Functions and Regulation of Meiotic HORMA-Domain Proteins
Abstract
:1. Introduction
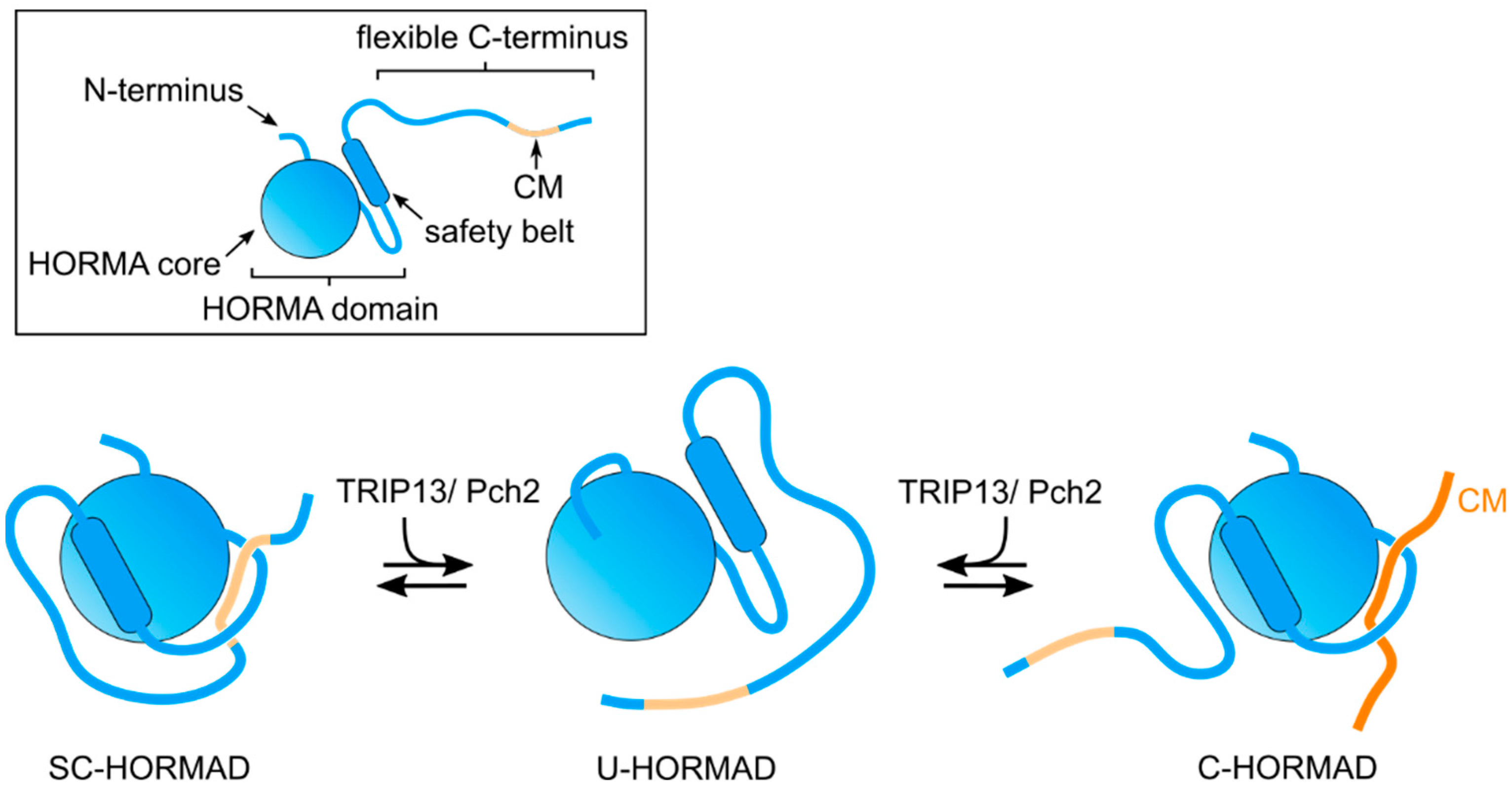
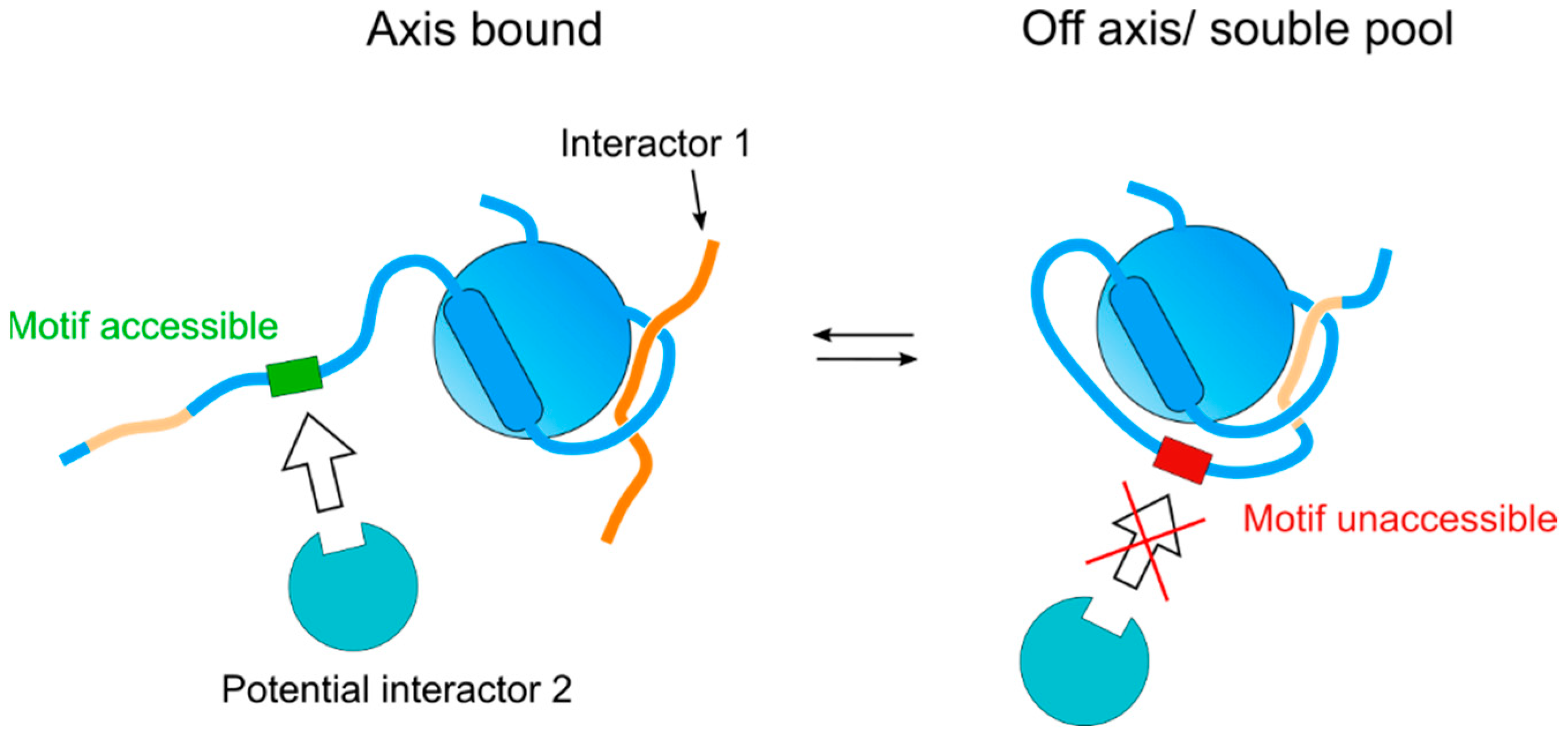
2. The HORMA Domain: A Protein-Protein Interaction Module for Regulatory Mechanisms
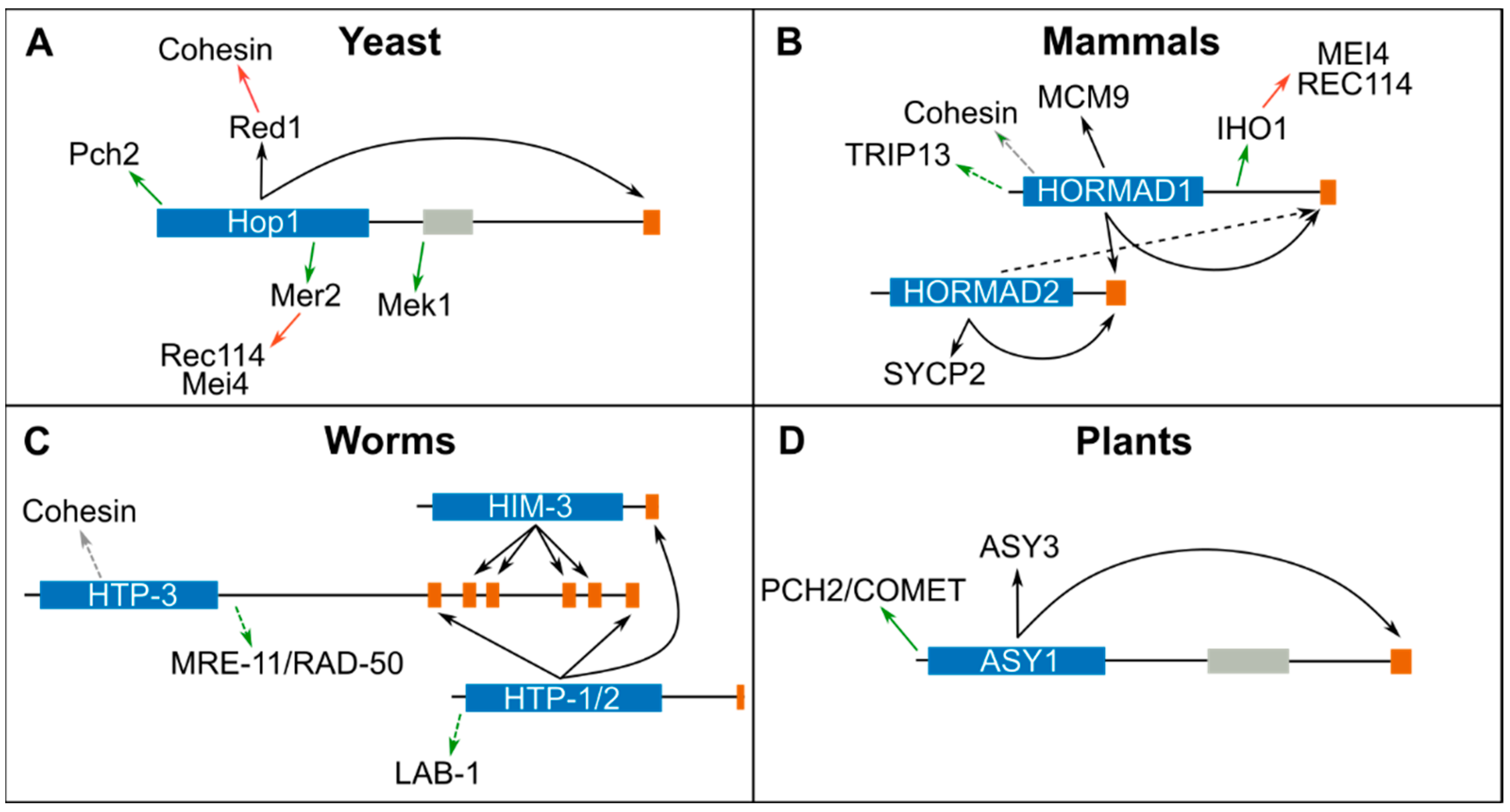
3. Protein Complexes Formed by Meiotic HORMADs
4. Binding of Meiotic HORMADs to Axial Elements
5. Pch2/TRIP13 Regulates the Assembly and Disassembly of Protein Complexes Containing Meiotic HORMADs
6. HORMADs Roles in Meiotic Recombination
7. HORMADs Involvement in SC Assembly
8. Checkpoint Regulation of Meiotic Prophase by Meiotic HORMADs
9. Regulation of Meiotic HORMADs by Post Translational Modifications
10. Conclusions
| . | Protein | Additional Structural Features | Interactors through Safety Belt-CM | Kd/μM | Interactors through an Alternate Interface | Interactors through an Uncharacterized Interface | References |
|---|---|---|---|---|---|---|---|
| H. sapiens | MAD2 | - | MAD1 | 1.04 | MAD2 | - | [25,37,98,99] |
| CDC20 | 0.1 | p31comet | [26,98,100] | ||||
| SGO2 | 0.69 | TRIP13 | [32,101] | ||||
| REV3 * | BUBR1 | [102,103] | |||||
| RIT1 * | WT1 | [104,105] | |||||
| H. sapiens | REV7 | - | SHLD3 | 0.013 ± 0.0004 | REV7 | CLTA | [28,36,106,107] |
| RAN | 1.85 | MAD2 | HCCA2 | [36,108,109] | |||
| REV3 | p31comet | PRCC | [35,36,110,111] | ||||
| IpaB | REV1 | SIM2 | [28,38,108,112] | ||||
| CAMP | SHLD2 | TCF4 | [28,39,113,114] | ||||
| ELK-1 * | TRIP13 | ADP/E3-11.6K | [39,115,116] | ||||
| MDC9 * | CDH1 | TF11-1 | [102,117,118] | ||||
| S. cerevisiae | Hop1 | C-ter CM | Hop1 * | 6.1 ± 1 ‡ | Pch2 | Mer2 | [27,60,71] |
| zinc finger | Red1 | 0.34 ± 0.03 ‡ | Mek1 † | PP4 # | [27,92,119] | ||
| S. pombe | Hop1 | C-ter CM | Rec10 * | Rec15 † | [43] | ||
| zinc finger | |||||||
| H. sapiens | HORMAD1 | C-ter CM | HORMAD1 * | [40] | |||
| HORMAD2 * | [40] | ||||||
| MCM9 * | [120] | ||||||
| M. musculus | HORMAD1 | C-ter CM | TRIP13 # | Cohesin # | [10,33,45] | ||
| IHO1 | [70] | ||||||
| HORMAD2 | C-ter CM | HORMAD2 * | 7.1 ± 0.5 ‡ | [44] | |||
| SYCP2 * | [44] | ||||||
| HORMAD1 * | [10] | ||||||
| C. elegans | HTP-1 | C-ter CM | HIM-3 | 0.7 | LAB-1 # | [19,40,88] | |
| HTP-3 motif #1 | 0.3 | [40,88] | |||||
| HTP-3 motif #6 * | 0.9 | [40,88] | |||||
| HTP-2 | C-ter CM | HIM-3 | 3.1 | [40] | |||
| HTP-3 motif #1 | 0.2 | [40] | |||||
| HTP-3 motif #6 | 0.3 | [40] | |||||
| HIM-3 | C-ter CM | HTP-3 motif #4 | 0.3 | [40,88] | |||
| HTP-3 motifs #2, 3, 5 * | [40,88] | ||||||
| HTP-3 | 6 C-ter CM | MRE-11/RAD-50 # | [14] | ||||
| Cohesin # | [40] | ||||||
| A. thaliana | ASY1 | C-ter CM | ASY1 * | COMET | [44,57,58] | ||
| SWIRM domain | ASY3 * | PCH2 # | [41,44] | ||||
| ASY2 | [121] | ||||||
| Oryza sativa | PAIR2 | C-ter CM SWIRM domain | PAIR3 * | CRC1 (PCH2) | [56,122] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grey, C.; de Massy, B. Chromosome Organization in Early Meiotic Prophase. Front. Cell Dev. Biol. 2021, 9, 688878. [Google Scholar] [CrossRef] [PubMed]
- Lake, C.M.; Hawley, R.S. Synaptonemal complex. Curr. Biol. 2021, 31, R225–R227. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, N.M.; Byers, B. HOP1: A yeast meiotic pairing gene. Genetics 1989, 121, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Schwacha, A.; Kleckner, N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell 1994, 76, 51–63. [Google Scholar] [CrossRef]
- Niu, H.; Wan, L.; Baumgartner, B.; Schaefer, D.; Loidl, J.; Hollingsworth, N.M. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 2005, 16, 5804–5818. [Google Scholar] [CrossRef] [Green Version]
- Woltering, D.; Baumgartner, B.; Bagchi, S.; Larkin, B.; Loidl, J.; de los Santos, T.; Hollingsworth, N.M. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 2000, 20, 6646–6658. [Google Scholar] [CrossRef] [Green Version]
