Genome-Wide Identification, Characterization, and Expression Profiling Analysis of SPL Gene Family during the Inflorescence Development in Trifolium repens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of TrSPL Genes
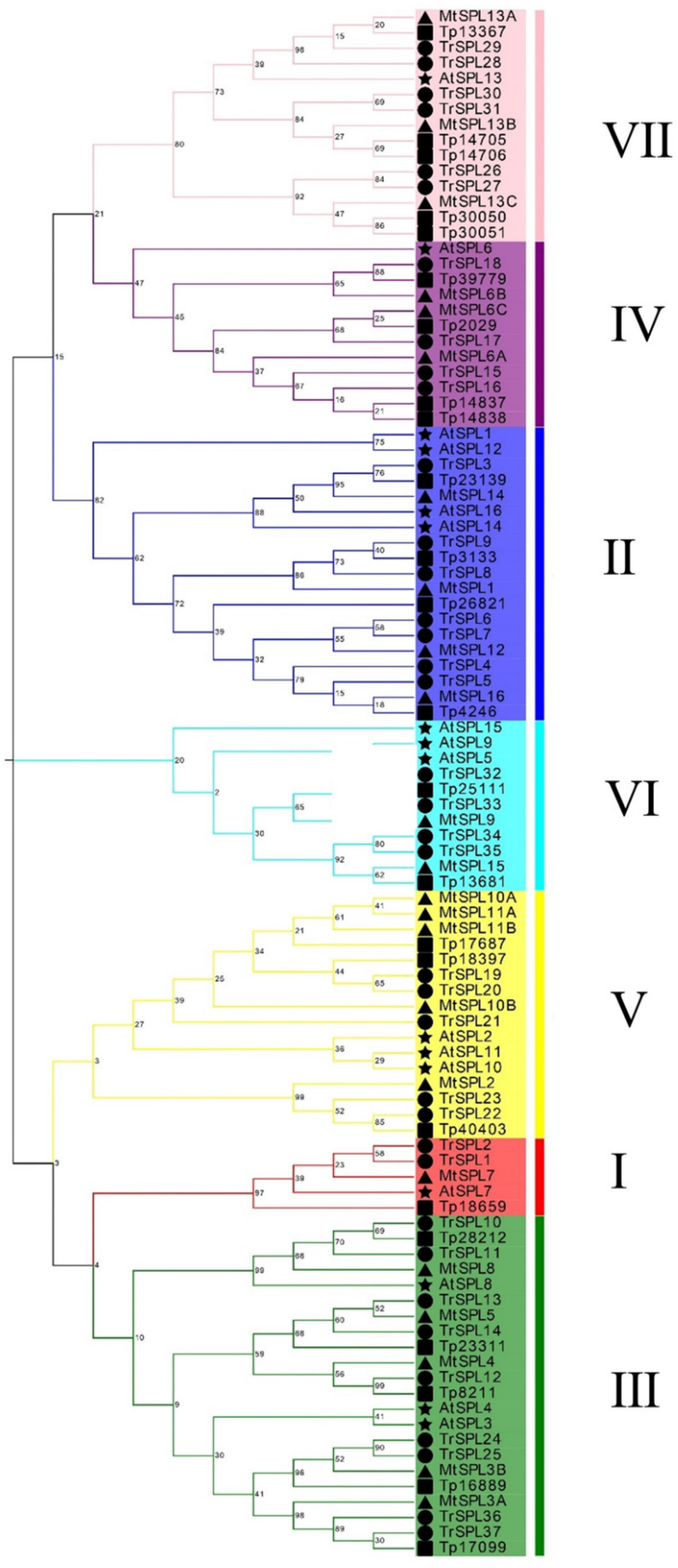
2.2. Phylogenetic Analyses and Classification of TrSPL Genes
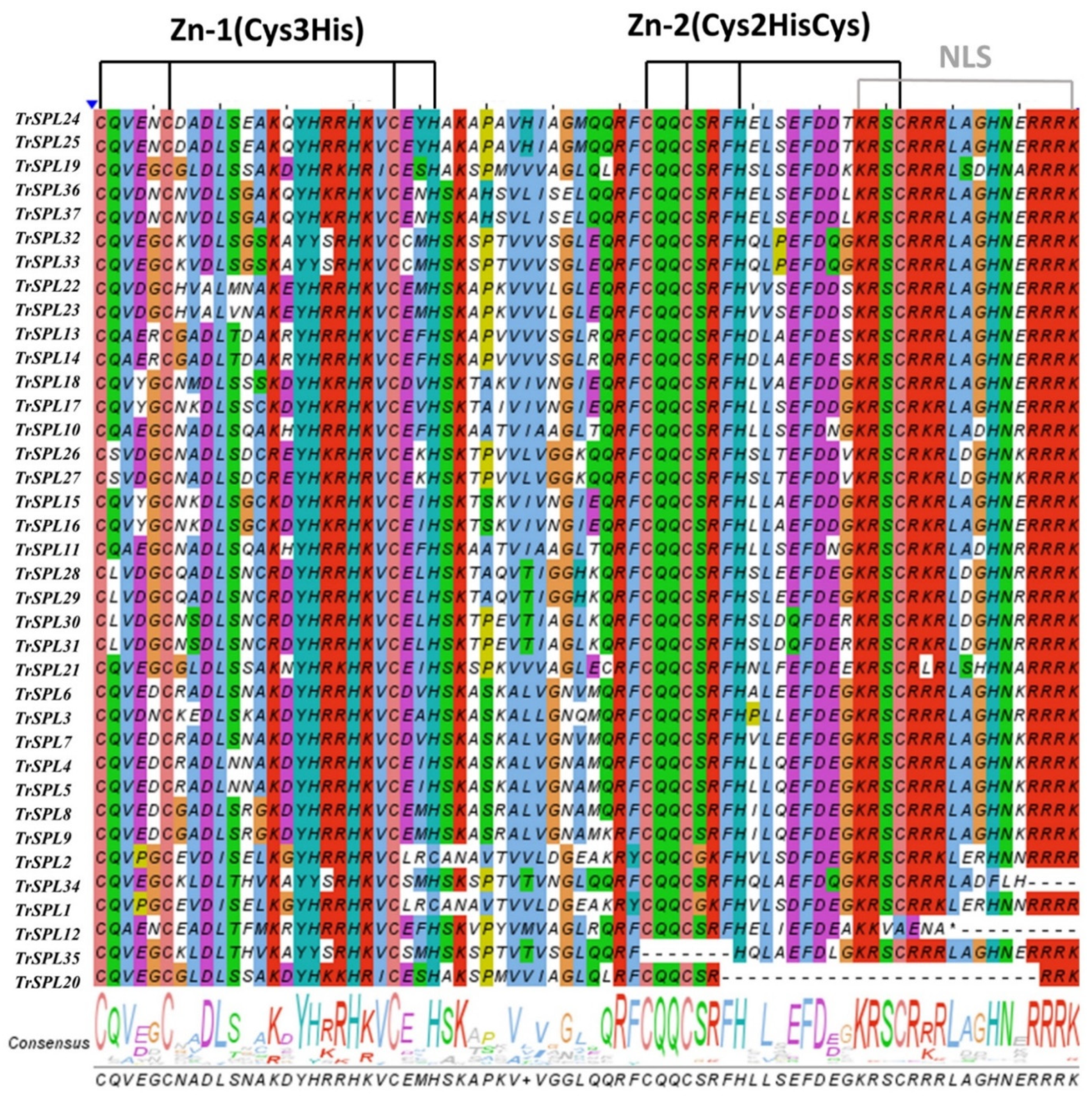
2.3. Gene Structure and Motif Analysis of TrSPL Genes
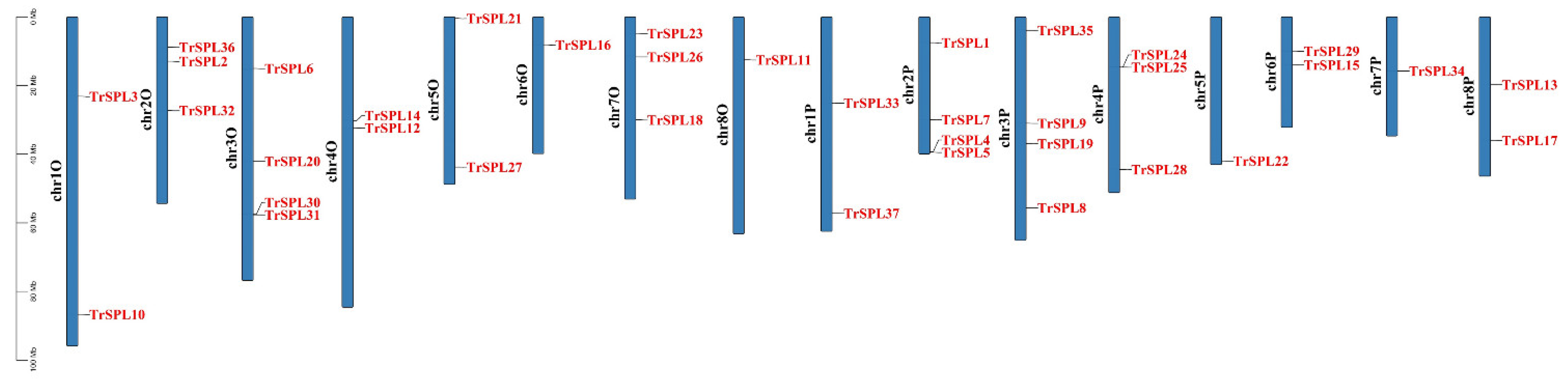
