A Comprehensive Review of Indel Detection Methods for Identification of Zebrafish Knockout Mutants Generated by Genome-Editing Nucleases
Abstract
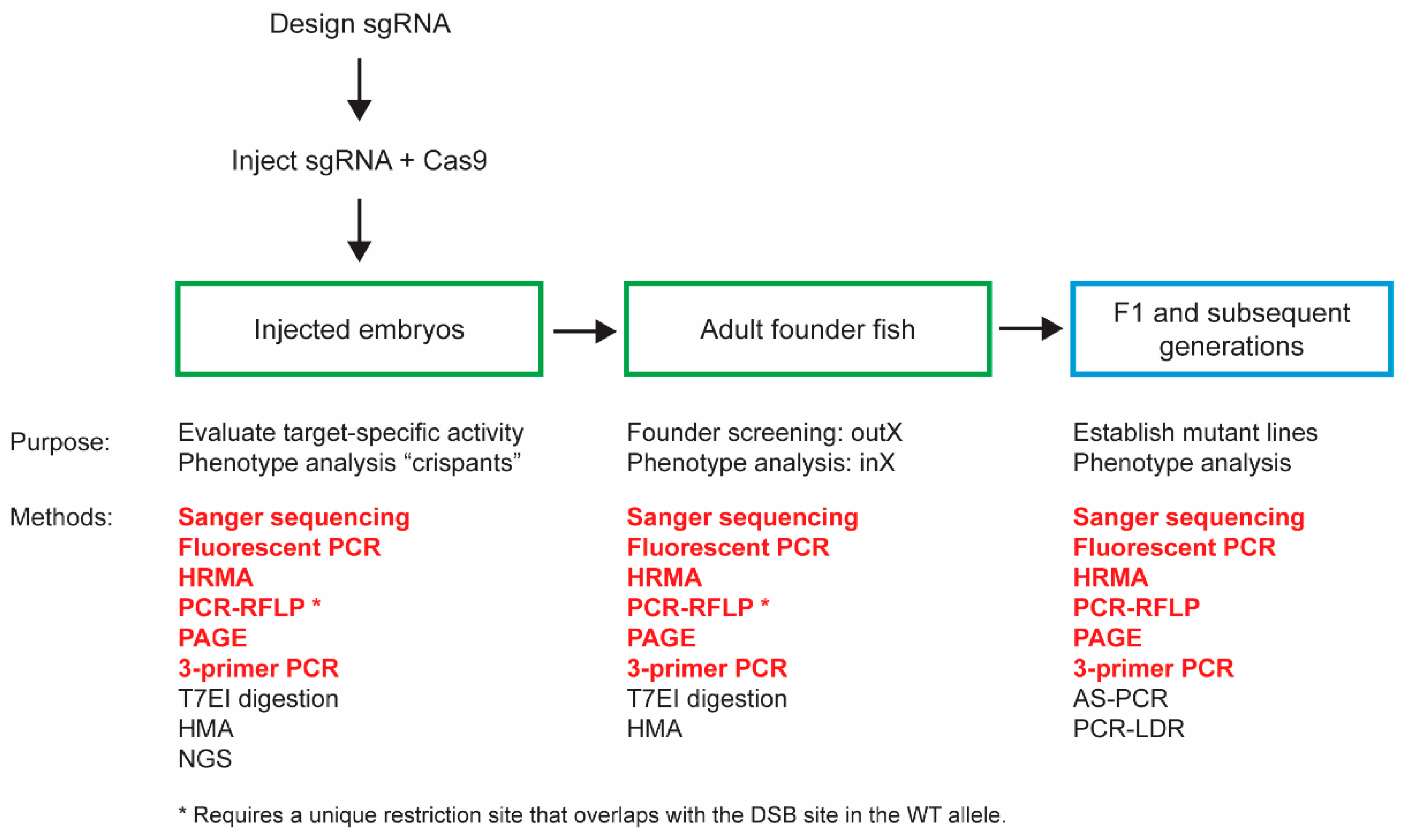
:1. Introduction
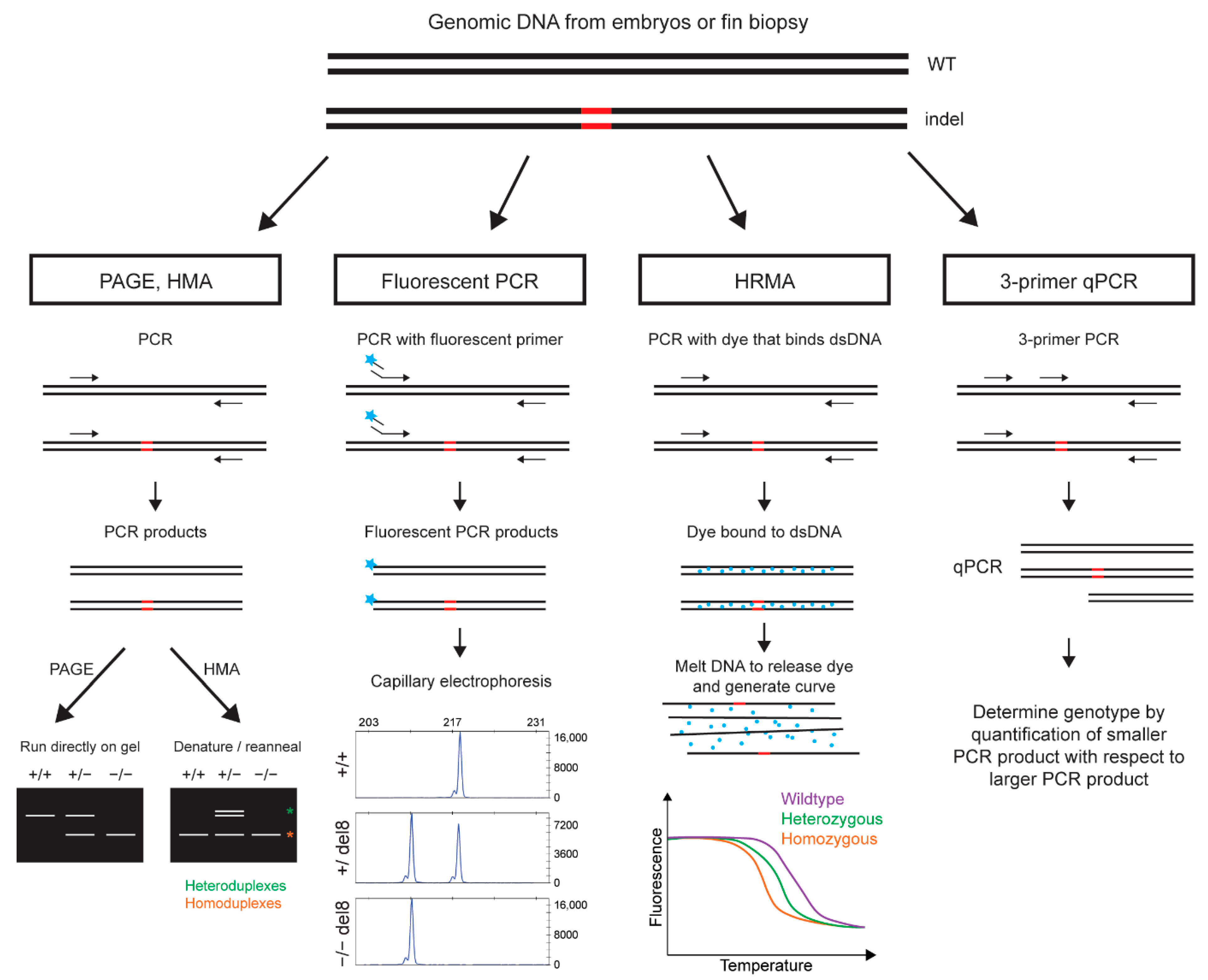
2. Methods Based on Direct Analysis of PCR Products
2.1. Indel Detection by Polyacrylamide Gel Electrophoresis (PAGE) of PCR Products
2.2. Indel Detection by Heteroduplex Mobility Assay (HMA)
2.3. Indel Detection by Fluorescent PCR and Capillary Electrophoresis
2.4. Indel Detection by High Resolution Melting Analysis (HRMA)
2.5. Indel Detection by 3-Primer Quantitative (q)PCR
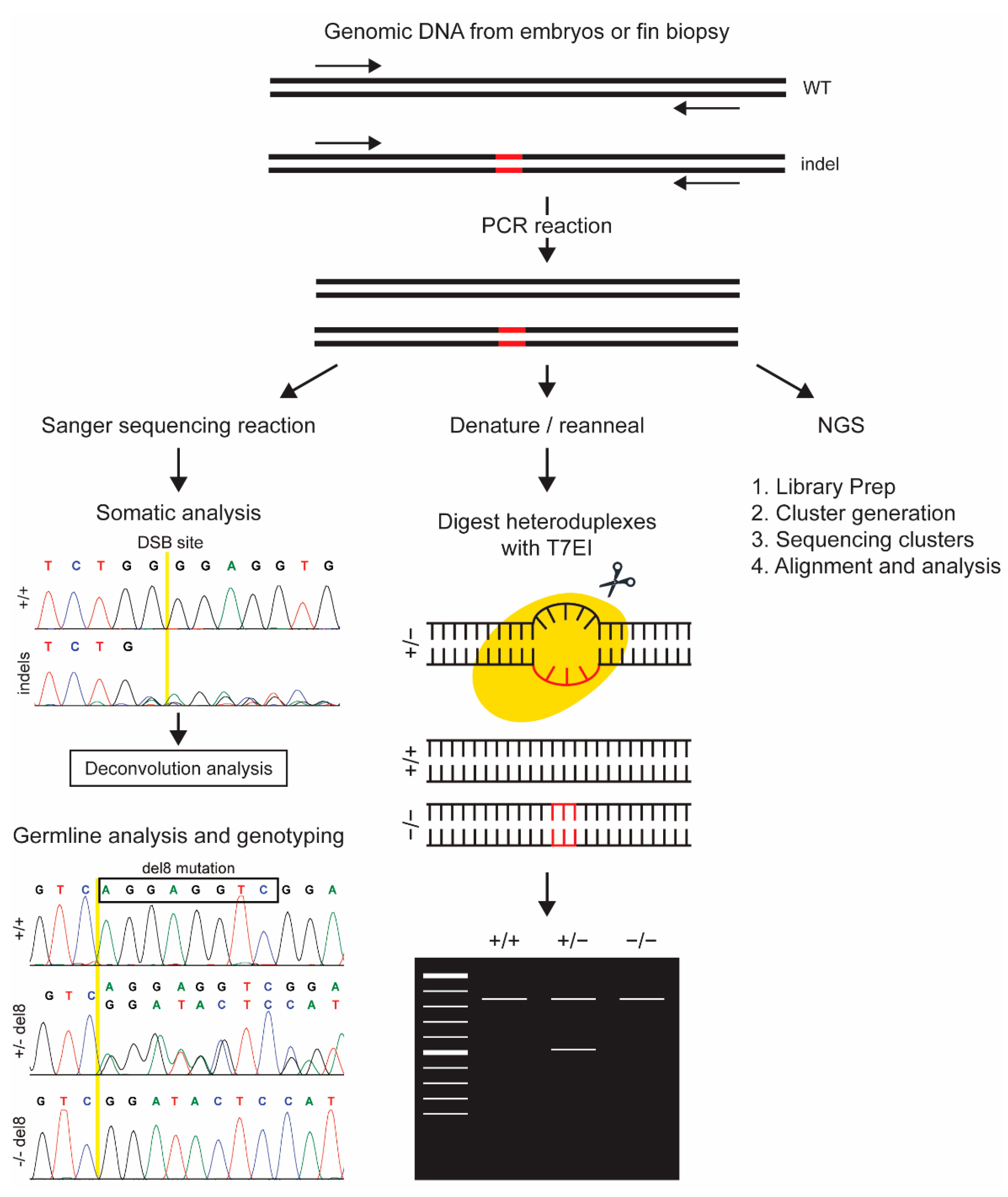
3. Methods Based on Analysis of Post Processing PCR Products
3.1. Indel Detection by Sanger Sequencing
3.1.1. Somatic Analysis by Deconvolution of Sanger Sequence Reads
3.1.2. Sanger Sequencing for Germline Indel Detection and Genotyping
3.2. Indel Detection by T7 Endonuclease I (T7EI) Digestion Assay
3.3. Next Generation Sequencing (NGS)
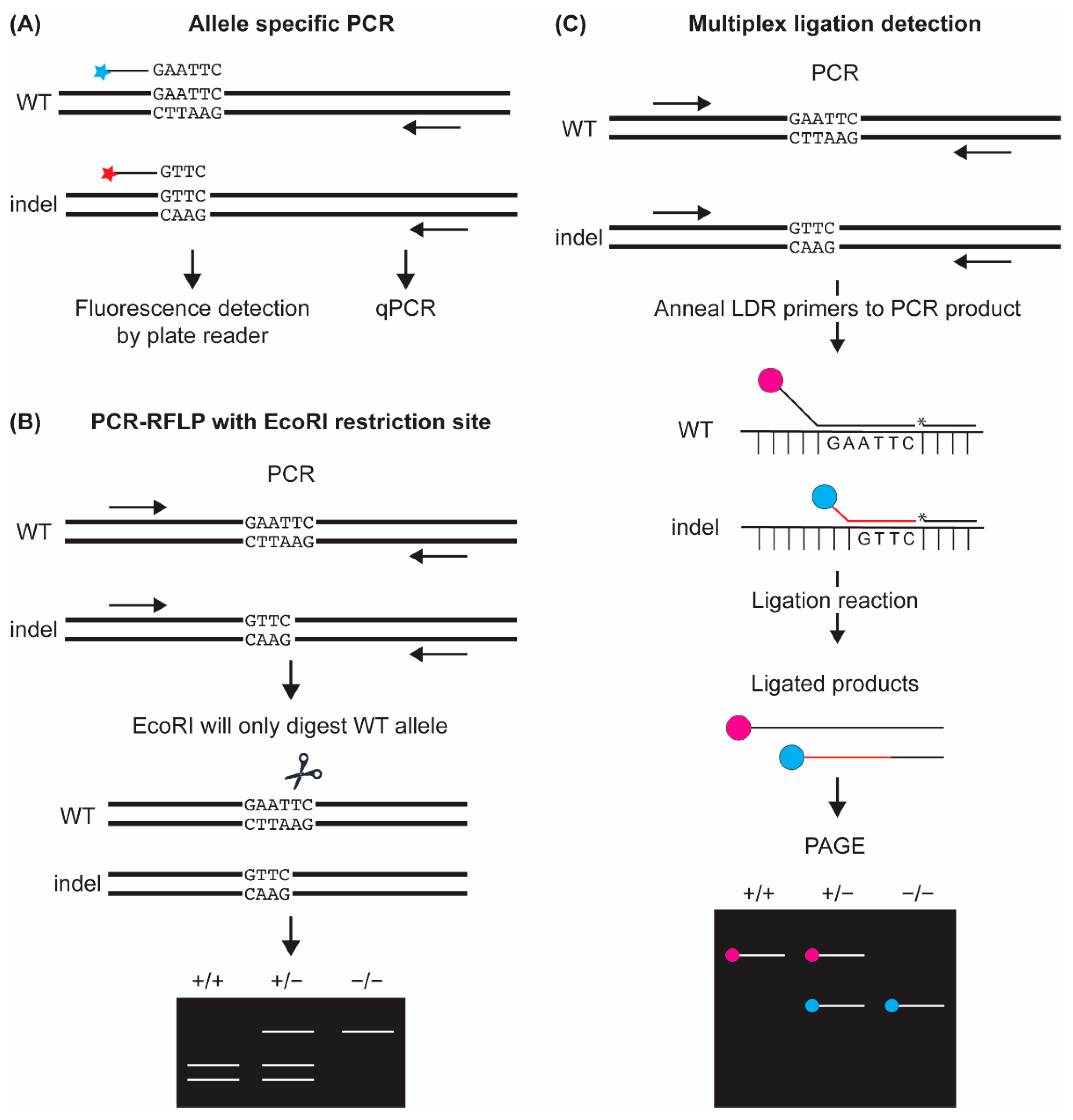
4. Indel Detection Methods That Require the Mutant Allele to Be Known
4.1. Indel Detection by Allele-Specific PCR (AS-PCR) Based Assays
4.2. Indel Detection by PCR-Restriction Fragment Length Polymorphism (PCR-RFLP) Assay
4.3. Indel Detection by Multiplex Ligation Detection Assay
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Amsterdam, A.; Burgess, S.; Golling, G.; Chen, W.; Sun, Z.; Townsend, K.; Farrington, S.; Haldi, M.; Hopkins, N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999, 13, 2713–2724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaiano, N.; Amsterdam, A.; Kawakami, K.; Allende, M.; Becker, T.; Hopkins, N. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature 1996, 383, 829–832. [Google Scholar] [CrossRef]
- Kettleborough, R.N.; Busch-Nentwich, E.M.; Harvey, S.A.; Dooley, C.M.; de Bruijn, E.; van Eeden, F.; Sealy, I.; White, R.J.; Herd, C.; Nijman, I.J.; et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 2013, 496, 494–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, N.D.; Wolfe, S.A. Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev. Cell 2011, 21, 48–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sood, R.; English, M.A.; Jones, M.; Mullikin, J.; Wang, D.M.; Anderson, M.; Wu, D.; Chandrasekharappa, S.C.; Yu, J.; Zhang, J.; et al. Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods 2006, 39, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Wienholds, E.; Schulte-Merker, S.; Walderich, B.; Plasterk, R.H. Target-selected inactivation of the zebrafish rag1 gene. Science 2002, 297, 99–102. [Google Scholar] [CrossRef]
- Wienholds, E.; van Eeden, F.; Kosters, M.; Mudde, J.; Plasterk, R.H.; Cuppen, E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003, 13, 2700–2707. [Google Scholar] [CrossRef] [Green Version]
- Varshney, G.K.; Burgess, S.M. Mutagenesis and phenotyping resources in zebrafish for studying development and human disease. Brief. Funct. Genom. 2014, 13, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Varshney, G.K.; Lu, J.; Gildea, D.E.; Huang, H.; Pei, W.; Yang, Z.; Huang, S.C.; Schoenfeld, D.; Pho, N.H.; Casero, D.; et al. A large-scale zebrafish gene knockout resource for the genome-wide study of gene function. Genome Res. 2013, 23, 727–735. [Google Scholar] [CrossRef] [Green Version]