- Latypov, V.; Rothenberg, M.; Lorenz, A.; Octobre, G.; Csutak, O.; Lehmann, E.; Loidl, J.; Kohli, J. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol. Cell. Biol. 2010, 30, 1570–1581. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.H.; Choi, Y.; Erdin, S.U.; Yatsenko, S.A.; Kloc, M.; Yang, F.; Wang, P.J.; Meistrich, M.L.; Rajkovic, A. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 2010, 6, e1001190. [Google Scholar] [CrossRef] [Green Version]
- Daniel, K.; Lange, J.; Hached, K.; Fu, J.; Anastassiadis, K.; Roig, I.; Cooke, H.J.; Stewart, A.F.; Wassmann, K.; Jasin, M.; et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat. Cell Biol. 2011, 13, 599–610. [Google Scholar] [CrossRef] [Green Version]
- Wojtasz, L.; Cloutier, J.M.; Baumann, M.; Daniel, K.; Varga, J.; Fu, J.; Anastassiadis, K.; Stewart, A.F.; Remenyi, A.; Turner, J.M.; et al. Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev. 2012, 26, 958–973. [Google Scholar] [CrossRef] [Green Version]
- Kogo, H.; Tsutsumi, M.; Inagaki, H.; Ohye, T.; Kiyonari, H.; Kurahashi, H. HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes Cells 2012, 17, 897–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Moran, E.; Santos, J.L.; Jones, G.H.; Franklin, F.C. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007, 21, 2220–2233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zetka, M.C.; Kawasaki, I.; Strome, S.; Muller, F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999, 13, 2258–2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodyer, W.; Kaitna, S.; Couteau, F.; Ward, J.D.; Boulton, S.J.; Zetka, M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 2008, 14, 263–274. [Google Scholar] [CrossRef] [Green Version]
- Couteau, F.; Zetka, M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005, 19, 2744–2756. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Perez, E.; Villeneuve, A.M. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005, 19, 2727–2743. [Google Scholar] [CrossRef] [Green Version]
- Severson, A.F.; Ling, L.; van Zuylen, V.; Meyer, B.J. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009, 23, 1763–1778. [Google Scholar] [CrossRef] [Green Version]
- Couteau, F.; Zetka, M. DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Dev. Cell 2011, 20, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Ferrandiz, N.; Barroso, C.; Telecan, O.; Shao, N.; Kim, H.M.; Testori, S.; Faull, P.; Cutillas, P.; Snijders, A.P.; Colaiacovo, M.P.; et al. Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 2018, 9, 834. [Google Scholar] [CrossRef]
- Martinez-Perez, E.; Schvarzstein, M.; Barroso, C.; Lightfoot, J.; Dernburg, A.F.; Villeneuve, A.M. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes Dev. 2008, 22, 2886–2901. [Google Scholar] [CrossRef] [Green Version]
- Aravind, L.; Koonin, E.V. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 1998, 23, 284–286. [Google Scholar] [CrossRef]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Krijger, I.; Boersma, V.; Jacobs, J.J.L. REV7: Jack of many trades. Trends Cell Biol. 2021, 31, 686–701. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Fang, G.; Coldiron, M.; Lin, Y.; Yu, H.; Kirschner, M.W.; Wagner, G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 2000, 7, 224–229. [Google Scholar] [CrossRef]
- Mapelli, M.; Massimiliano, L.; Santaguida, S.; Musacchio, A. The Mad2 conformational dimer: Structure and implications for the spindle assembly checkpoint. Cell 2007, 131, 730–743. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Tang, Z.; Rizo, J.; Yu, H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002, 9, 59–71. [Google Scholar] [CrossRef]
- West, A.M.V.; Komives, E.A.; Corbett, K.D. Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Res. 2018, 46, 279–292. [Google Scholar] [CrossRef] [Green Version]
