Uneven Levels of 5S and 45S rDNA Site Number and Loci Variations across Wild Chrysanthemum Accessions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Cytological Preparation and Multiplex Oligo-FISH
2.3. Karyotyping Analysis
3. Results
3.1. Chromosome Numbers and Karyotype Features
3.2. Geographic Distribution of Wild Chrysanthemum Genus Plants
3.3. Identification of 5S and 45S rDNA Sites in Chrysanthemum Accessions Using Oligo-FISH
3.4. Patterns of rDNA Sites
4. Discussion
4.1. Distribution Patterns of rDNA Sites in Wild Chrysanthemum Genus Plants
4.2. Relationship between Chromosome Number and Number of rDNA Sites in Wild Chrysanthemum Genus Plants
4.3. Potential Mechanism of rDNA Sites Variation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atri, M.; Chehregani, A.; Jalali, F.; Yousefi, S. New chromosome counts in some species of the genus Artemisia L. (Asteraceae) from Iran. Cytologia 2009, 74, 443–448. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lim, J.H.; Jung, J.A.; Kim, W.H.; Lim, K.-B.; Cabahug, R.A.M.; Hwang, Y.-J. Analysis of ploidy levels of Korean wild Asteraceae species using chromosome counting. Flower Res. J. 2019, 27, 278–284. [Google Scholar]
- Zandonadi, A.S.; Maia, C.; Barbosa, J.G.; Finger, F.L.; Grossi, J.A.S. Influence of long days on the production of cut chrysanthemum cultivars. Hortic. Bras. 2018, 36, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Hoang, T.K.; Wang, Y.; Hwang, Y.J.; Lim, J.H. Analysis of the morphological characteristics and karyomorphology of wild Chrysanthemum species in Korea. Hortic. Environ. Biotechnol. 2020, 61, 359–369. [Google Scholar] [CrossRef]
- He, J.; Lin, S.; Yu, Z.; Song, A.; Guan, Z.; Fang, W.; Chen, S.; Zhang, F.; Jiang, J.; Chen, F.; et al. Identification of 5S and 45S rDNA sites in Chrysanthemum species by using oligonucleotide fluorescence in situ hybridization (Oligo-FISH). Mol. Biol. Rep. 2021, 48, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jung, J.A.; Lim, K.-B.; Cabahug, R.A.M.; Hwang, Y.-J. Cytogenetic studies of Chrysanthemum: A Review. Flower Res. J. 2019, 27, 242–253. [Google Scholar]
- Flavell, R.B. Repetitive DNA and chromosome evolution in plants. Philos. Trans. R Soc. Lond. B Biol. Sci. 1986, 312, 227–242. [Google Scholar]
- Biscotti, M.A.; Olmo, E.; Heslop-Harrison, J.S. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015, 23, 415–420. [Google Scholar] [CrossRef]
