Comparative Analysis of Transcriptomes of Diploid and Tetraploid Miscanthus lutarioriparius under Drought Stress
Abstract
:1. Introduction
2. Material and Methods M. lutarioriparius
2.1. Plant Materials and Drought Stress Conditions
2.2. RNA Extraction, cDNA Library Preparation, and RNA-seq
2.3. De Novo Assembly of Transcripts
2.4. Analysis of Differential Gene Expression
2.5. GO Functional Annotation and KEGG Enrichment Analysis
3. Results
3.1. Phenotypes of Diploid and Tetraploid M. lutarioriparius under Drought Stress
3.2. De Novo Assembly and Evaluation of Transcriptomes
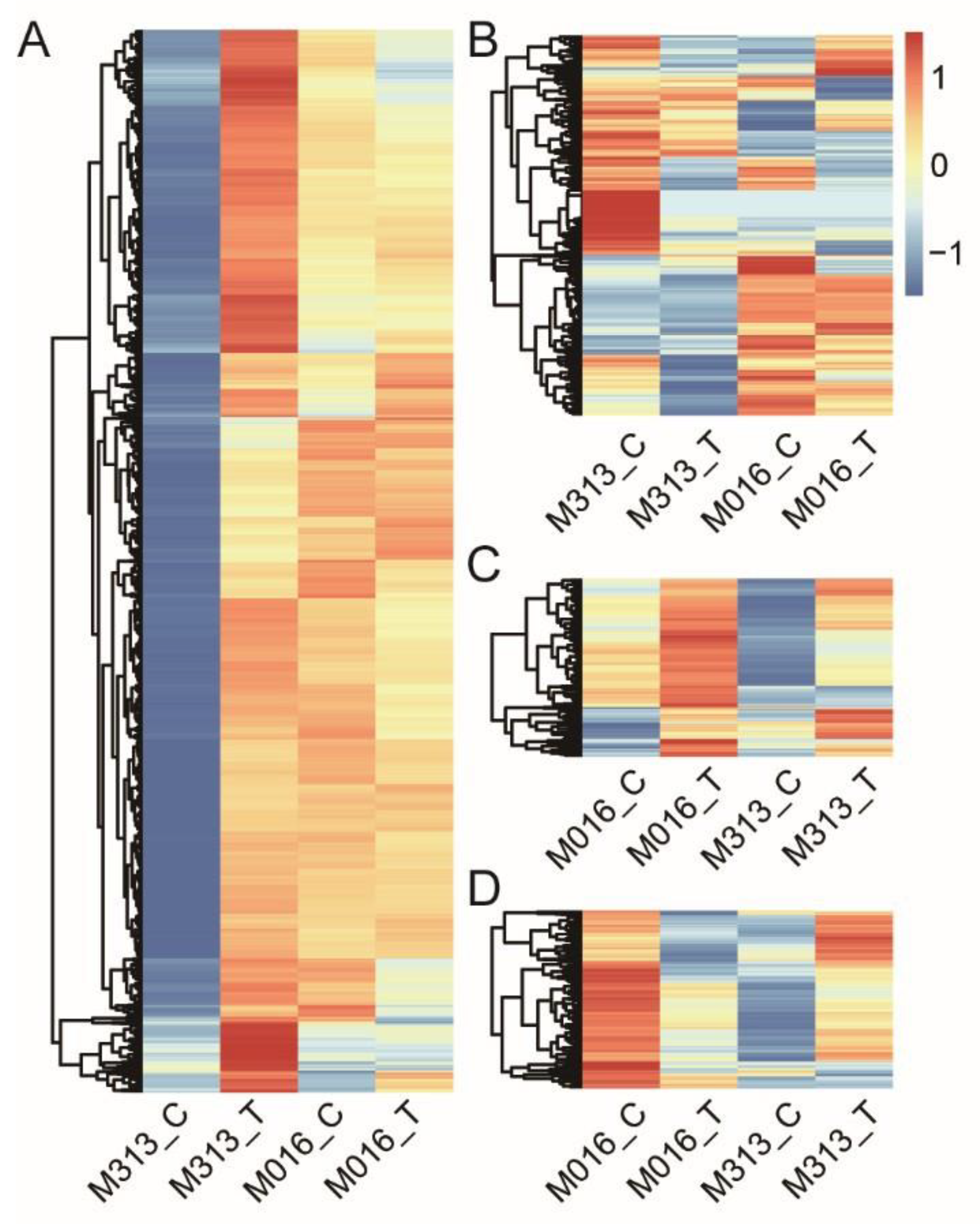
3.3. Screening of Differentially Expressed Genes
3.4. Analysis of Orthologous DEGs in Response to Drought Stress
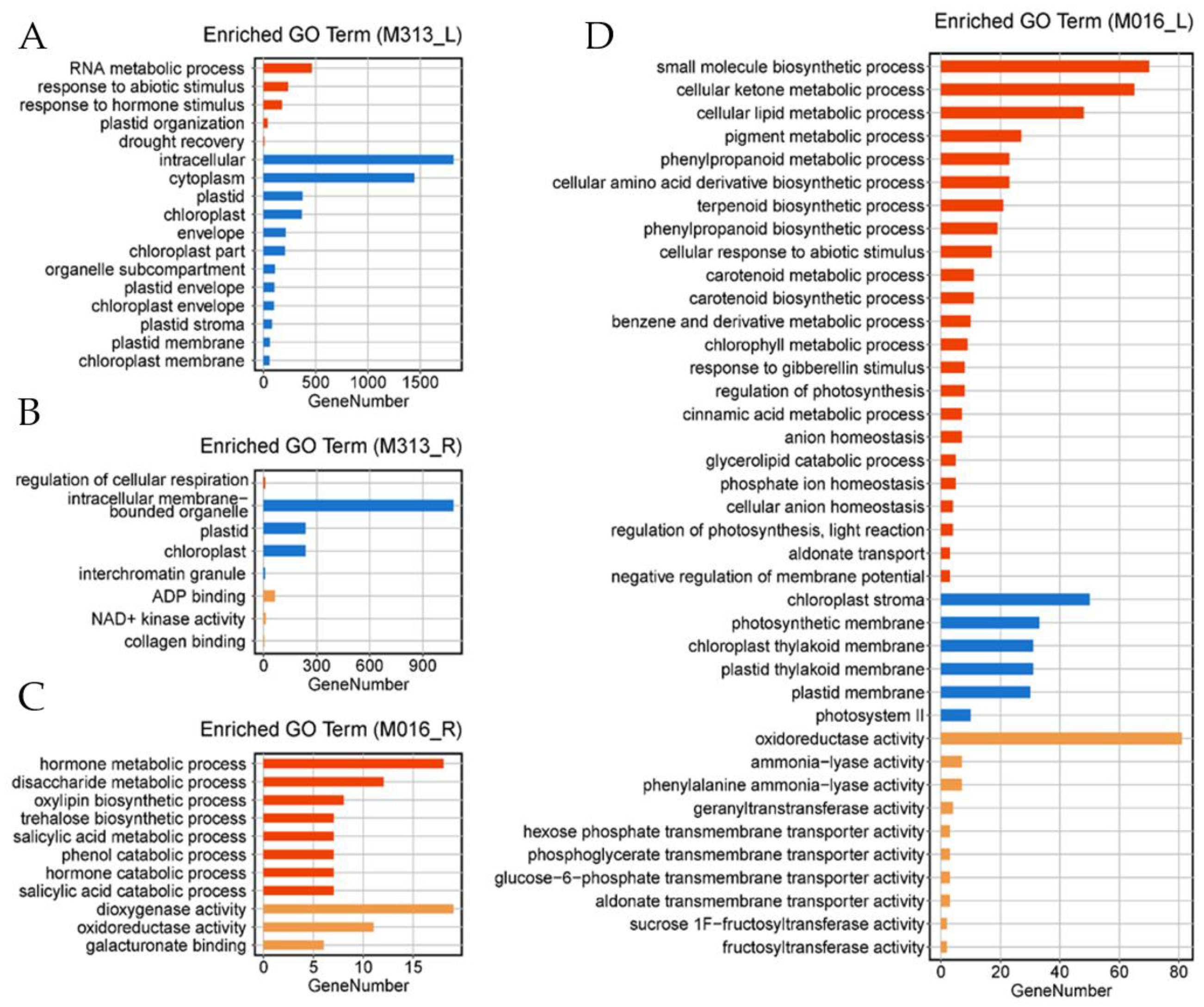
3.5. Functional Annotation of DEGs
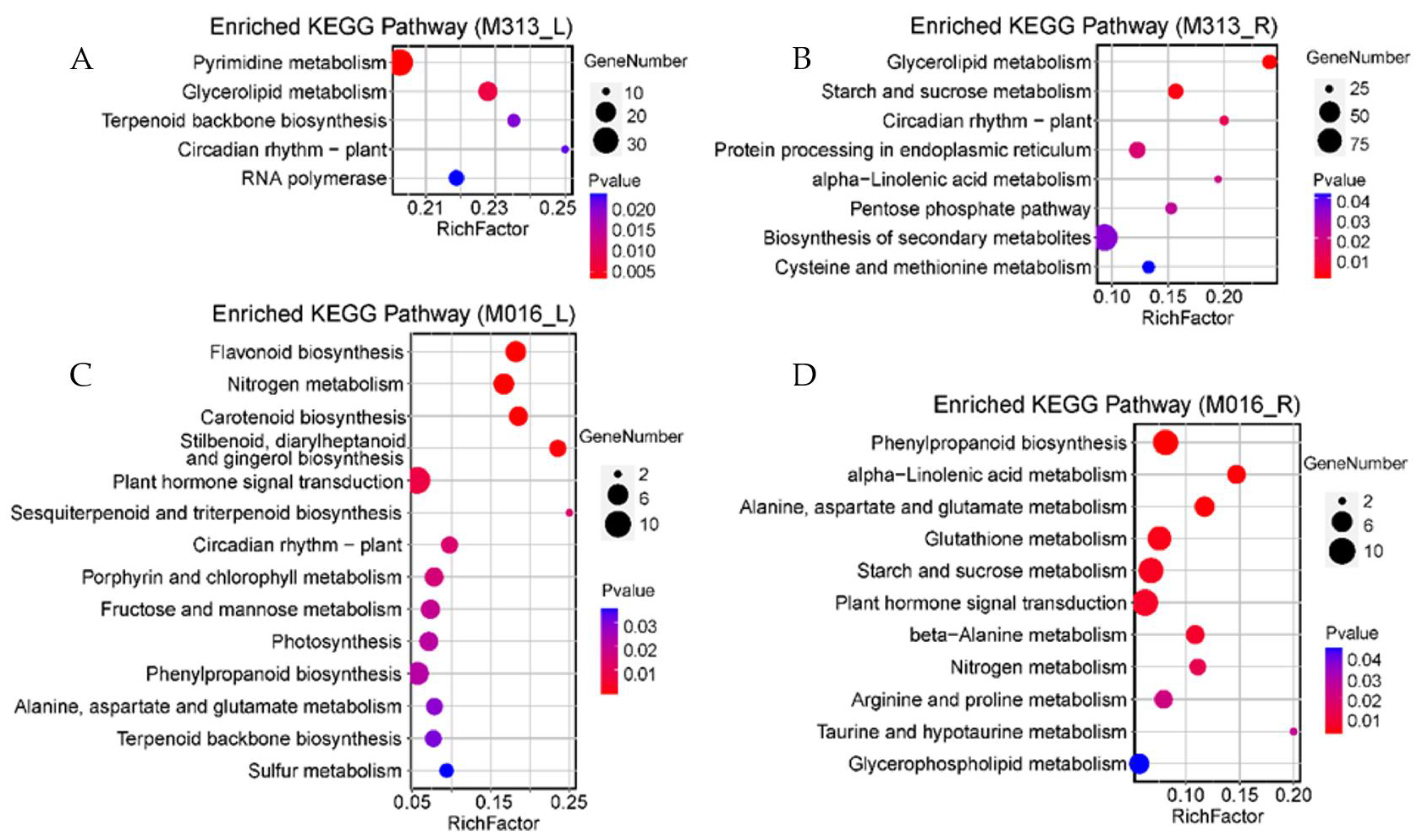
3.6. Enriched Differential Metabolic Pathways in Response to Drought Stress
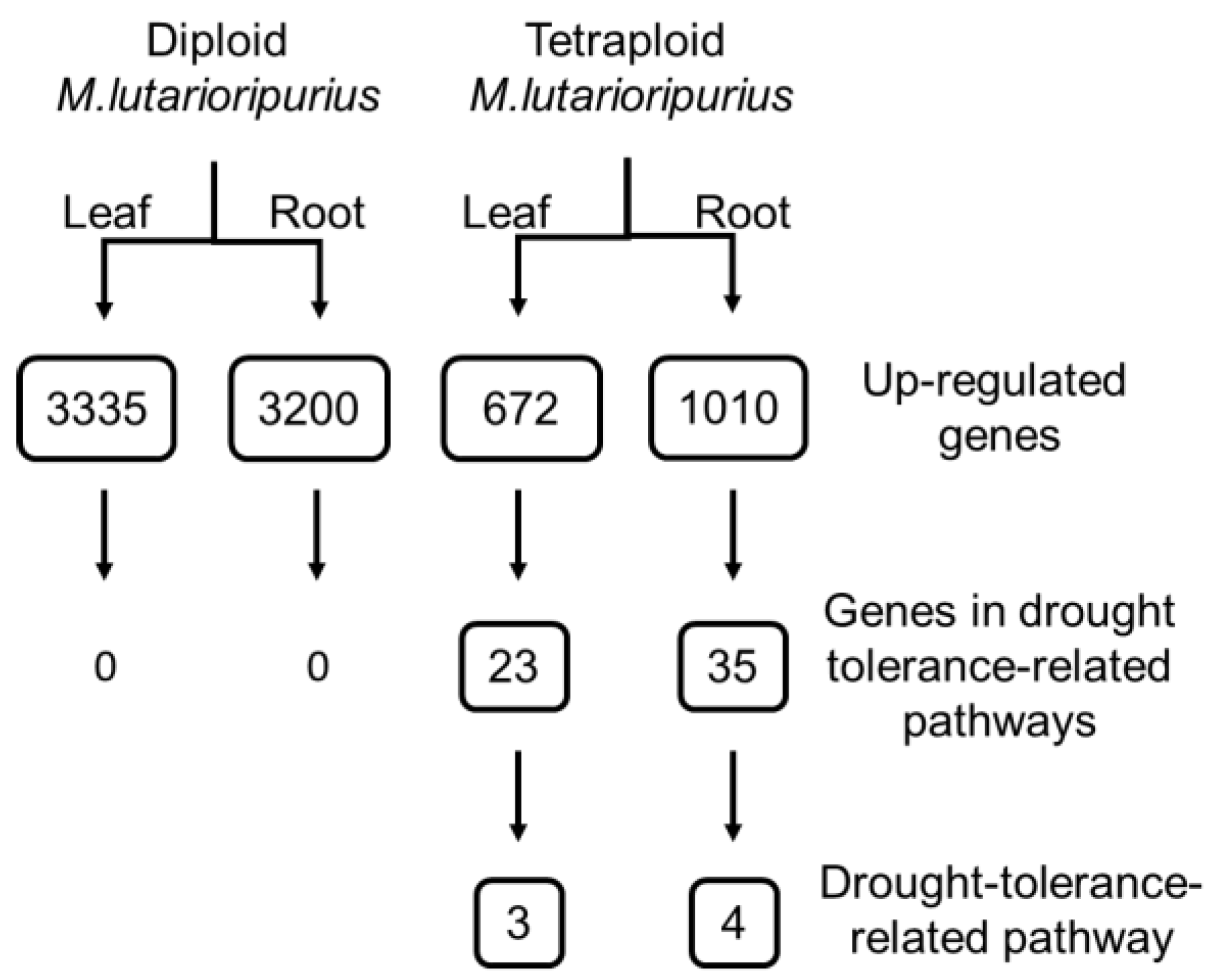
3.7. Enrichment of Induced Genes in Drought Tolerance Pathway
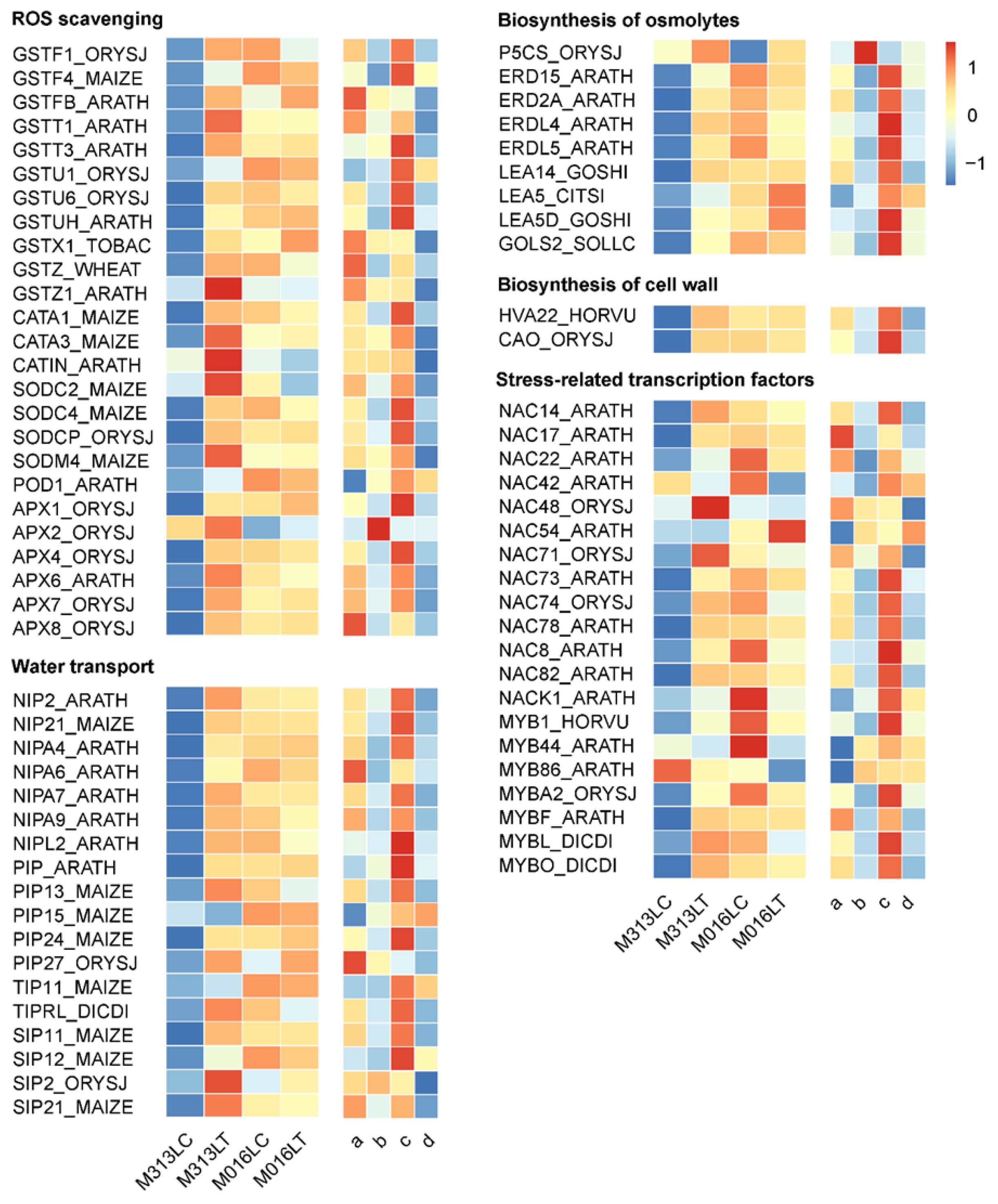