- Liang, L.; Feng, J.; Zuo, P.; Yang, J.; Lu, Y.; Yin, Y. Molecular basis for assembly of the shieldin complex and its implications for NHEJ. Nat. Commun. 2020, 11, 1972. [Google Scholar] [CrossRef]
- Gu, Y.; Desai, A.; Corbett, K.D. Evolutionary Dynamics and Molecular Mechanisms of HORMA Domain Protein Signaling. Annu. Rev. Biochem. 2022, 91. [Google Scholar] [CrossRef]
- Du Truong, C.; Craig, T.A.; Cui, G.; Botuyan, M.V.; Serkasevich, R.A.; Chan, K.Y.; Mer, G.; Chiu, P.L.; Kumar, R. Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta. J. Biol. Chem. 2021, 297, 100912. [Google Scholar] [CrossRef]
- Ye, Q.; Rosenberg, S.C.; Moeller, A.; Speir, J.A.; Su, T.Y.; Corbett, K.D. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife 2015, 4, e07367. [Google Scholar] [CrossRef] [PubMed]
- Alfieri, C.; Chang, L.; Barford, D. Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature 2018, 559, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Kim, D.H.; Dereli, I.; Rosenberg, S.C.; Hagemann, G.; Herzog, F.; Toth, A.; Cleveland, D.W.; Corbett, K.D. The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. EMBO J. 2017, 36, 2419–2434. [Google Scholar] [CrossRef] [PubMed]
- Clairmont, C.S.; Sarangi, P.; Ponnienselvan, K.; Galli, L.D.; Csete, I.; Moreau, L.; Adelmant, G.; Chowdhury, D.; Marto, J.A.; D’Andrea, A.D. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol. 2020, 22, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Sarangi, P.; Clairmont, C.S.; Galli, L.D.; Moreau, L.A.; D’Andrea, A.D. p31(comet) promotes homologous recombination by inactivating REV7 through the TRIP13 ATPase. Proc. Natl. Acad. Sci. USA 2020, 117, 26795–26803. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.A.; Vassel, F.M.; Chatterjee, N.; D’Souza, S.; Li, Y.; Hao, B.; Hemann, M.T.; Walker, G.C.; Korzhnev, D.M. Rev7 dimerization is important for assembly and function of the Rev1/Polzeta translesion synthesis complex. Proc. Natl. Acad. Sci. USA 2018, 115, E8191–E8200. [Google Scholar] [CrossRef] [Green Version]
- Mapelli, M.; Filipp, F.V.; Rancati, G.; Massimiliano, L.; Nezi, L.; Stier, G.; Hagan, R.S.; Confalonieri, S.; Piatti, S.; Sattler, M.; et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006, 25, 1273–1284. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, S.; Hara, K.; Shimizu, T.; Sato, M.; Hashimoto, H. Structural basis of recruitment of DNA polymerase zeta by interaction between REV1 and REV7 proteins. J. Biol. Chem. 2012, 287, 33847–33852. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Wang, S.; Wang, J.; de la Cruz, M.J.; Xu, G.; Scaltriti, M.; Patel, D.J. Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2024512118. [Google Scholar] [CrossRef]
- Kim, Y.; Rosenberg, S.C.; Kugel, C.L.; Kostow, N.; Rog, O.; Davydov, V.; Su, T.Y.; Dernburg, A.F.; Corbett, K.D. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev. Cell 2014, 31, 487–502. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Hu, B.; Portheine, S.M.; Chuenban, P.; Schnittger, A. State changes of the HORMA protein ASY1 are mediated by an interplay between its closure motif and PCH2. Nucleic Acids Res. 2020, 48, 11521–11535. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.V.; Roeder, G.S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997, 136, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Kariyazono, R.; Oda, A.; Yamada, T.; Ohta, K. Conserved HORMA domain-containing protein Hop1 stabilizes interaction between proteins of meiotic DNA break hotspots and chromosome axis. Nucleic Acids Res. 2019, 47, 10166–10180. [Google Scholar] [CrossRef] [PubMed]
- West, A.M.; Rosenberg, S.C.; Ur, S.N.; Lehmer, M.K.; Ye, Q.; Hagemann, G.; Caballero, I.; Uson, I.; MacQueen, A.J.; Herzog, F.; et al. A conserved filamentous assembly underlies the structure of the meiotic chromosome axis. Elife 2019, 8, e40372. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, Y.; Horisawa-Takada, Y.; Inoue, E.; Tani, N.; Shibuya, H.; Fujimura, S.; Kariyazono, R.; Sakata, T.; Ohta, K.; Araki, K.; et al. Meiotic cohesins mediate initial loading of HORMAD1 to the chromosomes and coordinate SC formation during meiotic prophase. PLoS Genet. 2020, 16, e1009048. [Google Scholar] [CrossRef]