- Garcia, S.; Kovarik, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the Plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef] [Green Version]
- She, C.W.; Jiang, X.H. Karyotype analysis of Lablab purpureus (L.) Sweet using fluorochrome banding and fluorescence in situ hybridization with rDNA probes. Czech J. Genet. Plant 2015, 51, 110–116. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.Y.; Zhang, F.; Guan, Z.Y.; Wang, H.B.; Jiang, J.F.; Chen, S.M.; Chen, F.D. Localization of 45S and 5S rDNA sites and karyotype of Chrysanthemum and its related genera by fluorescent in situ hybridization. Biochem. Syst. Ecol. 2015, 62, 164–172. [Google Scholar] [CrossRef]
- Abd El-Twab, M.H.; Kondo, K. Physical mapping of 5S and 45S rDNA in Chrysanthemum and related genera of the Anthemideae by FISH, and species relationships. J. Genet. 2012, 91, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Cuyacot, A.R.; Lim, K.B.; Kim, H.H.; Hwang, Y.J. Chromosomal characterization based on repetitive DNA distribution in a tetraploid cytotype of Chrysanthemum zawadskii. Hortic. Environ. Biotechnol. 2017, 58, 488–494. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative genome organization in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abd El-Twab, M.H.; Kondo, K. FISH physical mapping of 5S, 45S and Arabidopsis-type telomere sequence repeats in Chrysanthemum zawadskii showing intra-chromosomal variation and complexity in nature. Chromosome Bot. 2006, 1, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Yue, M.; Gautam, M.; Chen, Z.; Hou, J.; Zheng, X.; Hou, H.; Li, L. Histone acetylation of 45S rDNA correlates with disrupted nucleolar organization during heat stress response in Zea mays L. Physiol. Plant 2021, 172, 2079–2089. [Google Scholar] [CrossRef]
- Xu, C.M.; Bie, T.D.; Wang, C.M.; Zhou, B.; Chen, P.D. Distribution of 45S rDNA sequence on chromosomes of Triticum aestivum and its relative species. Yi Chuan 2007, 29, 1126–1130. [Google Scholar] [CrossRef]
- Liu, B.; Davis, T.M. Conservation and loss of ribosomal RNA gene sites in diploid and polyploid Fragaria (Rosaceae). BMC Plant Biol. 2011, 11, 157. [Google Scholar] [CrossRef] [Green Version]
- Wibowo, A.; Setiawan, A.; Purwantoro, A.; Kikuchi, S.; Koba, T. Cytological variation of rRNA genes and subtelomeric repeat sequences in Indonesian and Japanese cucumber accessions. Chromosome Sci. 2018, 21, 81–87. [Google Scholar]