- Varshney, G.K.; Sood, R.; Burgess, S.M. Understanding and Editing the Zebrafish Genome. Adv. Genet. 2015, 92, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Pei, W.; LaFave, M.C.; Idol, J.; Xu, L.; Gallardo, V.; Carrington, B.; Bishop, K.; Jones, M.; Li, M.; et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015, 25, 1030–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, B.; Zhang, Y.; Zhou, Y.; Zhang, B.; Krueger, C.J.; Bi, X.; Zhu, Z.; Tong, X.; Zhang, B. ErCas12a and T5exo-ErCas12a Mediate Simple and Efficient Genome Editing in Zebrafish. Biology 2022, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Luk, K.; Shin, M.; Idrizi, F.; Kwok, S.; Roscoe, B.; Mintzer, E.; Suresh, S.; Morrison, K.; Frazao, J.B.; et al. Enhanced Cas12a editing in mammalian cells and zebrafish. Nucleic Acids Res. 2019, 47, 4169–4180. [Google Scholar] [CrossRef] [Green Version]
- Meshalkina, D.A.; Glushchenko, A.S.; Kysil, E.V.; Mizgirev, I.V.; Frolov, A. SpCas9- and LbCas12a-Mediated DNA Editing Produce Different Gene Knockout Outcomes in Zebrafish Embryos. Genes 2020, 11, 740. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Fernandez, J.P.; Rouet, R.; Vejnar, C.E.; Lane, M.A.; Mis, E.; Khokha, M.K.; Doudna, J.A.; Giraldez, A.J. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 2017, 8, 2024. [Google Scholar] [CrossRef] [Green Version]
- Wierson, W.A.; Simone, B.W.; WareJoncas, Z.; Mann, C.; Welker, J.M.; Kar, B.; Emch, M.J.; Friedberg, I.; Gendron, W.A.C.; Barry, M.A.; et al. Expanding the CRISPR Toolbox with ErCas12a in Zebrafish and Human Cells. CRISPR J. 2019, 2, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Akhmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Ramanagoudr-Bhojappa, R.; Carrington, B.; Ramaswami, M.; Bishop, K.; Robbins, G.M.; Jones, M.; Harper, U.; Frederickson, S.C.; Kimble, D.C.; Sood, R.; et al. Multiplexed CRISPR/Cas9-mediated knockout of 19 Fanconi anemia pathway genes in zebrafish revealed their roles in growth, sexual development and fertility. PLoS Genet. 2018, 14, e1007821. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Davey, C.F.; Whitebirch, A.C.; Miller, A.C.; Moens, C.B. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods 2015, 12, 535–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, U.; Nakhro, K.; Oh, C.K.; Carrington, B.; Song, H.; Varshney, G.K.; Kim, Y.; Song, H.; Jeon, S.; Robbins, G.; et al. Large-scale generation and phenotypic characterization of zebrafish CRISPR mutants of DNA repair genes. DNA Repair 2021, 107, 103173. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Carrington, B.; Pei, W.; Bishop, K.; Chen, Z.; Fan, C.; Xu, L.; Jones, M.; LaFave, M.C.; Ledin, J.; et al. A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 2016, 11, 2357–2375. [Google Scholar] [CrossRef] [PubMed]
- Vejnar, C.E.; Moreno-Mateos, M.A.; Cifuentes, D.; Bazzini, A.A.; Giraldez, A.J. Optimized CRISPR-Cas9 System for Genome Editing in Zebrafish. Cold Spring Harb Protoc. 2016, 2016. [Google Scholar] [CrossRef]
- Ata, H.; Clark, K.J.; Ekker, S.C. The zebrafish genome editing toolkit. Methods Cell Biol 2016, 135, 149–170. [Google Scholar] [CrossRef]
- Pei, W.; Xu, L.; Huang, S.C.; Pettie, K.; Idol, J.; Rissone, A.; Jimenez, E.; Sinclair, J.W.; Slevin, C.; Varshney, G.K.; et al. Guided genetic screen to identify genes essential in the regeneration of hair cells and other tissues. NPJ Regen. Med. 2018, 3, 11. [Google Scholar] [CrossRef]
- Rafferty, S.A.; Quinn, T.A. A beginner’s guide to understanding and implementing the genetic modification of zebrafish. Prog. Biophys. Mol. Biol. 2018, 138, 3–19. [Google Scholar] [CrossRef]
- Sertori, R.; Trengove, M.; Basheer, F.; Ward, A.C.; Liongue, C. Genome editing in zebrafish: A practical overview. Brief. Funct. Genom. 2016, 15, 322–330. [Google Scholar] [CrossRef]
