Vibrio cholerae Chromosome Partitioning without Polar Anchoring by HubP
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids and Strains
2.2. Construction of MiniF-Plasmid Derivatives
2.3. Construction of Plasmids Used for Strain Constructions
2.4. Plasmid Stability Assay
2.5. Fluorescence Microscopy
3. Results
3.1. ParABS1 System Does Not Efficiently Stabilize a Mini-F in E. coli
3.2. HubP Is Dispensable for the ParABS1-Driven Stabilization of ch2 in V. cholerae
3.3. ParABS1-Driven Positioning of ch2 in V. cholerae in Absence of HubP
3.4. ParABS1 System Positions oriC1 in V. cholerae in Absence of HubP
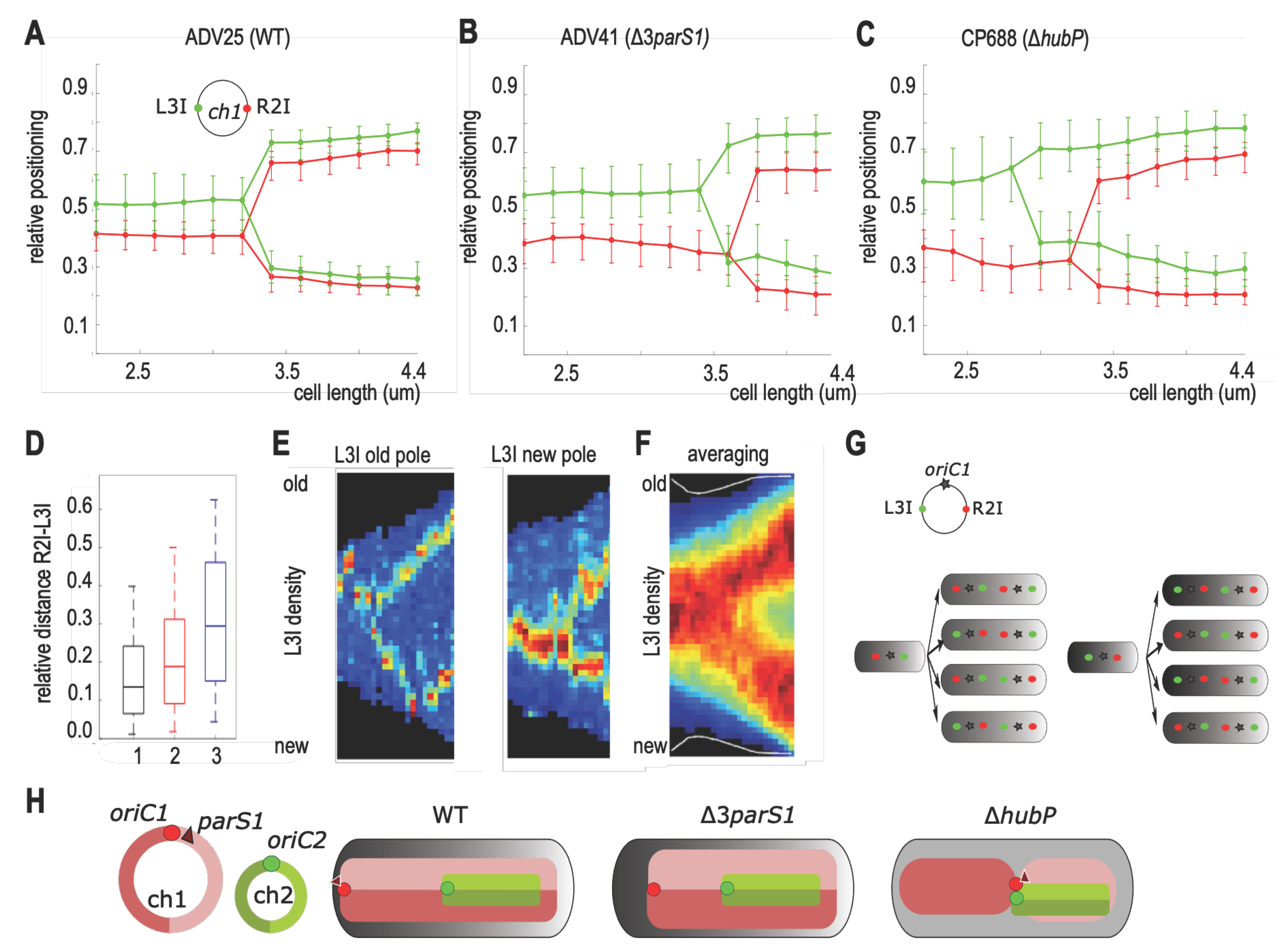
3.5. Chromosome I Rearrangement by Non-Anchored Partition System
4. Discussion
4.1. Intrinsic Adaptability of the Partition Systems
4.2. Positioning Interference of oriC1 and oriC2, the Two parS1-Containing Regions
4.3. Loss of ch1-Anchoring Leads to Transversal Organization of ch1
4.4. Role of the Polar Anchoring of the Origin of Replication
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| V. cholerae Chromosome 1 | ch1 |
| V. cholerae Chromosome 2 | ch2 |
| Origin of chromosome replication | oriC |
| Colony forming units | cfu |
| Wild type | WT |
| Replication terminus region | ter |
References
- Bouet, J.-Y.; Funnell, B.E. Plasmid Localization and Partition in Enterobacteriaceae. EcoSal Plus 2019, 8, ESP-0003-2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalal, A.S.B.; Le, T.B.K. Bacterial Chromosome Segregation by the ParABS System. Open Biol. 2020, 10, 200097. [Google Scholar] [CrossRef] [PubMed]
- Livny, J.; Yamaichi, Y.; Waldor, M.K. Distribution of Centromere-like ParS Sites in Bacteria: Insights from Comparative Genomics. J. Bacteriol. 2007, 189, 8693–8703. [Google Scholar] [CrossRef] [Green Version]
- Ogasawara, N.; Yoshikawa, H. Genes and Their Organization in the Replication Origin Region of the Bacterial Chromosome. Mol. Microbiol. 1992, 6, 629–634. [Google Scholar] [CrossRef]
- Toro, E.; Hong, S.-H.; McAdams, H.H.; Shapiro, L. Caulobacter Requires a Dedicated Mechanism to Initiate Chromosome Segregation. Proc. Natl. Acad. Sci. USA 2008, 105, 15435–15440. [Google Scholar] [CrossRef] [Green Version]
- David, A.; Demarre, G.; Muresan, L.; Paly, E.; Barre, F.-X.; Possoz, C. The Two Cis-Acting Sites, ParS1 and OriC1, Contribute to the Longitudinal Organisation of Vibrio Cholerae Chromosome I. PLoS Genet. 2014, 10, e1004448. [Google Scholar] [CrossRef] [Green Version]