- Su, D.; Chen, L.; Sun, J.; Zhang, L.; Gao, R.; Li, Q.; Han, Y.; Li, Z. Comparative chromosomal localization of 45S and 5S rDNA sites in 76 purple-fleshed sweet potato cultivars. Plants 2020, 9, 865. [Google Scholar] [CrossRef]
- Wang, Y.; Jung, J.A.; Kim, W.H.; Lim, K.B.; Hwang, Y.J. Morphological and rDNA fluorescence in situ hybridization analyses of Chrysanthemum cultivars from Korea. Hortic. Environ. Biotechnol. 2021, 62, 917–925. [Google Scholar] [CrossRef]
- Machackova, P.; Majesky, L.; Hrones, M.; Bilkova, L.; Hribova, E.; Vasut, R.J. New insights into ribosomal DNA variation in apomictic and sexual Taraxacum (Asteraceae). Bot. J. Linn. Soc. 2021, boab094. [Google Scholar] [CrossRef]
- Vozarova, R.; Herklotz, V.; Kovarik, A.; Tynkevich, Y.O.; Volkov, R.A.; Ritz, C.M.; Lunerova, J. Ancient origin of two 5S rDNA families dominating in the genus Rosa and their behavior in the Canina-type meiosis. Front. Plant Sci. 2021, 12, 643548. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Qiu, L.; Xiao, Z.; Fu, S.; Tang, Z. New oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 2016, 7, 118. [Google Scholar] [CrossRef] [Green Version]
- Beliveau, B.J.; Joyce, E.F.; Apostolopoulos, N.; Yilmaz, F.; Fonseka, C.Y.; McCole, R.B.; Chang, Y.; Li, J.B.; Senaratne, T.N.; Williams, B.R.; et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 2012, 109, 21301–21306. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Liu, J.; He, Z. Oligo-FISH can identify chromosomes and distinguish Hippophae rhamnoides L. Taxa. Genes 2022, 13, 195. [Google Scholar] [CrossRef]
- Jiang, J. Fluorescence in situ hybridization in plants: Recent developments and future applications. Chromosome Res. 2019, 27, 153–165. [Google Scholar] [CrossRef]
- Roa, F.; Guerra, M. Distribution of 45S rDNA sites in chromosomes of plants: Structural and evolutionary implications. BMC Evol. Biol. 2012, 12, 225. [Google Scholar] [CrossRef] [Green Version]
- Garcia, S.; Galvez, F.; Gras, A.; Kovarik, A.; Garnatje, T. Plant rDNA database: Update and new features. Database 2014, 2014, bau063. [Google Scholar] [CrossRef] [Green Version]
- Kovarik, A.; Dadejova, M.; Lim, Y.K.; Chase, M.W.; Clarkson, J.J.; Knapp, S.; Leitch, A.R. Evolution of rDNA in Nicotiana allopolyploids: A potential link between rDNA homogenization and epigenetics. Ann. Bot. 2008, 101, 815–823. [Google Scholar] [CrossRef] [Green Version]