- Buglo, E.; Sarmiento, E.; Martuscelli, N.B.; Sant, D.W.; Danzi, M.C.; Abrams, A.J.; Dallman, J.E.; Zuchner, S. Genetic compensation in a stable slc25a46 mutant zebrafish: A case for using F0 CRISPR mutagenesis to study phenotypes caused by inherited disease. PLoS ONE 2020, 15, e0230566. [Google Scholar] [CrossRef] [Green Version]
- Burger, A.; Lindsay, H.; Felker, A.; Hess, C.; Anders, C.; Chiavacci, E.; Zaugg, J.; Weber, L.M.; Catena, R.; Jinek, M.; et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 2016, 143, 2025–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshijima, K.; Jurynec, M.J.; Klatt Shaw, D.; Jacobi, A.M.; Behlke, M.A.; Grunwald, D.J. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev. Cell 2019, 51, 645–657.e4. [Google Scholar] [CrossRef] [PubMed]
- Kague, E.; Turci, F.; Newman, E.; Yang, Y.; Brown, K.R.; Aglan, M.S.; Otaify, G.A.; Temtamy, S.A.; Ruiz-Perez, V.L.; Cross, S.; et al. 3D assessment of intervertebral disc degeneration in zebrafish identifies changes in bone density that prime disc disease. Bone Res. 2021, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Klatt Shaw, D.; Mokalled, M.H. Efficient CRISPR/Cas9 mutagenesis for neurobehavioral screening in adult zebrafish. G3 2021, 11, jkab089. [Google Scholar] [CrossRef]
- Shah, A.N.; Moens, C.B.; Miller, A.C. Targeted candidate gene screens using CRISPR/Cas9 technology. Methods Cell Biol. 2016, 135, 89–106. [Google Scholar] [CrossRef]
- Wu, R.S.; Lam, I.I.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell 2018, 46, 112–125.e4. [Google Scholar] [CrossRef] [Green Version]
- Meeker, N.D.; Hutchinson, S.A.; Ho, L.; Trede, N.S. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 2007, 43, 610, 612, 614. [Google Scholar] [CrossRef]
- Sood, R.; Carrington, B.; Bishop, K.; Jones, M.; Rissone, A.; Candotti, F.; Chandrasekharappa, S.C.; Liu, P. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: Data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS ONE 2013, 8, e57239. [Google Scholar] [CrossRef]
- VanLeuven, A.J.; Park, S.; Menke, D.B.; Lauderdale, J.D. A PAGE screening approach for identifying CRISPR-Cas9-induced mutations in zebrafish. Biotechniques 2018, 64, 275–278. [Google Scholar] [CrossRef] [Green Version]
- Ota, S.; Hisano, Y.; Muraki, M.; Hoshijima, K.; Dahlem, T.J.; Grunwald, D.J.; Okada, Y.; Kawahara, A. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells 2013, 18, 450–458. [Google Scholar] [CrossRef]
- Prykhozhij, S.V.; Steele, S.L.; Razaghi, B.; Berman, J.N. A rapid and effective method for screening, sequencing and reporter verification of engineered frameshift mutations in zebrafish. Dis. Models Mech. 2017, 10, 811–822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, X.; Wang, T.; Li, Z.; Guan, G.; Hong, Y. Efficient detection, quantification and enrichment of subtle allelic alterations. DNA Res. 2012, 19, 423–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, S.D.; Glover, S.R.; Turner, A.N.; Chatti, K.; Challa, A.K. A mixing heteroduplex mobility assay (mHMA) to genotype homozygous mutants with small indels generated by CRISPR-Cas9 nucleases. MethodsX 2019, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kakui, H.; Yamazaki, M.; Shimizu, K.K. PRIMA: A rapid and cost-effective genotyping method to detect single-nucleotide differences using probe-induced heteroduplexes. Sci. Rep. 2021, 11, 20741. [Google Scholar] [CrossRef]