- Vecchiarelli, A.G.; Mizuuchi, K.; Funnell, B.E. Surfing Biological Surfaces: Exploiting the Nucleoid for Partition and Transport in Bacteria. Mol. Microbiol. 2012, 86, 513–523. [Google Scholar] [CrossRef]
- Sherratt, D. Plasmid Partition: Sisters Drifting Apart. EMBO J. 2013, 32, 1208–1210. [Google Scholar] [CrossRef] [Green Version]
- Rodionov, O.; Lobocka, M.; Yarmolinsky, M. Silencing of Genes Flanking the P1 Plasmid Centromere. Science 1999, 283, 546–549. [Google Scholar] [CrossRef] [Green Version]
- Lynch, A.S.; Wang, J.C. SopB Protein-Mediated Silencing of Genes Linked to the SopC Locus of Escherichia Coli F Plasmid. Proc. Natl. Acad. Sci. USA 1995, 92, 1896–1900. [Google Scholar] [CrossRef] [Green Version]
- Murray, H.; Ferreira, H.; Errington, J. The Bacterial Chromosome Segregation Protein Spo0J Spreads along DNA from ParS Nucleation Sites. Mol. Microbiol. 2006, 61, 1352–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breier, A.M.; Grossman, A.D. Whole-Genome Analysis of the Chromosome Partitioning and Sporulation Protein Spo0J (ParB) Reveals Spreading and Origin-Distal Sites on the Bacillus Subtilis Chromosome. Mol. Microbiol. 2007, 64, 703–718. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Valeriano, M.; Altegoer, F.; Steinchen, W.; Urban, S.; Liu, Y.; Bange, G.; Thanbichler, M. ParB-Type DNA Segregation Proteins Are CTP-Dependent Molecular Switches. Cell 2019, 179, 1512–1524.e15. [Google Scholar] [CrossRef] [PubMed]
- Soh, Y.-M.; Davidson, I.F.; Zamuner, S.; Basquin, J.; Bock, F.P.; Taschner, M.; Veening, J.-W.; De Los Rios, P.; Peters, J.-M.; Gruber, S. Self-Organization of ParS Centromeres by the ParB CTP Hydrolase. Science 2019, 366, 1129–1133. [Google Scholar] [CrossRef]
- Jalal, A.S.; Tran, N.T.; Le, T.B. ParB Spreading on DNA Requires Cytidine Triphosphate in Vitro. eLife 2020, 9, e53515. [Google Scholar] [CrossRef]
- Osorio-Valeriano, M.; Altegoer, F.; Das, C.K.; Steinchen, W.; Panis, G.; Connolley, L.; Giacomelli, G.; Feddersen, H.; Corrales-Guerrero, L.; Giammarinaro, P.I.; et al. The CTPase Activity of ParB Determines the Size and Dynamics of Prokaryotic DNA Partition Complexes. Mol. Cell 2021, 81, 3992–4007.e10. [Google Scholar] [CrossRef]
- Dubarry, N.; Pasta, F.; Lane, D. ParABS Systems of the Four Replicons of Burkholderia Cenocepacia: New Chromosome Centromeres Confer Partition Specificity. J. Bacteriol. 2006, 188, 1489–1496. [Google Scholar] [CrossRef] [Green Version]
- Kahng, L.S.; Shapiro, L. Polar Localization of Replicon Origins in the Multipartite Genomes of Agrobacterium Tumefaciens and Sinorhizobium Meliloti. J. Bacteriol. 2003, 185, 3384–3391. [Google Scholar] [CrossRef] [Green Version]