- Pedrosa-Harand, A.; de Almeida, C.C.; Mosiolek, M.; Blair, M.W.; Schweizer, D.; Guerra, M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Appl. Genet. 2006, 112, 924–933. [Google Scholar] [CrossRef] [PubMed]
- El-Twab, M.H.; Kondo, K. Physical mapping of 5S rDNA in chromosomes of Dendranthema by fluorescence in situ hybridization. Chromosome Sci. 2002, 6, 13–16. [Google Scholar]
- El-Twab, M.H.; Kondo, K. Physical mapping of 45S rDNA loci by fluorescent in situ hybridization and evolution among polyploid Dendranthema species. Chromosome Sci. 2003, 7, 71–76. [Google Scholar]
- Huang, X.; Zhu, M.; Zhuang, L.; Zhang, S.; Wang, J.; Chen, X.; Wang, D.; Chen, J.; Bao, Y.; Guo, J.; et al. Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Appl. Genet. 2018, 131, 1967–1986. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Du, P.; Zhuang, L.; Chu, C.; Zhao, H.; Qi, Z. A simple and efficient non-denaturing FISH method for maize chromosome differentiation using single-strand oligonucleotide probes. Genome 2017, 60, 657–664. [Google Scholar] [CrossRef]
- He, J.; Yu, Z.; Jiang, J.; Chen, S.; Fang, W.; Guan, Z.; Liao, Y.; Wang, Z.; Chen, F.; Wang, H. An eruption of LTR retrotransposons in the autopolyploid genomes of Chrysanthemum nankingense (Asteraceae). Plants 2022, 11, 315. [Google Scholar] [CrossRef]
- Waminal, N.E.; Pellerin, R.J.; Kim, N.S.; Jayakodi, M.; Park, J.Y.; Yang, T.J.; Kim, H.H. Rapid and efficient FISH using pre-labeled oligomer probes. Sci. Rep. 2018, 8, 8224. [Google Scholar] [CrossRef] [Green Version]
- Mahmoudi, S.; Mirzaghaderi, G. Tools for drawing informative idiograms. bioRxiv 2021. [Google Scholar] [CrossRef]
- He, Q.; Cai, Z.; Hu, T.; Liu, H.; Bao, C.; Mao, W.; Jin, W. Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.). BMC Plant Biol. 2015, 15, 105. [Google Scholar] [CrossRef] [Green Version]
- Shirabe, K.; Ryota, E.; Kaori, S.; Akio, K.; Scoles, G. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 2013, 56, 131–137. [Google Scholar]
- Luo, X.; Liu, J.; Wang, J.; Gong, W.; Chen, L.; Wan, W. FISH analysis of Zanthoxylum armatum based on oligonucleotides for 5S rDNA and (GAA)6. Genome 2018, 61, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Zhou, M.Q.; Hu, Z.L. Chromosomal localization of the 5S and 45S rDNA sites and 5S rDNA sequence analysis in Nelumbo Species. Agric. Sci. China 2011, 10, 679–685. [Google Scholar] [CrossRef]