- Brownstein, M.J.; Carpten, J.D.; Smith, J.R. Modulation of non-templated nucleotide addition by Taq DNA polymerase: Primer modifications that facilitate genotyping. Biotechniques 1996, 20, 1004–1006, 1008–1010. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Steentoft, C.; Hauge, C.; Hansen, L.; Thomsen, A.L.; Niola, F.; Vester-Christensen, M.B.; Frodin, M.; Clausen, H.; Wandall, H.H.; et al. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 2015, 43, e59. [Google Scholar] [CrossRef] [Green Version]
- Ramlee, M.K.; Yan, T.; Cheung, A.M.; Chuah, C.T.; Li, S. High-throughput genotyping of CRISPR/Cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci. Rep. 2015, 5, 15587. [Google Scholar] [CrossRef]
- McCafferty, J.; Reid, R.; Spencer, M.; Hamp, T.; Fodor, A. Peak Studio: A tool for the visualization and analysis of fragment analysis files. Environ. Microbiol. Rep. 2012, 4, 556–561. [Google Scholar] [CrossRef]
- Bresciani, E.; Carrington, B.; Wincovitch, S.; Jones, M.; Gore, A.V.; Weinstein, B.M.; Sood, R.; Liu, P.P. CBFbeta and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood 2014, 124, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Burke, E.A.; Sturgeon, M.; Zastrow, D.B.; Fernandez, L.; Prybol, C.; Marwaha, S.; Frothingham, E.P.; Ward, P.A.; Eng, C.M.; Fresard, L.; et al. Compound heterozygous KCTD7 variants in progressive myoclonus epilepsy. J. Neurogenet. 2021, 35, 74–83. [Google Scholar] [CrossRef]
- Hong, S.; Hu, P.; Jang, J.H.; Carrington, B.; Sood, R.; Berger, S.I.; Roessler, E.; Muenke, M. Functional analysis of Sonic Hedgehog variants associated with holoprosencephaly in humans using a CRISPR/Cas9 zebrafish model. Hum. Mutat. 2020, 41, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- McElderry, J.; Carrington, B.; Bishop, K.; Kim, E.; Pei, W.; Chen, Z.; Ramanagoudr-Bhojappa, R.; Prakash, A.; Burgess, S.M.; Liu, P.P.; et al. Splicing factor DHX15 affects tp53 and mdm2 expression via alternate splicing and promoter usage. Hum. Mol. Genet. 2019, 28, 4173–4185. [Google Scholar] [CrossRef] [PubMed]
- Nagai-Tanima, M.; Hong, S.; Hu, P.; Carrington, B.; Sood, R.; Roessler, E.; Muenke, M. Rare hypomorphic human variation in the heptahelical domain of SMO contributes to holoprosencephaly phenotypes. Hum. Mutat. 2020, 41, 2105–2118. [Google Scholar] [CrossRef] [PubMed]
- Pei, W.; Xu, L.; Varshney, G.K.; Carrington, B.; Bishop, K.; Jones, M.; Huang, S.C.; Idol, J.; Pretorius, P.R.; Beirl, A.; et al. Additive reductions in zebrafish PRPS1 activity result in a spectrum of deficiencies modeling several human PRPS1-associated diseases. Sci. Rep. 2016, 6, 29946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillay, L.M.; Yano, J.J.; Davis, A.E.; Butler, M.G.; Ezeude, M.O.; Park, J.S.; Barnes, K.A.; Reyes, V.L.; Castranova, D.; Gore, A.V.; et al. In vivo dissection of Rhoa function in vascular development using zebrafish. Angiogenesis 2022. [Google Scholar] [CrossRef]
- Rissone, A.; Weinacht, K.G.; la Marca, G.; Bishop, K.; Giocaliere, E.; Jagadeesh, J.; Felgentreff, K.; Dobbs, K.; Al-Herz, W.; Jones, M.; et al. Reticular dysgenesis-associated AK2 protects hematopoietic stem and progenitor cell development from oxidative stress. J. Exp. Med. 2015, 212, 1185–1202. [Google Scholar] [CrossRef] [Green Version]
- Sloan, J.L.; Achilly, N.P.; Arnold, M.L.; Catlett, J.L.; Blake, T.; Bishop, K.; Jones, M.; Harper, U.; English, M.A.; Anderson, S.; et al. The vitamin B12 processing enzyme, mmachc, is essential for zebrafish survival, growth and retinal morphology. Hum. Mol. Genet. 2020, 29, 2109–2123. [Google Scholar] [CrossRef]
- Unal Eroglu, A.; Mulligan, T.S.; Zhang, L.; White, D.T.; Sengupta, S.; Nie, C.; Lu, N.Y.; Qian, J.; Xu, L.; Pei, W.; et al. Multiplexed CRISPR/Cas9 Targeting of Genes Implicated in Retinal Regeneration and Degeneration. Front. Cell Dev. Biol. 2018, 6, 88. [Google Scholar] [CrossRef] [Green Version]