3.8. DEGs Involved in Key Drought-Tolerance-Related Biological Process
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ambavaram, M.M.; Basu, S.; Krishnan, A.; Ramegowda, V.; Batlang, U.; Rahman, L.; Baisakh, N.; Pereira, A. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 2014, 5, 5302. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Dias, P.C.; Araujo, W.L.; Moraes, G.A.; Barros, R.S.; DaMatta, F.M. Morphological and physiological responses of two coffee progenies to soil water availability. J. Plant Physiol. 2007, 164, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Wegener, C.; Jansen, G. Antioxidants in different potato genotypes: Effect of drought and wounding stress. Agriculture 2013, 3, 131–146. [Google Scholar] [CrossRef] [Green Version]
- Ahuja, I.; De Vos, R.C.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci. 2010, 15, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Zinselmeier, C.; Westgate, M.E.; Schussler, J.R.; Jones, R.J. Low water potential disrupts carbohydrate metabolism in maize (Zea mays L.) ovaries. Plant Physiol. 1995, 107, 385–391. [Google Scholar] [CrossRef]
- Pinheiro, C.; Chaves, M.M. Photosynthesis and drought: Can we make metabolic connections from available data? J. Exp. Bot. 2011, 62, 869–882. [Google Scholar] [CrossRef] [Green Version]
- Ramachandra Reddy, A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance im-provement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- He, X.; Xu, L.; Pan, C.; Gong, C.; Wang, Y.; Liu, X.; Yu, Y. Drought resistance of Camellia oleifera under drought stress: Changes in physiology and growth characteristics. PLoS ONE 2020, 15, e0235795. [Google Scholar] [CrossRef]
- Rai, A.N.; Penna, S. Molecular evolution of plant P5CS gene involved in proline biosynthesis. Mol. Biol. Rep. 2013, 40, 6429–6435. [Google Scholar] [CrossRef] [PubMed]
- Alexandersson, E.; Fraysse, L.; Sjovall-Larsen, S.; Gustavsson, S.; Fellert, M.; Karlsson, M.; Johanson, U.; Kjellbom, P. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 2005, 59, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, A.; Murata, N. The role of glycine betaine in the protection of plants from stress: Clues from transgenic plants. Plant Cell Environ. 