- Woglar, A.; Yamaya, K.; Roelens, B.; Boettiger, A.; Kohler, S.; Villeneuve, A.M. Quantitative cytogenetics reveals molecular stoichiometry and longitudinal organization of meiotic chromosome axes and loops. PLoS Biol. 2020, 18, e3000817. [Google Scholar] [CrossRef]
- Sakuno, T.; Watanabe, Y. Phosphorylation of cohesin Rec11/SA3 by casein kinase 1 promotes homologous recombination by assembling the meiotic chromosome axis. Dev. Cell 2015, 32, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Kironmai, K.M.; Muniyappa, K.; Friedman, D.B.; Hollingsworth, N.M.; Byers, B. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol. 1998, 18, 1424–1435. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Huang, L.; Markowitz, T.E.; Blitzblau, H.G.; Chen, D.; Klein, F.; Hochwagen, A. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife 2015, 4, e07424. [Google Scholar] [CrossRef]
- Heldrich, J.; Milano, C.R.; Markowitz, T.E.; Ur, S.N.; Vale-Silva, L.A.; Corbett, K.D.; Hochwagen, A. Two pathways drive meiotic chromosome axis assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2022, gkac227. [Google Scholar] [CrossRef]
- Lambing, C.; Tock, A.J.; Topp, S.D.; Choi, K.; Kuo, P.C.; Zhao, X.; Osman, K.; Higgins, J.D.; Franklin, F.C.H.; Henderson, I.R. Interacting Genomic Landscapes of REC8-Cohesin, Chromatin, and Meiotic Recombination in Arabidopsis. Plant Cell 2020, 32, 1218–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San-Segundo, P.A.; Roeder, G.S. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 1999, 97, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Bhalla, N.; Dernburg, A.F. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 2005, 310, 1683–1686. [Google Scholar] [CrossRef]
- Wojtasz, L.; Daniel, K.; Roig, I.; Bolcun-Filas, E.; Xu, H.; Boonsanay, V.; Eckmann, C.R.; Cooke, H.J.; Jasin, M.; Keeney, S.; et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet. 2009, 5, e1000702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambing, C.; Osman, K.; Nuntasoontorn, K.; West, A.; Higgins, J.D.; Copenhaver, G.P.; Yang, J.; Armstrong, S.J.; Mechtler, K.; Roitinger, E.; et al. Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers. PLoS Genet. 2015, 11, e1005372. [Google Scholar] [CrossRef] [Green Version]
- Miao, C.; Tang, D.; Zhang, H.; Wang, M.; Li, Y.; Tang, S.; Yu, H.; Gu, M.; Cheng, Z. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell 2013, 25, 2998–3009. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Sofroni, K.; Wijnker, E.; Hamamura, Y.; Carstens, L.; Harashima, H.; Stolze, S.C.; Vezon, D.; Chelysheva, L.; Orban-Nemeth, Z.; et al. The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. EMBO J. 2020, 39, e101625. [Google Scholar] [CrossRef] [PubMed]
- Balboni, M.; Yang, C.; Komaki, S.; Brun, J.; Schnittger, A. COMET Functions as a PCH2 Cofactor in Regulating the HORMA Domain Protein ASY1. Curr. Biol. 2020, 30, 4113–4127.e6. [Google Scholar] [CrossRef]
- Ji, J.; Tang, D.; Shen, Y.; Xue, Z.; Wang, H.; Shi, W.; Zhang, C.; Du, G.; Li, Y.; Cheng, Z. P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice. Proc. Natl. Acad. Sci. USA 2016, 113, 10577–10582. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Jomaa, A.; Ortega, J.; Alani, E.E. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc. Natl. Acad. Sci. USA 2014, 111, E44–E53. [Google Scholar] [CrossRef] [Green Version]
- Herruzo, E.; Lago-Maciel, A.; Baztan, S.; Santos, B.; Carballo, J.A.; San-Segundo, P.A. Pch2 orchestrates the meiotic recombination checkpoint from the cytoplasm. PLoS Genet. 2021, 17, e1009560. [Google Scholar] [CrossRef] [PubMed]