- Liu, D.; Thakur, S.; Kumar, U.; Malik, R.; Bisht, D.; Balyan, P.; Mir, R.R.; Kumar, S. Physical localization of 45S rDNA in Cymbopogon and the analysis of differential distribution of rDNA in homologous chromosomes of Cymbopogon winterianus. PLoS ONE 2021, 16, e0257115. [Google Scholar]
- Pontes, O.; Neves, N.; Silva, M.; Lewis, M.S.; Madlung, A.; Comai, L.; Viegas, W.; Pikaard, C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 2004, 101, 18240–18245. [Google Scholar] [CrossRef] [Green Version]
- Li, K.P.; Wu, Y.X.; Zhao, H.; Wang, Y.; Lu, X.M.; Wang, J.M.; Xu, Y.; Li, Z.Y.; Han, Y.H. Cytogenetic relationships among Citrullus species in comparison with some genera of the tribe Benincaseae (Cucurbitaceae) as inferred from rDNA distribution patterns. BMC Evol. Biol. 2016, 16, 85. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.G.P.M., Jr. Conservative distribution of 5S rDNA loci in Schizodon (Pisces, Anostomidae) chromosomes. Chromosome Res. 2000, 8, 353–355. [Google Scholar] [CrossRef]
- Kamisugi, Y.; Nakayama, S.; O’Neil, C.M.; Mathias, R.J.; Trick, M.; Fukui, K. Visualization of the Brassica self-incompatibility S-locus on identified oilseed rape chromosomes. Plant Mol. Biol. 1998, 38, 1081–1087. [Google Scholar] [CrossRef]
- Taketa, S.; Harrison, G.E.; Heslop-Harrison, J.S. Comparative physical mapping of the 5S and 18S-25S rDNA in nine wild Hordeum species and cytotypes. Appl. Genet. 1999, 98, 1–9. [Google Scholar] [CrossRef]
- Abd El-Twab, M.H.; Kondo, K. Genome Mutation revealed by artificial hybridization between Chrysanthemum yoshinaganthum and Chrysanthemum vestitum assessed by FISH and GISH. J. Bot. 2012, 2012, 480310. [Google Scholar] [CrossRef] [Green Version]
- Matoba, H.; Uchiyama, H. Physical mapping of 5S rDNA, 18S rDNA and telomere sequences in three species of the genus Artemisia (Asteraceae) with distinct basic chromosome numbers. Cytologia 2009, 74, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gituru, W.R.; Wang, Q.F. Distribution of basic diploid and polyploid species of Isoetes in East Asia. J. Biogeogr. 2004, 31, 1239–1250. [Google Scholar] [CrossRef]
- Dai, X.K.; Yang, Y.J.; Liu, X. Transplanting experiment and transcriptome sequencing reveal the potential ecological adaptation to plateau environments in the allopolyploid Isoetessinensis. Aquat. Bot. 2021, 172, 103394. [Google Scholar] [CrossRef]