- Bresciani, E.; Carrington, B.; Yu, K.; Kim, E.M.; Zhen, T.; Guzman, V.S.; Broadbridge, E.; Bishop, K.; Kirby, M.; Harper, U.; et al. Redundant mechanisms driven independently by RUNX1 and GATA2 for hematopoietic development. Blood Adv. 2021, 5, 4949–4962. [Google Scholar] [CrossRef]
- Han, C.R.; Holmsen, E.; Carrington, B.; Bishop, K.; Zhu, Y.J.; Starost, M.; Meltzer, P.; Sood, R.; Liu, P.; Cheng, S.Y. Generation of Novel Genetic Models to Dissect Resistance to Thyroid Hormone Receptor alpha in Zebrafish. Thyroid 2020, 30, 314–328. [Google Scholar] [CrossRef]
- Trivellin, G.; Tirosh, A.; Hernandez-Ramirez, L.C.; Gupta, T.; Tsai-Morris, C.H.; Faucz, F.R.; Burgess, H.A.; Feldman, B.; Stratakis, C.A. The X-linked acrogigantism-associated gene gpr101 is a regulator of early embryonic development and growth in zebrafish. Mol. Cell. Endocrinol. 2021, 520, 111091. [Google Scholar] [CrossRef] [PubMed]
- Golenberg, N.; Squirrell, J.M.; Bennin, D.A.; Rindy, J.; Pistono, P.E.; Eliceiri, K.W.; Shelef, M.A.; Kang, J.; Huttenlocher, A. Citrullination regulates wound responses and tissue regeneration in zebrafish. J. Cell Biol. 2020, 219, e201908164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petree, C.; Varshney, G.K. MultiFRAGing: Rapid and Simultaneous Genotyping of Multiple Alleles in a Single Reaction. Sci. Rep. 2020, 10, 3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunt, L.H.; Begg, K.; Kague, E.; Cross, S.; Hammond, C.L. Wnt signalling controls the response to mechanical loading during zebrafish joint development. Development 2017, 144, 2798–2809. [Google Scholar] [CrossRef] [Green Version]
- Carrington, B.; Varshney, G.K.; Burgess, S.M.; Sood, R. CRISPR-STAT: An easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 2015, 43, e157. [Google Scholar] [CrossRef] [Green Version]
- DiNapoli, S.E.; Martinez-McFaline, R.; Gribbin, C.K.; Wrighton, P.J.; Balgobin, C.A.; Nelson, I.; Leonard, A.; Maskin, C.R.; Shwartz, A.; Quenzer, E.D.; et al. Synthetic CRISPR/Cas9 reagents facilitate genome editing and homology directed repair. Nucleic Acids Res. 2020, 48, e38. [Google Scholar] [CrossRef]
- Lopez-Cuevas, P.; Deane, L.; Yang, Y.; Hammond, C.L.; Kague, E. Transformed notochordal cells trigger chronic wounds destabilizing the vertebral column and bone homeostasis. Dis. Models Mech. 2021, 14, dmm047001. [Google Scholar] [CrossRef]
- Waldmann, L.; Leyhr, J.; Zhang, H.; Ohman-Magi, C.; Allalou, A.; Haitina, T. The broad role of Nkx3.2 in the development of the zebrafish axial skeleton. PLoS ONE 2021, 16, e0255953. [Google Scholar] [CrossRef]
- D’Agostino, Y.; Locascio, A.; Ristoratore, F.; Sordino, P.; Spagnuolo, A.; Borra, M.; D’Aniello, S. A Rapid and Cheap Methodology for CRISPR/Cas9 Zebrafish Mutant Screening. Mol. Biotechnol. 2016, 58, 73–78. [Google Scholar] [CrossRef] [Green Version]
- Dahlem, T.J.; Hoshijima, K.; Jurynec, M.J.; Gunther, D.; Starker, C.G.; Locke, A.S.; Weis, A.M.; Voytas, D.F.; Grunwald, D.J. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012, 8, e1002861. [Google Scholar] [CrossRef] [Green Version]
- Parant, J.M.; George, S.A.; Pryor, R.; Wittwer, C.T.; Yost, H.J. A rapid and efficient method of genotyping zebrafish mutants. Dev. Dyn. 2009, 238, 3168–3174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samarut, E.; Lissouba, A.; Drapeau, P. A simplified method for identifying early CRISPR-induced indels in zebrafish embryos using High Resolution Melting analysis. BMC Genomics 2016, 17, 547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, H.R.; Percival, S.M.; Yoder, B.K.; Parant, J.M. High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PLoS ONE 2014, 9, e114632. [Google Scholar] [CrossRef] [PubMed]