2002, 25, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Kim, D.G.; Kim, Y.O.; Kim, J.S.; Kang, H. An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana. Plant Mol. Biol. 2004, 54, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; Van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Byrt, C.S.; Grof, C.P.; Furbank, R.T. C4 plants as biofuel feedstocks: Optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integr. Plant Biol. 2011, 53, 120–135. [Google Scholar] [CrossRef] [PubMed]
- Van der Weijde, T.; Alvim Kamei, C.L.; Torres, A.F.; Vermerris, W.; Dolstra, O.; Visser, R.G.; Trindade, L.M. The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 2013, 4, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Cheng, S.; Han, Y.; Zhao, D.; Li, H.; Wang, Y.; Zhang, G.; Chen, C. Natural variation of lignocellulosic components in miscanthus biomass in china. Front. Chem. 2020, 8, 595143. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, B.; He, J.; Yang, J.; Pan, L.; Sun, D.; Peng, J. Genetic diversity and population structure of Miscanthus sinensis germplasm in China. PLoS ONE 2013, 8, e75672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greef, J.M.; Deuter, M.; Jung, C.; Schondelmaier, J. Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genet. Resour. Crop Evol. 1997, 44, 185–195. [Google Scholar] [CrossRef]
- Godfree, R.C.; Marshall, D.J.; Young, A.G.; Miller, C.H.; Mathews, S. Empirical evidence of fixed and homeostatic patterns of polyploid advantage in a keystone grass exposed to drought and heat stress. R. Soc. Open Sci. 2017, 4, 170934. [Google Scholar] [CrossRef] [Green Version]
- Bei, X.; Shahid, M.Q.; Wu, J.; Chen, Z.; Wang, L.; Liu, X. Resequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neotetraploid rice. PLoS ONE 2019, 14, e0214953. [Google Scholar] [CrossRef] [Green Version]
- Tu, S.; Luan, L.; Liu, Y.; Long, W.; Kong, F.; He, T.; Xu, Q.; Yan, W.; Yu, M. Production and heterosis analysis of rice autotetraploid hybrids. Crop Sci. 2007, 47, 2356–2363. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.W.; Hu, C.Y.; Shahid, M.Q.; Guo, H.B.; Zeng, Y.X.; Liu, X.D.; Lu, Y.G. Analysis on genetic diversification and heterosis in autotetraploid rice. SpringerPlus 2013, 2, 439. [Google Scholar] [CrossRef] [Green Version]