- Huang, Y.; An, Y.M.; Meng, S.Y.; Guo, Y.P.; Rao, G.Y. Taxonomic status and phylogenetic position of Phaeostigma in the subtribe Artemisiinae (Asteraceae). J. Syst. Evol. 2017, 55, 426–436. [Google Scholar] [CrossRef]
- Chen, X.; Wang, H.; Yang, X.; Jiang, J.; Ren, G.; Wang, Z.; Dong, X.; Chen, F. Small-scale alpine topography at low latitudes and high altitudes: Refuge areas of the genus Chrysanthemum and its allies. Hortic. Res. 2020, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Bernardes, E.C.; Benko-Iseppon, A.M.; Vasconcelos, S.; Carvalho, R.; Brasileiro-Vidal, A.C. Intra- and interspecific chromosome polymorphisms in cultivated Cichorium L. species (Asteraceae). Genet. Mol. Biol. 2013, 36, 357–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Melo, N.F.; Guerra, M. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann. Bot. 2003, 92, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Chiarini, F.E. Variation in rDNA loci of polyploid Solanum elaeagnifolium (Solanaceae). N. Z. J. Bot. 2014, 52, 277–284. [Google Scholar] [CrossRef]
- Roa, F.; Guerra, M. Non-Random Distribution of 5S rDNA sites and its association with 45S rDNA in plant chromosomes. Cytogenet. Genome Res. 2015, 146, 243–249. [Google Scholar] [CrossRef]
- Rosato, M.; Alvarez, I.; Nieto Feliner, G.; Rossello, J.A. High and uneven levels of 45S rDNA site-number variation across wild populations of a diploid plant genus (Anacyclus, Asteraceae). PLoS ONE 2017, 12, e0187131. [Google Scholar] [CrossRef] [Green Version]
- Mazzella, C.; Rodriguez, M.; Vaio, M.; Gaiero, P.; Lopez-Carro, B.; Santinaque, F.F.; Folle, G.A.; Guerra, M. Karyological Features of Achyrocline (Asteraceae, Gnaphalieae): Stable karyotypes, low DNA content variation, and linkage of rRNA genes. Cytogenet. Genome Res. 2010, 128, 169–176. [Google Scholar] [CrossRef]
- Matoba, H.; Mizutani, T.; Nagano, K.; Hoshi, Y.; Uchiyama, H. Chromosomal study of lettuce and its allied species (Lactuca spp., Asteraceae) by means of karyotype analysis and fluorescence in situ hybridization. Hereditas 2007, 144, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Dydak, M.; Kolano, B.; Nowak, T.; Siwinska, D.; Maluszynska, J. Cytogenetic studies of three European species of Centaurea L. (Asteraceae). Hereditas 2009, 146, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ganley, A.R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 2005, 309, 1581–1584. [Google Scholar] [CrossRef] [PubMed]