- Fogel, M.A.; Waldor, M.K. Distinct Segregation Dynamics of the Two Vibrio Cholerae Chromosomes. Mol. Microbiol. 2005, 55, 125–136. [Google Scholar] [CrossRef]
- Viollier, P.H.; Thanbichler, M.; McGrath, P.T.; West, L.; Meewan, M.; McAdams, H.H.; Shapiro, L. Rapid and Sequential Movement of Individual Chromosomal Loci to Specific Subcellular Locations during Bacterial DNA Replication. Proc. Natl. Acad. Sci. USA 2004, 101, 9257–9262. [Google Scholar] [CrossRef] [Green Version]
- Donovan, C.; Schwaiger, A.; Krämer, R.; Bramkamp, M. Subcellular Localization and Characterization of the ParAB System from Corynebacterium Glutamicum. J. Bacteriol. 2010, 192, 3441–3451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.C.; Surovtsev, I.V.; Beltran, B.G.; Huang, F.; Bewersdorf, J.; Jacobs-Wagner, C. Evidence for a DNA-Relay Mechanism in ParABS-Mediated Chromosome Segregation. eLife 2014, 3, e02758. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.R.; Comolli, L.R.; Zhu, J.; Eckart, M.; Koenig, M.; Downing, K.H.; Moerner, W.E.; Earnest, T.; Shapiro, L. A Polymeric Protein Anchors the Chromosomal Origin/ParB Complex at a Bacterial Cell Pole. Cell 2008, 134, 945–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebersbach, G.; Briegel, A.; Jensen, G.J.; Jacobs-Wagner, C. A Self-Associating Protein Critical for Chromosome Attachment, Division, and Polar Organization in Caulobacter. Cell 2008, 134, 956–968. [Google Scholar] [CrossRef] [Green Version]
- Badrinarayanan, A.; Le, T.B.K.; Laub, M.T. Bacterial Chromosome Organization and Segregation. Annu. Rev. Cell Dev. Biol. 2015, 31, 171–199. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.; Johnson, S.L.; Davenport, K.W.; Rajavel, M.; Waldminghaus, T.; Detter, J.C.; Chain, P.S.; Sozhamannan, S. Exception to the Rule: Genomic Characterization of Naturally Occurring Unusual Vibrio Cholerae Strains with a Single Chromosome. Int. J. Genomics 2017, 2017, 8724304. [Google Scholar] [CrossRef] [Green Version]
- Yamaichi, Y.; Niki, H. Active Segregation by the Bacillus Subtilis Partitioning System in Escherichia Coli. Proc. Natl. Acad. Sci. USA 2000, 97, 14656–14661. [Google Scholar] [CrossRef] [Green Version]
- Yamaichi, Y.; Fogel, M.A.; McLeod, S.M.; Hui, M.P.; Waldor, M.K. Distinct Centromere-like ParS Sites on the Two Chromosomes of Vibrio spp. J. Bacteriol. 2007, 189, 5314–5324. [Google Scholar] [CrossRef] [Green Version]