- Dwight, Z.; Palais, R.; Wittwer, C.T. uMELT: Prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics 2011, 27, 1019–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, E.S.; Vetsigian, K.H. DesignSignatures: A tool for designing primers that yields amplicons with distinct signatures. Bioinformatics 2016, 32, 1565–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaFave, M.C.; Varshney, G.K.; Vemulapalli, M.; Mullikin, J.C.; Burgess, S.M. A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics 2014, 198, 167–170. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Zhang, Y.; Yao, S.; Wei, Y. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS ONE 2014, 9, e98282. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef]
- Dehairs, J.; Talebi, A.; Cherifi, Y.; Swinnen, J.V. CRISP-ID: Decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep. 2016, 6, 28973. [Google Scholar] [CrossRef] [Green Version]
- Bloh, K.; Kanchana, R.; Bialk, P.; Banas, K.; Zhang, Z.; Yoo, B.C.; Kmiec, E.B. Deconvolution of Complex DNA Repair (DECODR): Establishing a Novel Deconvolution Algorithm for Comprehensive Analysis of CRISPR-Edited Sanger Sequencing Data. CRISPR J. 2021, 4, 120–131. [Google Scholar] [CrossRef]
- Conant, D.; Hsiau, T.; Rossi, N.; Oki, J.; Maures, T.; Waite, K.; Yang, J.; Joshi, S.; Kelso, R.; Holden, K.; et al. Inference of CRISPR Edits from Sanger Trace Data. CRISPR J. 2022, 5, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Su, Y.C.; Smith, M.; Yost, H.J. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 2014, 243, 1632–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jie, H.; Li, Z.; Wang, P.; Zhao, L.; Zhang, Q.; Yao, X.; Song, X.; Zhao, Y.; Yao, S. A simple method based on Sanger sequencing and MS Word wildcard searching to identify Cas9-induced frameshift mutations. Lab. Investig. 2017, 97, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.L.; Mulligan, T.S.; Shen, M.C.; Wang, H.; Scahill, C.M.; Tan, F.J.; Du, S.J.; Busch-Nentwich, E.M.; Farber, S.A. mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 2017, 13, e1007105. [Google Scholar] [CrossRef] [PubMed]
- de Vos, I.; Wong, A.S.W.; Taslim, J.; Ong, S.L.M.; Syder, N.C.; Goggi, J.L.; Carney, T.J.; van Steensel, M.A.M. The novel zebrafish model pretzel demonstrates a central role for SH3PXD2B in defective collagen remodelling and fibrosis in Frank-Ter Haar syndrome. Biol. Open 2020, 9, bio054270. [Google Scholar] [CrossRef] [PubMed]
- Facchinello, N.; Skobo, T.; Meneghetti, G.; Colletti, E.; Dinarello, A.; Tiso, N.; Costa, R.; Gioacchini, G.; Carnevali, O.; Argenton, F.; et al. nr3c1 null mutant zebrafish are viable and reveal DNA-binding-independent activities of the glucocorticoid receptor. Sci. Rep. 2017, 7, 4371. [Google Scholar] [CrossRef] [Green Version]
- Lou, B.; Boger, M.; Bennewitz, K.; Sticht, C.; Kopf, S.; Morgenstern, J.; Fleming, T.; Hell, R.; Yuan, Z.; Nawroth, P.P.; et al. Elevated 4-hydroxynonenal induces hyperglycaemia via Aldh3a1 loss in zebrafish and associates with diabetes progression in humans. Redox Biol. 2020, 37, 101723. [Google Scholar] [CrossRef]
- Wohlfart, D.P.; Lou, B.; Middel, C.S.; Morgenstern, J.; Fleming, T.; Sticht, C.; Hausser, I.; Hell, R.; Hammes, H.P.; Szendrodi, J.; et al. Accumulation of acetaldehyde in aldh2.1(-/-) zebrafish causes increased retinal angiogenesis and impaired glucose metabolism. Redox Biol. 2022, 50, 102249. [Google Scholar] [CrossRef]