- Raina, V.B.; Vader, G. Homeostatic Control of Meiotic Prophase Checkpoint Function by Pch2 and Hop1. Curr. Biol. 2020, 30, 4413–4424.e5. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, V.V.; MacQueen, A.J.; Vader, G.; Shinohara, M.; Sanchez, A.; Borde, V.; Shinohara, A.; Hochwagen, A. Chromosome Synapsis Alleviates Mek1-Dependent Suppression of Meiotic DNA Repair. PLoS Biol. 2016, 14, e1002369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso da Silva, R.; Vader, G. Getting there: Understanding the chromosomal recruitment of the AAA+ ATPase Pch2/TRIP13 during meiosis. Curr. Genet. 2021, 67, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Deshong, A.J.; Ye, A.L.; Lamelza, P.; Bhalla, N. A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genet. 2014, 10, e1004291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacopazzi, S.; Vong, D.; Devigne, A.; Bhalla, N. PCH-2 collaborates with CMT-1 to proofread meiotic homolog interactions. PLoS Genet. 2020, 16, e1008904. [Google Scholar] [CrossRef]
- Keeney, S.; Giroux, C.N.; Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 1997, 88, 375–384. [Google Scholar] [CrossRef] [Green Version]
- Panizza, S.; Mendoza, M.A.; Berlinger, M.; Huang, L.; Nicolas, A.; Shirahige, K.; Klein, F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 2011, 146, 372–383. [Google Scholar] [CrossRef] [Green Version]
- Tesse, S.; Bourbon, H.M.; Debuchy, R.; Budin, K.; Dubois, E.; Liangran, Z.; Antoine, R.; Piolot, T.; Kleckner, N.; Zickler, D.; et al. Asy2/Mer2: An evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 2017, 31, 1880–1893. [Google Scholar] [CrossRef] [Green Version]
- Stanzione, M.; Baumann, M.; Papanikos, F.; Dereli, I.; Lange, J.; Ramlal, A.; Trankner, D.; Shibuya, H.; de Massy, B.; Watanabe, Y.; et al. Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 2016, 18, 1208–1220. [Google Scholar] [CrossRef]
- Rousova, D.; Nivsarkar, V.; Altmannova, V.; Raina, V.B.; Funk, S.K.; Liedtke, D.; Janning, P.; Muller, F.; Reichle, H.; Vader, G.; et al. Novel mechanistic insights into the role of Mer2 as the keystone of meiotic DNA break formation. Elife 2021, 10, e72330. [Google Scholar] [CrossRef] [PubMed]
- Dernburg, A.F.; McDonald, K.; Moulder, G.; Barstead, R.; Dresser, M.; Villeneuve, A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 1998, 94, 387–398. [Google Scholar] [CrossRef] [Green Version]
- Castellano-Pozo, M.; Pacheco, S.; Sioutas, G.; Jaso-Tamame, A.L.; Dore, M.H.; Karimi, M.M.; Martinez-Perez, E. Surveillance of cohesin-supported chromosome structure controls meiotic progression. Nat. Commun. 2020, 11, 4345. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Wan, L.; Busygina, V.; Kwon, Y.; Allen, J.A.; Li, X.; Kunz, R.C.; Kubota, K.; Wang, B.; Sung, P.; et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell. 2009, 36, 393–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballo, J.A.; Johnson, A.L.; Sedgwick, S.G.; Cha, R.S. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 2008, 132, 758–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambing, C.; Kuo, P.C.; Tock, A.J.; Topp, S.D.; Henderson, I.R. ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 13647–13658. [Google Scholar] [CrossRef]
- Qiao, H.; Rao, H.; Yun, Y.; Sandhu, S.; Fong, J.H.; Sapre, M.; Nguyen, M.; Tham, A.; Van, B.W.; Chng, T.Y.H.; et al. Impeding DNA Break Repair Enables Oocyte Quality Control. Mol. Cell. 2018, 72, 211–221.e3. [Google Scholar] [CrossRef] [Green Version]
- Carofiglio, F.; Sleddens-Linkels, E.; Wassenaar, E.; Inagaki, A.; van Cappellen, W.A.; Grootegoed, J.A.; Toth, A.; Baarends, W.M. Repair of exogenous DNA double-strand breaks promotes chromosome synapsis in SPO11-mutant mouse meiocytes, and is altered in the absence of HORMAD1. DNA Repair 2018, 63, 25–38. [Google Scholar] [CrossRef]