- Raskina, O.; Barber, J.C.; Nevo, E.; Belyayev, A. Repetitive DNA and chromosomal rearrangements: Speciation-related events in plant genomes. Cytogenet. Genome Res. 2008, 120, 351–357. [Google Scholar] [CrossRef]
- Hanson, R.E.; Islam-Faridi, M.N.; Percival, E.A.; Crane, C.F.; Ji, Y.; McKnight, T.D.; Stelly, D.M.; Price, H.J. Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 1996, 105, 55–61. [Google Scholar] [CrossRef]
- Huang, M.; Li, H.; Zhang, L.; Gao, F.; Wang, P.; Hu, Y.; Yan, S.; Zhao, L.; Zhang, Q.; Tan, J.; et al. Plant 45S rDNA clusters are fragile sites, and their instability is associated with epigenetic alterations. PLoS ONE 2012, 7, e35139. [Google Scholar] [CrossRef] [Green Version]
- Rosato, M.; Moreno-Saiz, J.C.; Galian, J.A.; Rossello, J.A. Evolutionary site-number changes of ribosomal DNA loci during speciation: Complex scenarios of ancestral and more recent polyploid events. AoB Plants 2015, 7, plv135. [Google Scholar] [CrossRef] [Green Version]
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148. [Google Scholar] [CrossRef]
- Srisuwan, S.; Sihachakr, D.; Siljak-Yakovlev, S. The origin and evolution of sweet potato (Ipomoea batatas Lam.) and its wild relatives through the cytogenetic approaches. Plant Sci. 2006, 171, 424–433. [Google Scholar] [CrossRef]
- Pikaard, C.S. Nucleolar dominance and silencing of transcription. Trends Plant Sci. 1999, 4, 478–483. [Google Scholar] [CrossRef]
- Chen, Z.J. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 2007, 58, 377–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
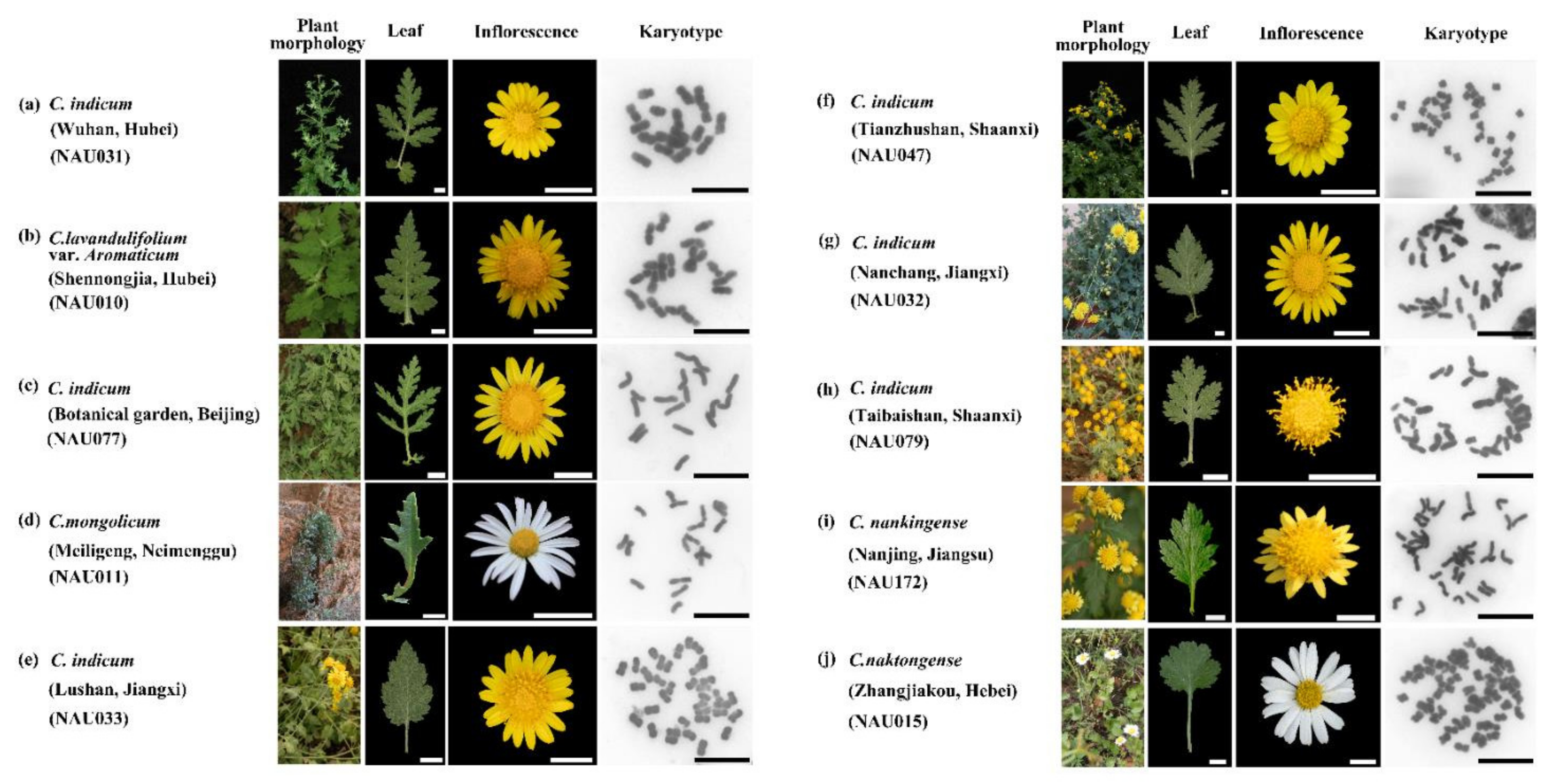
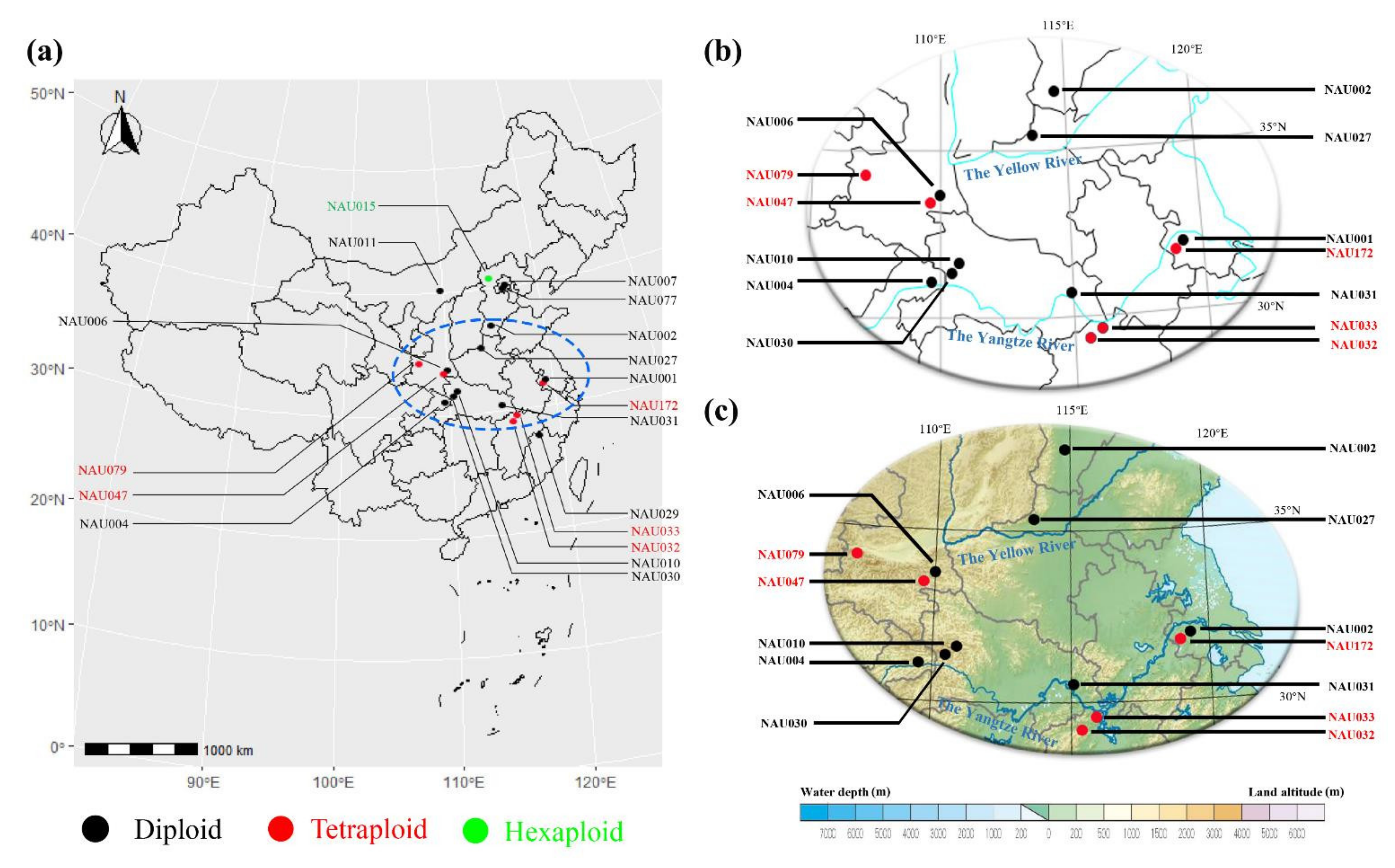
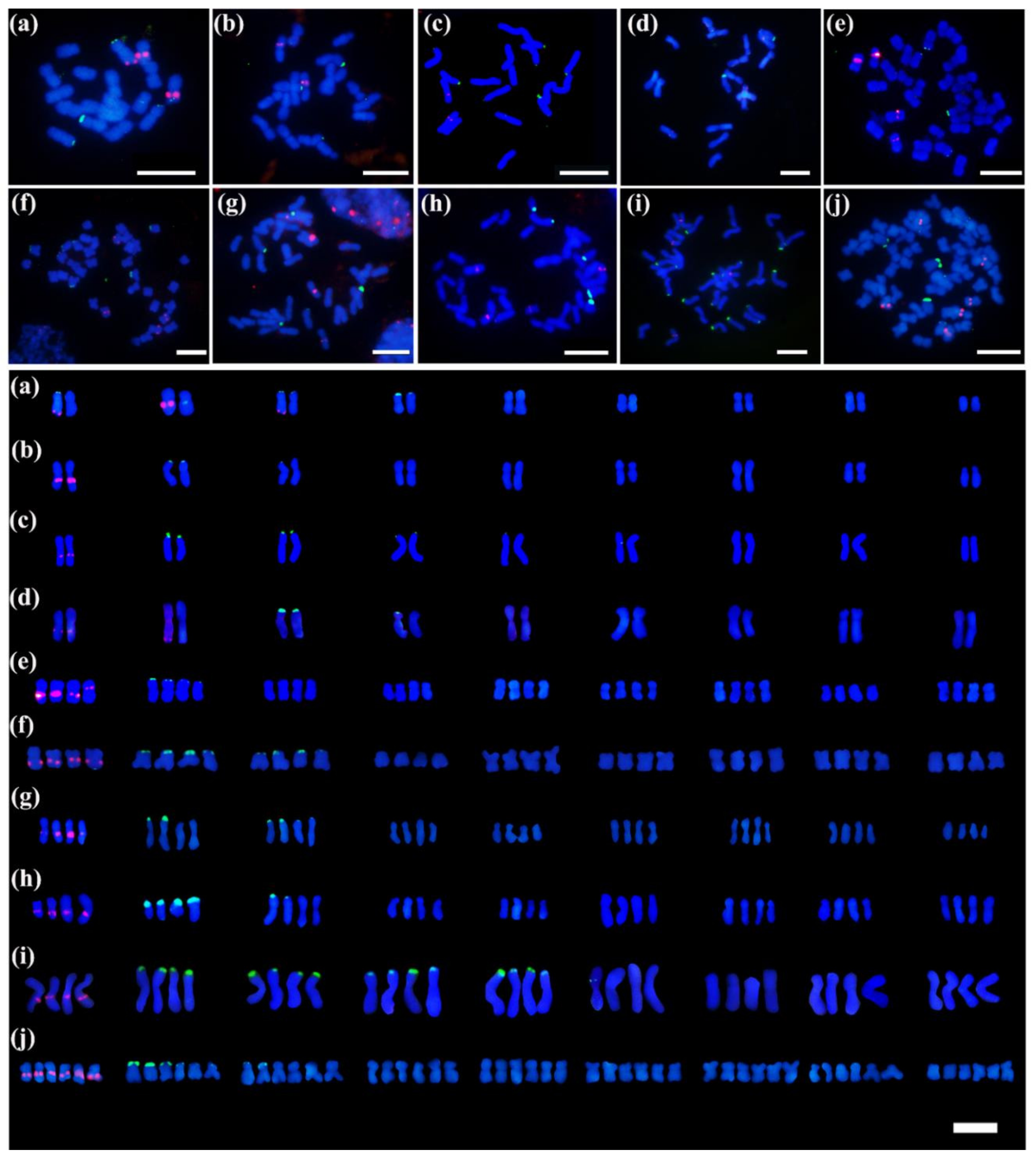
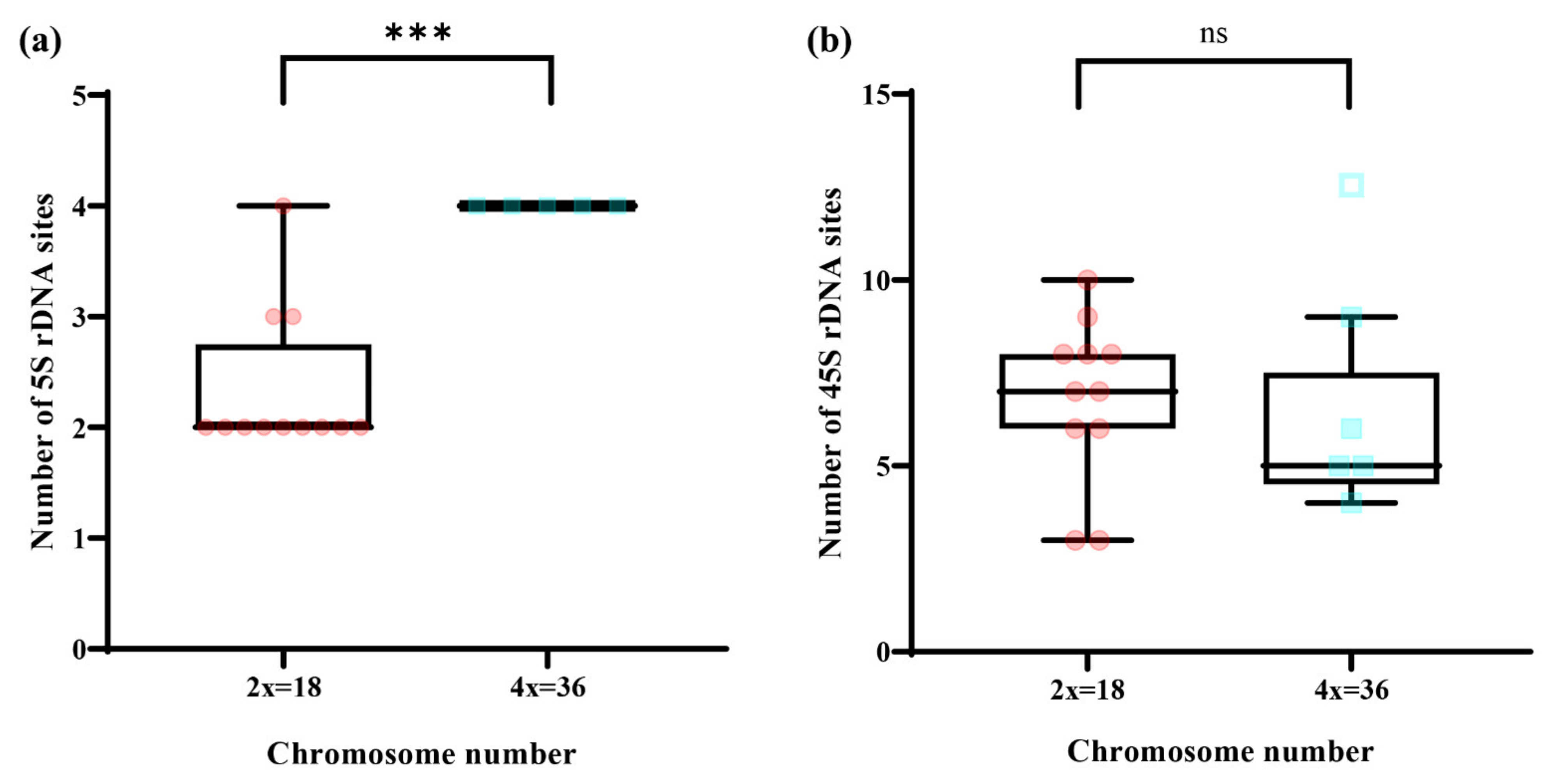

| Accession Number | Taxon | Source | No. of 5S rDNA Sites | No. of 45S rDNA Sites | Chromosome Number | Source of Data |
|---|---|---|---|---|---|---|
| NAU001 | Chrysanthemum nankingense | Nanjing, Jiangsu | 2 | 7 | 2n = 2x = 18 | He et al. [5] |
| NAU002 | C. dichrum | Xingtai, Hebei | 2 | 8 | 2n = 2x = 18 | He et al. [5] |
| NAU004 | C. rhombifolium | Wushan, Chongqing | 4 | 5 | 2n = 2x = 18 | He et al. [5] |
| NAU006 | C. potentilloides | Tianzhushan, Shaanxi | 2 | 9 | 2n = 2x = 18 | He et al. [5] |