- Hu, P.; Zhao, X.; Zhang, Q.; Li, W.; Zu, Y. Comparison of Various Nuclear Localization Signal-Fused Cas9 Proteins and Cas9 mRNA for Genome Editing in Zebrafish. G3 2018, 8, 823–831. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.H.; Zhang, G. Generating Stable Knockout Zebrafish Lines by Deleting Large Chromosomal Fragments Using Multiple gRNAs. G3 2020, 10, 1029–1037. [Google Scholar] [CrossRef] [Green Version]
- Sung, Y.H.; Kim, J.M.; Kim, H.T.; Lee, J.; Jeon, J.; Jin, Y.; Choi, J.H.; Ban, Y.H.; Ha, S.J.; Kim, C.H.; et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2014, 24, 125–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, P.; Shandilya, H.; D’Alessio, J.M.; O’Connor, K.; Durocher, J.; Gerard, G.F. Mutation detection using Surveyor nuclease. Biotechniques 2004, 36, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Brocal, I.; White, R.J.; Dooley, C.M.; Carruthers, S.N.; Clark, R.; Hall, A.; Busch-Nentwich, E.M.; Stemple, D.L.; Kettleborough, R.N. Efficient identification of CRISPR/Cas9-induced insertions/deletions by direct germline screening in zebrafish. BMC Genom. 2016, 17, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Mateos, M.A.; Vejnar, C.E.; Beaudoin, J.D.; Fernandez, J.P.; Mis, E.K.; Khokha, M.K.; Giraldez, A.J. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 2015, 12, 982–988. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.B.; Schwab, T.L.; Koleilat, A.; Ata, H.; Daby, C.L.; Cervera, R.L.; McNulty, M.S.; Bostwick, H.S.; Clark, K.J. Allele-Specific Quantitative PCR for Accurate, Rapid, and Cost-Effective Genotyping. Hum. Gene Ther. 2016, 27, 425–435. [Google Scholar] [CrossRef]
- Bando, H.; Gergics, P.; Bohnsack, B.L.; Toolan, K.P.; Richter, C.E.; Shavit, J.A.; Camper, S.A. Otx2b mutant zebrafish have pituitary, eye and mandible defects that model mammalian disease. Hum. Mol. Genet. 2020, 29, 1648–1657. [Google Scholar] [CrossRef]
- Dupret, B.; Volkel, P.; Vennin, C.; Toillon, R.A.; Le Bourhis, X.; Angrand, P.O. The histone lysine methyltransferase Ezh2 is required for maintenance of the intestine integrity and for caudal fin regeneration in zebrafish. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1079–1093. [Google Scholar] [CrossRef]
- Keatinge, M.; Tsarouchas, T.M.; Munir, T.; Porter, N.J.; Larraz, J.; Gianni, D.; Tsai, H.H.; Becker, C.G.; Lyons, D.A.; Becker, T. CRISPR gRNA phenotypic screening in zebrafish reveals pro-regenerative genes in spinal cord injury. PLoS Genet. 2021, 17, e1009515. [Google Scholar] [CrossRef]
- Trubiroha, A.; Gillotay, P.; Giusti, N.; Gacquer, D.; Libert, F.; Lefort, A.; Haerlingen, B.; De Deken, X.; Opitz, R.; Costagliola, S. A Rapid CRISPR/Cas-based Mutagenesis Assay in Zebrafish for Identification of Genes Involved in Thyroid Morphogenesis and Function. Sci. Rep. 2018, 8, 5647. [Google Scholar] [CrossRef]
- Chen, X.-K.; Yi, Z.-N.; Lau, J.J.-Y.; Ma, A.C.-H. Distinct roles of core autophagy-related genes (ATGs) in zebrafish definitive hematopoiesis. bioRxiv 2022. [Google Scholar] [CrossRef]