- Rinaldi, V.D.; Bolcun-Filas, E.; Kogo, H.; Kurahashi, H.; Schimenti, J.C. The DNA Damage Checkpoint Eliminates Mouse Oocytes with Chromosome Synapsis Failure. Mol. Cell. 2017, 67, 1026–1036.e2. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.H.; McGuire, M.M.; Rajkovic, A. Mouse HORMAD1 is a meiosis i checkpoint protein that modulates DNA double- strand break repair during female meiosis. Biol. Reprod. 2013, 89, 29. [Google Scholar] [CrossRef]
- Couteau, F.; Nabeshima, K.; Villeneuve, A.; Zetka, M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004, 14, 585–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, S.; Wojcik, M.; Xu, K.; Dernburg, A.F. Superresolution microscopy reveals the three-dimensional organization of meiotic chromosome axes in intact Caenorhabditis elegans tissue. Proc. Natl. Acad. Sci. USA 2017, 114, E4734–E4743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeney, S.; Lange, J.; Mohibullah, N. Self-organization of meiotic recombination initiation: General principles and molecular pathways. Annu. Rev. Genet. 2014, 48, 187–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, V.V.; Hochwagen, A. The meiotic checkpoint network: Step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 2014, 6, a016675. [Google Scholar] [CrossRef] [PubMed]
- Prugar, E.; Burnett, C.; Chen, X.; Hollingsworth, N.M. Coordination of Double Strand Break Repair and Meiotic Progression in Yeast by a Mek1-Ndt80 Negative Feedback Loop. Genetics 2017, 206, 497–512. [Google Scholar] [CrossRef] [Green Version]
- Stamper, E.L.; Rodenbusch, S.E.; Rosu, S.; Ahringer, J.; Villeneuve, A.M.; Dernburg, A.F. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 2013, 9, e1003679. [Google Scholar] [CrossRef]
- Rosu, S.; Zawadzki, K.A.; Stamper, E.L.; Libuda, D.E.; Reese, A.L.; Dernburg, A.F.; Villeneuve, A.M. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 2013, 9, e1003674. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Kostow, N.; Dernburg, A.F. The Chromosome Axis Mediates Feedback Control of CHK-2 to Ensure Crossover Formation in C. elegans. Dev. Cell 2015, 35, 247–261. [Google Scholar] [CrossRef] [Green Version]
- Bohr, T.; Ashley, G.; Eggleston, E.; Firestone, K.; Bhalla, N. Synaptonemal Complex Components Are Required for Meiotic Checkpoint Function in Caenorhabditis elegans. Genetics 2016, 204, 987–997. [Google Scholar] [CrossRef] [Green Version]
- Silva, N.; Ferrandiz, N.; Barroso, C.; Tognetti, S.; Lightfoot, J.; Telecan, O.; Encheva, V.; Faull, P.; Hanni, S.; Furger, A.; et al. The fidelity of synaptonemal complex assembly is regulated by a signaling mechanism that controls early meiotic progression. Dev. Cell 2014, 31, 503–511. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Sun, H.; Ball, H.L.; Wassmann, K.; Luo, X.; Yu, H. Phosphorylation of the spindle checkpoint protein Mad2 regulates its conformational transition. Proc. Natl. Acad. Sci. USA 2010, 107, 19772–19777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Shinohara, M. Meiotic DSB-independent role of protein phosphatase 4 in Hop1 assembly to promote meiotic chromosome axis formation in budding yeast. bioRxiv 2022. [Google Scholar] [CrossRef]
- Das, D.; Chen, S.Y.; Arur, S. ERK phosphorylates chromosomal axis component HORMA domain protein HTP-1 to regulate oocyte numbers. Sci. Adv. 2020, 6, eabc5580. [Google Scholar] [CrossRef] [PubMed]
- Sato-Carlton, A.; Nakamura-Tabuchi, C.; Li, X.; Boog, H.; Lehmer, M.K.; Rosenberg, S.C.; Barroso, C.; Martinez-Perez, E.; Corbett, K.D.; Carlton, P.M. Phosphoregulation of HORMA domain protein HIM-3 promotes asymmetric synaptonemal complex disassembly in meiotic prophase in Caenorhabditis elegans. PLoS Genet. 2020, 16, e1008968. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Pratto, F.; Schimenti, J.C.; Turner, J.M.; Camerini-Otero, R.D.; Hoog, C. Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis. PLoS Genet. 2012, 8, e1002485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogo, H.; Tsutsumi, M.; Ohye, T.; Inagaki, H.; Abe, T.; Kurahashi, H. HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes Cells 2012, 17, 439–454. [Google Scholar] [CrossRef]
