2750–2768. [Google Scholar] [CrossRef]

| Species | ID | Term | Category | FDR (<0.05) |
|---|---|---|---|---|
| Northern brown kiwi | GO:0004984 | olfactory receptor activity | Molecular Function | 5.59 × 10−9 |
| GO:0005549 | odorant binding | Molecular Function | 9.52 × 10−3 | |
| GO:0050911 | detection of chemical stimulus involved in sensory | Biological Process | 5.59 × 10−9 | |
| GO:0007608 | sensory perception of smell | Biological Process | 3.66 × 10−2 | |
| Mallard | GO:0004984 | olfactory receptor activity | Molecular Function | 2.29 × 10−71 |
| Crested ibis | GO:0032426 | stereocilium tip | Cellular Component | 1.07 × 10−3 |
| GO:0048839 | inner ear development | Biological Process | 3.94 × 10−3 | |
| GO:0007605 | sensory perception of sound | Biological Process | 2.60 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Sun, L.; Wan, Q.-H.; Fang, S.-G. Comparative Genomics Provides Insights into Adaptive Evolution in Tactile-Foraging Birds. Genes 2022, 13, 678. https://doi.org/10.3390/genes13040678
Wang L, Sun L, Wan Q-H, Fang S-G. Comparative Genomics Provides Insights into Adaptive Evolution in Tactile-Foraging Birds. Genes. 2022; 13(4):678. https://doi.org/10.3390/genes13040678
Chicago/Turabian StyleWang, Li, Li Sun, Qiu-Hong Wan, and Sheng-Guo Fang. 2022. "Comparative Genomics Provides Insights into Adaptive Evolution in Tactile-Foraging Birds" Genes 13, no. 4: 678. https://doi.org/10.3390/genes13040678
APA StyleWang, L., Sun, L., Wan, Q.-H., & Fang, S.-G. (2022). Comparative Genomics Provides Insights into Adaptive Evolution in Tactile-Foraging Birds. Genes, 13(4), 678. https://doi.org/10.3390/genes13040678