- Yamaichi, Y.; Bruckner, R.; Ringgaard, S.; Möll, A.; Cameron, D.E.; Briegel, A.; Jensen, G.J.; Davis, B.M.; Waldor, M.K. A Multidomain Hub Anchors the Chromosome Segregation and Chemotactic Machinery to the Bacterial Pole. Genes Dev. 2012, 26, 2348–2360. [Google Scholar] [CrossRef] [Green Version]
- Saint-Dic, D.; Frushour, B.P.; Kehrl, J.H.; Kahng, L.S. A ParA Homolog Selectively Influences Positioning of the Large Chromosome Origin in Vibrio Cholerae. J. Bacteriol. 2006, 188, 5626–5631. [Google Scholar] [CrossRef] [Green Version]
- Yamaichi, Y.; Fogel, M.A.; Waldor, M.K. Par Genes and the Pathology of Chromosome Loss in Vibrio Cholerae. Proc. Natl. Acad. Sci. USA 2007, 104, 630–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohl, D.A.; Gober, J.W. Cell Cycle-Dependent Polar Localization of Chromosome Partitioning Proteins in Caulobacter Crescentus. Cell 1997, 88, 675–684. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Vecchiarelli, A.G.; Mizuuchi, K.; Neuman, K.C.; Liu, J. Brownian Ratchet Mechanism for Faithful Segregation of Low-Copy-Number Plasmids. Biophys. J. 2017, 112, 1489–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meibom, K.L.; Blokesch, M.; Dolganov, N.A.; Wu, C.-Y.; Schoolnik, G.K. Chitin Induces Natural Competence in Vibrio Cholerae. Science 2005, 310, 1824–1827. [Google Scholar] [CrossRef]
- Niki, H.; Hiraga, S. Subcellular Distribution of Actively Partitioning F Plasmid during the Cell Division Cycle in E. coli. Cell 1997, 90, 951–957. [Google Scholar] [CrossRef] [Green Version]
- Sliusarenko, O.; Heinritz, J.; Emonet, T.; Jacobs-Wagner, C. High-Throughput, Subpixel Precision Analysis of Bacterial Morphogenesis and Intracellular Spatio-Temporal Dynamics. Mol. Microbiol. 2011, 80, 612–627. [Google Scholar] [CrossRef] [Green Version]
- Demarre, G.; Galli, E.; Muresan, L.; Paly, E.; David, A.; Possoz, C.; Barre, F.-X. Differential Management of the Replication Terminus Regions of the Two Vibrio Cholerae Chromosomes during Cell Division. PLoS Genet. 2014, 10, e1004557. [Google Scholar] [CrossRef]
- Godfrin-Estevenon, A.-M.; Pasta, F.; Lane, D. The ParAB Gene Products of Pseudomonas Putida Exhibit Partition Activity in Both P. putida and Escherichia coli. Mol. Microbiol. 2002, 43, 39–49. [Google Scholar] [CrossRef]
- Jaskólska, M.; Adams, D.W.; Blokesch, M. Two Defence Systems Eliminate Plasmids from Seventh Pandemic Vibrio Cholerae. Nature 2022, 604, 323–329. [Google Scholar] [CrossRef]
- Baek, J.H.; Rajagopala, S.V.; Chattoraj, D.K. Chromosome Segregation Proteins of Vibrio Cholerae as Transcription Regulators. mBio 2014, 5, e01061-14. [Google Scholar] [CrossRef] [Green Version]