- Bhagwat, N.R.; Owens, S.N.; Ito, M.; Boinapalli, J.V.; Poa, P.; Ditzel, A.; Kopparapu, S.; Mahalawat, M.; Davies, O.R.; Collins, S.R.; et al. SUMO is a pervasive regulator of meiosis. Elife 2021, 10, e57720. [Google Scholar] [CrossRef]
- Sironi, L.; Mapelli, M.; Knapp, S.; De Antoni, A.; Jeang, K.T.; Musacchio, A. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002, 21, 2496–2506. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Li, B.; Liu, C.-J.; Tomchick, D.R.; Machius, M.; Rizo, J.; Yu, H.; Luo, X. Insights into Mad2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric Mad2 Dimer. PLoS Biol. 2008, 6, e50. [Google Scholar] [CrossRef]
- Yang, M.; Li, B.; Tomchick, D.R.; Machius, M.; Rizo, J.; Yu, H.; Luo, X. p31comet blocks Mad2 activation through structural mimicry. Cell 2007, 131, 744–755. [Google Scholar] [CrossRef] [Green Version]
- Orth, M.; Mayer, B.; Rehm, K.; Rothweiler, U.; Heidmann, D.; Holak, T.A.; Stemmann, O. Shugoshin is a Mad1/Cdc20-like interactor of Mad2. EMBO J. 2011, 30, 2868–2880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanafusa, T.; Habu, T.; Tomida, J.; Ohashi, E.; Murakumo, Y.; Ohmori, H. Overlapping in short motif sequences for binding to human REV7 and MAD2 proteins. Genes Cells 2010, 15, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Tipton, A.R.; Wang, K.; Link, L.; Bellizzi, J.J.; Huang, H.; Yen, T.; Liu, S.-T. BUBR1 and Closed MAD2 (C-MAD2) Interact Directly to Assemble a Functional Mitotic Checkpoint Complex. J. Biol. Chem. 2011, 286, 21173–21179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuevas-Navarro, A.; Van, R.; Cheng, A.; Urisman, A.; Castel, P.; McCormick, F. The RAS GTPase RIT1 compromises mitotic fidelity through spindle assembly checkpoint suppression. Curr. Biol. 2021, 31, 3915–3924.e3919. [Google Scholar] [CrossRef]
- Shandilya, J.; Toska, E.; Richard, D.J.; Medler, K.F.; Roberts, S.G.E. WT1 interacts with MAD2 and regulates mitotic checkpoint function. Nat. Commun. 2014, 5, 4903. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Zhang, F.; Wang, L.; Shan, S.; Gong, Z.; Zhou, Z. Structural basis for shieldin complex subunit 3–mediated recruitment of the checkpoint protein REV7 during DNA double-strand break repair. J. Biol. Chem. 2020, 295, 250–262. [Google Scholar] [CrossRef]
- Medendorp, K.; Vreede, L.; van Groningen, J.J.M.; Hetterschijt, L.; Brugmans, L.; Jansen, P.A.M.; van den Hurk, W.H.; de Bruijn, D.R.H.; van Kessel, A.G. The Mitotic Arrest Deficient Protein MAD2B Interacts with the Clathrin Light Chain A during Mitosis. PLoS ONE 2010, 5, e15128. [Google Scholar] [CrossRef]
- Wang, X.; Pernicone, N.; Pertz, L.; Hua, D.; Zhang, T.; Listovsky, T.; Xie, W. REV7 has a dynamic adaptor region to accommodate small GTPase RAN/Shigella IpaB ligands, and its activity is regulated by the RanGTP/GDP switch. J. Biol. Chem. 2019, 294, 15733–15742. [Google Scholar] [CrossRef]
- Li, L.; Shi, Y.; Wu, H.; Wan, B.; Li, P.; Zhou, L.; Shi, H.; Huo, K. Hepatocellular carcinoma-associated gene 2 interacts with MAD2L2. Mol. Cell. Biochem. 2007, 304, 297–304. [Google Scholar] [CrossRef]
- Hara, K.; Hashimoto, H.; Murakumo, Y.; Kobayashi, S.; Kogame, T.; Unzai, S.; Akashi, S.; Takeda, S.; Shimizu, T.; Sato, M. Crystal Structure of Human REV7 in Complex with a Human REV3 Fragment and Structural Implication of the Interaction between DNA Polymerase ζ and REV1. J. Biol. Chem. 2010, 285, 12299–12307. [Google Scholar] [CrossRef] [Green Version]