- Bedell, V.M.; Wang, Y.; Campbell, J.M.; Poshusta, T.L.; Starker, C.G.; Krug, R.G., 2nd; Tan, W.; Penheiter, S.G.; Ma, A.C.; Leung, A.Y.; et al. In vivo genome editing using a high-efficiency TALEN system. Nature 2012, 491, 114–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DuVal, M.G.; Allison, W.T. Photoreceptor Progenitors Depend Upon Coordination of gdf6a, thrbeta, and tbx2b to Generate Precise Populations of Cone Photoreceptor Subtypes. Investig. Ophthalmol. Vis. Sci. 2018, 59, 6089–6101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, A.C.; Lee, H.B.; Clark, K.J.; Ekker, S.C. High efficiency In Vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PLoS ONE 2013, 8, e65259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelinka, C.P.; Sotolongo-Lopez, M.; Fadool, J.M. Targeted disruption of the endogenous zebrafish rhodopsin locus as models of rapid rod photoreceptor degeneration. Mol. Vis. 2018, 24, 587–602. [Google Scholar] [PubMed]
- Kc, R.; Srivastava, A.; Wilkowski, J.M.; Richter, C.E.; Shavit, J.A.; Burke, D.T.; Bielas, S.L. Detection of nucleotide-specific CRISPR/Cas9 modified alleles using multiplex ligation detection. Sci. Rep. 2016, 6, 32048. [Google Scholar] [CrossRef] [PubMed]
| Method | Throughput | Specific Requirements | Limitations | |
|---|---|---|---|---|
| Equipment | Reagents * | |||
| Fluorescent PCR | High | • Capillary electrophoresis instrument | • Fluorescently labeled primers • Size standard (e.g., ROX400) | • Access to equipment |
| HRMA | High | • Lightscanner or qPCR instrument | • dsDNA binding dye | • Amplicon size • Access to equipment • Sensitive to SNPs in amplicon |
| 3-primer qPCR | High | • qPCR instrument | • qPCR reagents | • Access to equipment |
| NGS | High | • Illumina or Ion Torrent instrument | • Appropriate NGS kit | • Data analysis • Expensive • Access to equipment • Cannot be used for founder screening or genotyping |
| AS-PCR | High | • Plate reader or qPCR instrument | • Depends on chosen platform | • Cannot be used for somatic analysis or founder screening • Access to equipment |
| Sanger sequencing | Low/High ** | • Capillary electrophoresis instrument | • Post-PCR clean up kit • Dye terminator kit • Sequence reaction cleanup kit | • Data analysis • Expensive • Access to equipment |
| PAGE | Low | • None | • Polyacrylamide gels | • Laborious • Resolution of smaller indels |
| HMA | Low | • None | • Polyacrylamide gels | • Laborious • Resolution of smaller indels • Sensitive to SNPs in amplicon |
| T7EI digestion | Low | • None | • Post-PCR clean up kit • T7 endonuclease I • Gels | • Laborious • Sensitive to SNPs in amplicon • Cannot be used for genotyping |
| PCR-RFLP | Low | • None | • Post-PCR clean up kit • Restriction enzyme • Gels | • Requirement for indel to cause loss or creation of a unique restriction site |
| PCR-LDR | Low | • Gel imager capable of detecting fluorescence | • LDR primers • Protease • DNA ligase • Polyacrylamide gels | • Laborious • Access to equipment |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrington, B.; Bishop, K.; Sood, R. A Comprehensive Review of Indel Detection Methods for Identification of Zebrafish Knockout Mutants Generated by Genome-Editing Nucleases. Genes 2022, 13, 857. https://doi.org/10.3390/genes13050857
Carrington B, Bishop K, Sood R. A Comprehensive Review of Indel Detection Methods for Identification of Zebrafish Knockout Mutants Generated by Genome-Editing Nucleases. Genes. 2022; 13(5):857. https://doi.org/10.3390/genes13050857
Chicago/Turabian StyleCarrington, Blake, Kevin Bishop, and Raman Sood. 2022. "A Comprehensive Review of Indel Detection Methods for Identification of Zebrafish Knockout Mutants Generated by Genome-Editing Nucleases" Genes 13, no. 5: 857. https://doi.org/10.3390/genes13050857
APA StyleCarrington, B., Bishop, K., & Sood, R. (2022). A Comprehensive Review of Indel Detection Methods for Identification of Zebrafish Knockout Mutants Generated by Genome-Editing Nucleases. Genes, 13(5), 857. https://doi.org/10.3390/genes13050857