- Murray, H.; Errington, J. Dynamic Control of the DNA Replication Initiation Protein DnaA by Soj/ParA. Cell 2008, 135, 74–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadoya, R.; Baek, J.H.; Sarker, A.; Chattoraj, D.K. Participation of Chromosome Segregation Protein ParAI of Vibrio Cholerae in Chromosome Replication. J. Bacteriol. 2011, 193, 1504–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thanbichler, M.; Shapiro, L. MipZ, a Spatial Regulator Coordinating Chromosome Segregation with Cell Division in Caulobacter. Cell 2006, 126, 147–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Gall, A.; Cattoni, D.I.; Guilhas, B.; Mathieu-Demazière, C.; Oudjedi, L.; Fiche, J.-B.; Rech, J.; Abrahamsson, S.; Murray, H.; Bouet, J.-Y.; et al. Bacterial Partition Complexes Segregate within the Volume of the Nucleoid. Nat. Commun. 2016, 7, 12107. [Google Scholar] [CrossRef]
- Ah-Seng, Y.; Rech, J.; Lane, D.; Bouet, J.-Y. Defining the Role of ATP Hydrolysis in Mitotic Segregation of Bacterial Plasmids. PLoS Genet. 2013, 9, e1003956. [Google Scholar] [CrossRef] [Green Version]
- Tran, N.T.; Stevenson, C.E.; Som, N.F.; Thanapipatsiri, A.; Jalal, A.S.B.; Le, T.B.K. Permissive Zones for the Centromere-Binding Protein ParB on the Caulobacter Crescentus Chromosome. Nucleic Acids Res. 2018, 46, 1196–1209. [Google Scholar] [CrossRef] [Green Version]
- Lagage, V.; Boccard, F.; Vallet-Gely, I. Regional Control of Chromosome Segregation in Pseudomonas Aeruginosa. PLoS Genet. 2016, 12, e1006428. [Google Scholar] [CrossRef]
- Wang, X.; Possoz, C.; Sherratt, D.J. Dancing around the Divisome: Asymmetric Chromosome Segregation in Escherichia coli. Genes Dev. 2005, 19, 2367–2377. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, X.; Possoz, C.; Sherratt, D.J. The Two Escherichia coli Chromosome Arms Locate to Separate Cell Halves. Genes Dev. 2006, 20, 1727–1731. [Google Scholar] [CrossRef] [Green Version]
- Danilova, O.; Reyes-Lamothe, R.; Pinskaya, M.; Sherratt, D.; Possoz, C. MukB Colocalizes with the OriC Region and Is Required for Organization of the Two Escherichia coli Chromosome Arms into Separate Cell Halves. Mol. Microbiol. 2007, 65, 1485–1492. [Google Scholar] [CrossRef] [Green Version]
- Mäkelä, J.; Sherratt, D.J. Organization of the Escherichia coli Chromosome by a MukBEF Axial Core. Mol. Cell 2020, 78, 250–260.e5. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, J.; Uphoff, S.; Sherratt, D.J. Nonrandom Segregation of Sister Chromosomes by Escherichia coli MukBEF. Proc. Natl. Acad. Sci. USA 2021, 118, e2022078118. [Google Scholar] [CrossRef] [PubMed]