- Weterman, M.A.J.; van Groningen, J.J.M.; Tertoolen, L.; van Kessel, A.G. Impairment of MAD2B–PRCC interaction in mitotic checkpoint defective t(X;1)-positive renal cell carcinomas. Proc. Natl. Acad. Sci. USA 2001, 98, 13808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Tian, X.; Wang, X.; Gao, P.; Zhang, C. A novel binding protein of single-minded 2: The mitotic arrest-deficient protein MAD2B. Neurogenetics 2012, 13, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Taharazako, S.; Ikeda, M.; Fujita, H.; Mikami, Y.; Kikuchi, S.; Hishiki, A.; Yokoyama, H.; Ishikawa, Y.; Kanno, S.-I.; et al. Dynamic feature of mitotic arrest deficient 2–like protein 2 (MAD2L2) and structural basis for its interaction with chromosome alignment–maintaining phosphoprotein (CAMP). J. Biol. Chem. 2017, 292, 17658–17667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, C.-F.; Chou, Y.-T.; Lin, Y.-S.; Wu, C.-W. MAD2B, a Novel TCF4-binding Protein, Modulates TCF4-mediated Epithelial-Mesenchymal Transdifferentiation. J. Biol. Chem. 2009, 284, 19613–19622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, B.; Wold, W.S.M. Adenovirus ADP protein (E3-11.6K), which is required for efficient cell lysis and virus release, interacts with human MAD2B. Virology 2003, 313, 224–234. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yang, S.-H.; Sharrocks, A.D. Rev7/MAD2B links c-Jun N-terminal protein kinase pathway signaling to activation of the transcription factor Elk-1. Mol. Cell. Biol. 2007, 27, 2861–2869. [Google Scholar] [CrossRef] [Green Version]
- Pernicone, N.; Grinshpon, S.; Listovsky, T. CDH1 binds MAD2L2 in a Rev1-like pattern. Biochem. Biophys. Res. Commun. 2020, 531, 566–572. [Google Scholar] [CrossRef]
- Fattah, F.J.; Hara, K.; Fattah, K.R.; Yang, C.; Wu, N.; Warrington, R.; Chen, D.J.; Zhou, P.; Boothman, D.A.; Yu, H. The transcription factor TFII-I promotes DNA translesion synthesis and genomic stability. PLoS Genet. 2014, 10, e1004419. [Google Scholar] [CrossRef]
- Xie, C.; He, C.; Jiang, Y.; Yu, H.; Cheng, L.; Nshogoza, G.; Ala, M.S.; Tian, C.; Wu, J.; Shi, Y.; et al. Structural insights into the recognition of phosphorylated Hop1 by Mek1. Acta Crystallogr. D Struct. Biol. 2018, 74, 1027–1038. [Google Scholar] [CrossRef]
- Liu, K.; Wang, Y.; Zhu, Q.; Li, P.; Chen, J.; Tang, Z.; Shen, Y.; Cheng, X.; Lu, L.-Y.; Liu, Y. Aberrantly expressed HORMAD1 disrupts nuclear localization of MCM8–MCM9 complex and compromises DNA mismatch repair in cancer cells. Cell Death Dis. 2020, 11, 519. [Google Scholar] [CrossRef]
- Caryl, A.P.; Armstrong, S.J.; Jones, G.H.; Franklin, F.C.H. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 2000, 109, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, M.; Tang, D.; Shen, Y.; Qin, B.; Li, M.; Cheng, Z. PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Mol. Biol. Cell 2011, 22, 12–19. [Google Scholar] [CrossRef] [PubMed]


| S. cerevisiae | Homo sapiens/ Mus musculus | C. elegans | Arabidopsis thaliana | |
|---|---|---|---|---|
| HORMADs | Hop1 | HORMAD1 | HTP-1 | ASY1 |
| HORMAD2 | HTP-2 | ASY2 | ||
| HTP-3 | ||||
| HIM-3 | ||||
| HORMAD interactor on axis | Red1 | SYCP2 | - | ASY3 |
| Cohesin | Cohesin | |||
| HORMAD regulation | Pch2 | TRIP13 | PCH-2 | PCH2 |
| p31COMET | CMT-1 | COMET |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prince, J.P.; Martinez-Perez, E. Functions and Regulation of Meiotic HORMA-Domain Proteins. Genes 2022, 13, 777. https://doi.org/10.3390/genes13050777
Prince JP, Martinez-Perez E. Functions and Regulation of Meiotic HORMA-Domain Proteins. Genes. 2022; 13(5):777. https://doi.org/10.3390/genes13050777
Chicago/Turabian StylePrince, Josh P., and Enrique Martinez-Perez. 2022. "Functions and Regulation of Meiotic HORMA-Domain Proteins" Genes 13, no. 5: 777. https://doi.org/10.3390/genes13050777
APA StylePrince, J. P., & Martinez-Perez, E. (2022). Functions and Regulation of Meiotic HORMA-Domain Proteins. Genes, 13(5), 777. https://doi.org/10.3390/genes13050777