| NAU007 | C. lavandulifolium | Botanical garden, Beijing | 2 | 7 | 2n = 2x = 18 | He et al. [5] |
| NAU010 | C.lavandulifolium var. aromaticum | Shennongjia, Hubei | 2 | 3 | 2n = 2x = 18 | This study |
| NAU011 | C.mongolicum | Meiligeng, Neimenggu | 3 | 3 | 2n = 2x = 18 | This study |
| NAU015 | C.naktongense | Zhangjiakou, Hebei | 6 | 8 | 2n = 6x = 54 | This study |
| NAU027 | indicum | Yuntaishan, Henan | 2 | 8 | 2n = 2x = 18 | He et al. [5] |
| NAU029 | C. indicum | Wuyishan, Fujian | 2 | 6 | 2n = 2x = 18 | He et al. [5] |
| NAU030 | C. indicum | Shennongjia, Hubei | 2 | 10 | 2n = 2x = 18 | He et al. [5] |
| NAU031 | C. indicum | Wuhan, Hubei | 3 | 6 | 2n = 2x = 18 | This study |
| NAU032 | C. indicum | Nanchang, Jiangxi | 4 | 6 | 2n = 4x = 36 | This study |
| NAU033 | C. indicum | Lushan, Jiangxi | 4 | 4 | 2n = 4x = 36 | This study |
| NAU047 | C. indicum | Tianzhushan, Shaanxi | 4 | 9 | 2n = 4x = 36 | This study |
| NAU077 | C. indicum | Botanical garden, Beijing | 2 | 8 | 2n = 2x = 18 | This study |
| NAU079 | C. indicum | Taibaishan, Shaanxi | 4 | 5 | 2n = 4x = 36 | This study |
| NAU172 | C. nankingense | Nanjing, Jiangsu | 4 | 17 | 2n = 4x = 36 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Zhao, Y.; Zhang, S.; He, Y.; Jiang, J.; Chen, S.; Fang, W.; Guan, Z.; Liao, Y.; Wang, Z.; et al. Uneven Levels of 5S and 45S rDNA Site Number and Loci Variations across Wild Chrysanthemum Accessions. Genes 2022, 13, 894. https://doi.org/10.3390/genes13050894
He J, Zhao Y, Zhang S, He Y, Jiang J, Chen S, Fang W, Guan Z, Liao Y, Wang Z, et al. Uneven Levels of 5S and 45S rDNA Site Number and Loci Variations across Wild Chrysanthemum Accessions. Genes. 2022; 13(5):894. https://doi.org/10.3390/genes13050894
Chicago/Turabian StyleHe, Jun, Yong Zhao, Shuangshuang Zhang, Yanze He, Jiafu Jiang, Sumei Chen, Weimin Fang, Zhiyong Guan, Yuan Liao, Zhenxing Wang, and et al. 2022. "Uneven Levels of 5S and 45S rDNA Site Number and Loci Variations across Wild Chrysanthemum Accessions" Genes 13, no. 5: 894. https://doi.org/10.3390/genes13050894
APA StyleHe, J., Zhao, Y., Zhang, S., He, Y., Jiang, J., Chen, S., Fang, W., Guan, Z., Liao, Y., Wang, Z., Chen, F., & Wang, H. (2022). Uneven Levels of 5S and 45S rDNA Site Number and Loci Variations across Wild Chrysanthemum Accessions. Genes, 13(5), 894. https://doi.org/10.3390/genes13050894