- Chao, D.Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids exhibit higher potas-sium uptake and salinity tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Chen, J.; Sun, M.; Yan, H.; Feng, G.; Wu, B.; Zhang, X.; Wang, X.; Huang, L. Comparative transcriptome study of switchgrass (Panicum virgatum L.) homologous autopolyploid and its parental amphidiploid responding to consistent drought stress. Biotechnol. Biofuels 2020, 13, 170. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.C.; Ramirez-Parra, E. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant Cell Environ. 2014, 37, 2722–2737. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Fan, G.; Niu, S.; Zhao, Z.; Deng, M.; Dong, Y. Transcriptome-wide profiling and expression analysis of diploid and autotetraploid Paulownia tomentosa × Paulownia fortunei under drought stress. PLoS ONE 2014, 9, e113313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Lechner, M.; Findeiss, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (co-)orthologs inlargescale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef] [Green Version]

| Total Unigenes | N50 (bp) | Percent GC (%) | Average Mapped Ratio (%) | |
|---|---|---|---|---|
| M016 | 84,329 | 1211 | 50.07 | 89.70 |
| M313 | 95,974 | 1092 | 48.85 | 88.29 |
| Average | 90,152 | 1152 | 49.46 | 89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Wang, S.; Han, Y.; Wang, Y.; Xu, P.; Chen, C.; Zhang, G. Comparative Analysis of Transcriptomes of Diploid and Tetraploid Miscanthus lutarioriparius under Drought Stress. Genes 2022, 13, 873. https://doi.org/10.3390/genes13050873
Xu X, Wang S, Han Y, Wang Y, Xu P, Chen C, Zhang G. Comparative Analysis of Transcriptomes of Diploid and Tetraploid Miscanthus lutarioriparius under Drought Stress. Genes. 2022; 13(5):873. https://doi.org/10.3390/genes13050873
Chicago/Turabian StyleXu, Xitong, Shukai Wang, Yanbin Han, Yancui Wang, Pingping Xu, Cuixia Chen, and Guobin Zhang. 2022. "Comparative Analysis of Transcriptomes of Diploid and Tetraploid Miscanthus lutarioriparius under Drought Stress" Genes 13, no. 5: 873. https://doi.org/10.3390/genes13050873