- Badrinarayanan, A.; Lesterlin, C.; Reyes-Lamothe, R.; Sherratt, D. The Escherichia coli SMC Complex, MukBEF, Shapes Nucleoid Organization Independently of DNA Replication. J. Bacteriol. 2012, 194, 4669–4676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nolivos, S.; Upton, A.L.; Badrinarayanan, A.; Müller, J.; Zawadzka, K.; Wiktor, J.; Gill, A.; Arciszewska, L.; Nicolas, E.; Sherratt, D. MatP Regulates the Coordinated Action of Topoisomerase IV and MukBEF in Chromosome Segregation. Nat. Commun. 2016, 7, 10466. [Google Scholar] [CrossRef] [PubMed]
- Lioy, V.S.; Junier, I.; Lagage, V.; Vallet, I.; Boccard, F. Distinct Activities of Bacterial Condensins for Chromosome Management in Pseudomonas Aeruginosa. Cell Rep. 2020, 33, 108344. [Google Scholar] [CrossRef]
- Zhao, H.; Bhowmik, B.K.; Petrushenko, Z.M.; Rybenkov, V.V. Alternating Dynamics of OriC, SMC, and MksBEF in Segregation of Pseudomonas Aeruginosa Chromosome. mSphere 2020, 5, e00238-20. [Google Scholar] [CrossRef]
- Lee, P.S.; Grossman, A.D. The Chromosome Partitioning Proteins Soj (ParA) and Spo0J (ParB) Contribute to Accurate Chromosome Partitioning, Separation of Replicated Sister Origins, and Regulation of Replication Initiation in Bacillus Subtilis. Mol. Microbiol. 2006, 60, 853–869. [Google Scholar] [CrossRef]
- Galli, E.; Poidevin, M.; Le Bars, R.; Desfontaines, J.-M.; Muresan, L.; Paly, E.; Yamaichi, Y.; Barre, F.-X. Cell Division Licensing in the Multi-Chromosomal Vibrio Cholerae Bacterium. Nat. Microbiol. 2016, 1, 16094. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Possoz, C.; Yamaichi, Y.; Galli, E.; Ferat, J.-L.; Barre, F.-X. Vibrio cholerae Chromosome Partitioning without Polar Anchoring by HubP. Genes 2022, 13, 877. https://doi.org/10.3390/genes13050877
Possoz C, Yamaichi Y, Galli E, Ferat J-L, Barre F-X. Vibrio cholerae Chromosome Partitioning without Polar Anchoring by HubP. Genes. 2022; 13(5):877. https://doi.org/10.3390/genes13050877
Chicago/Turabian StylePossoz, Christophe, Yoshiharu Yamaichi, Elisa Galli, Jean-Luc Ferat, and Francois-Xavier Barre. 2022. "Vibrio cholerae Chromosome Partitioning without Polar Anchoring by HubP" Genes 13, no. 5: 877. https://doi.org/10.3390/genes13050877
APA StylePossoz, C., Yamaichi, Y., Galli, E., Ferat, J.-L., & Barre, F.-X. (2022). Vibrio cholerae Chromosome Partitioning without Polar Anchoring by HubP. Genes, 13(5), 877. https://doi.org/10.3390/genes13050877