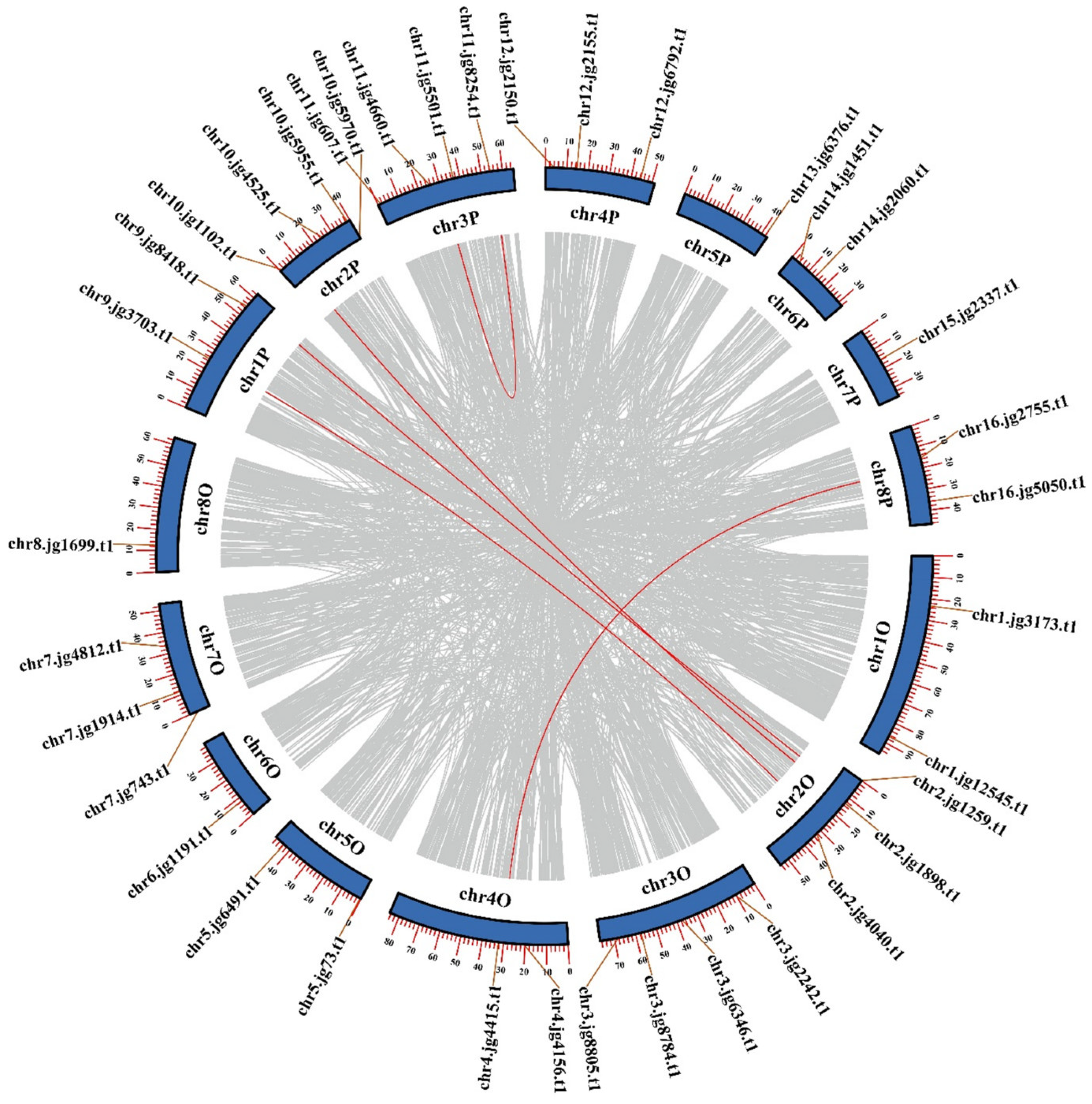
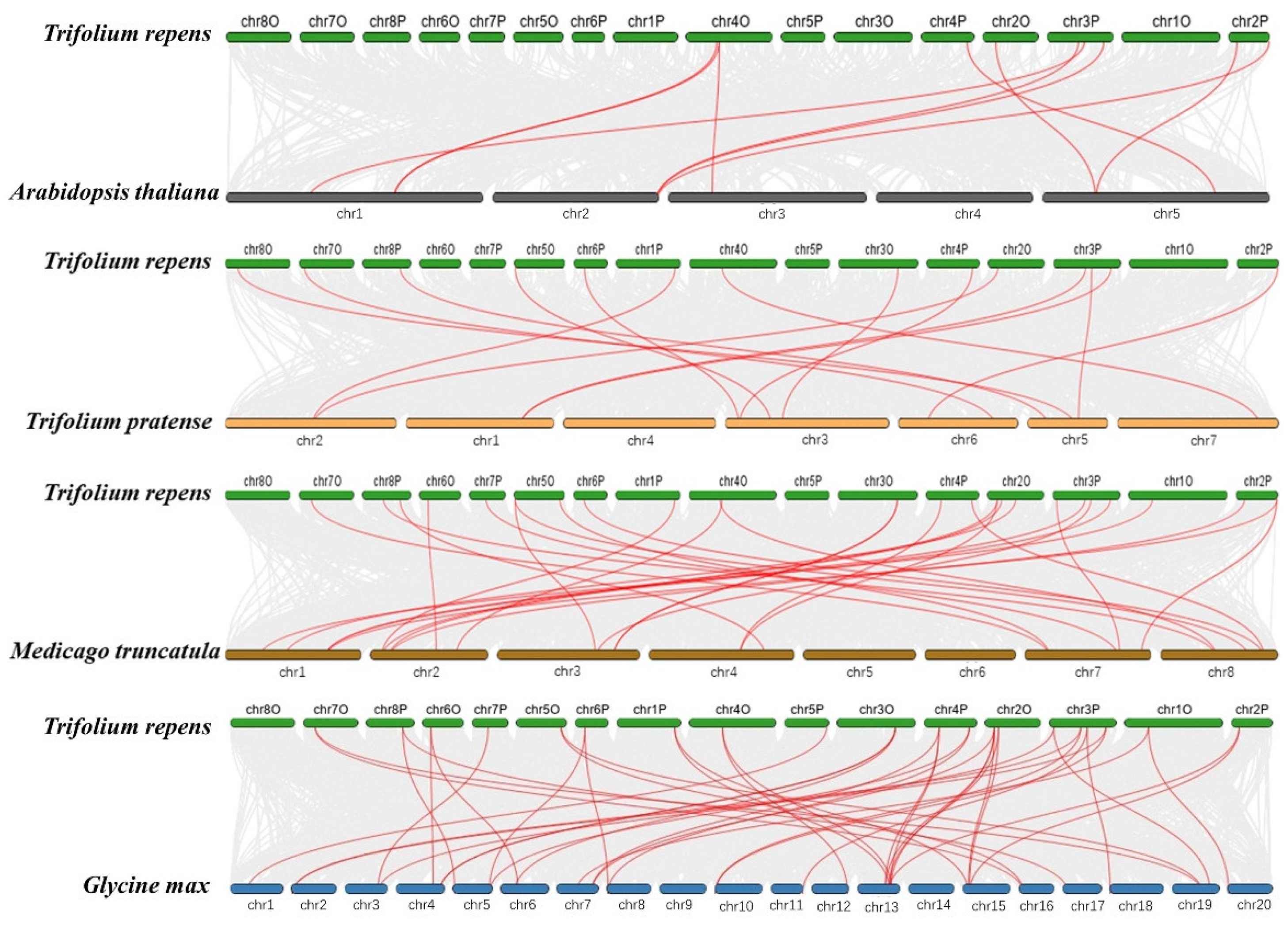
2.4. Chromosomal Locations and Synteny Analysis of TrSPL Genes
2.5. Material Culture, Sampling and qRT-PCR
3. Results
3.1. Genome-Wide Identification of T. repens SPL Genes
3.2. Phylogenetic Analyses and Classification of the TrSPL Gene Family
3.3. Sequence Feature and Gene Structure of TrSPL Genes
3.4. Chromosomal Locations and Synteny Analysis of TrSPL Genes
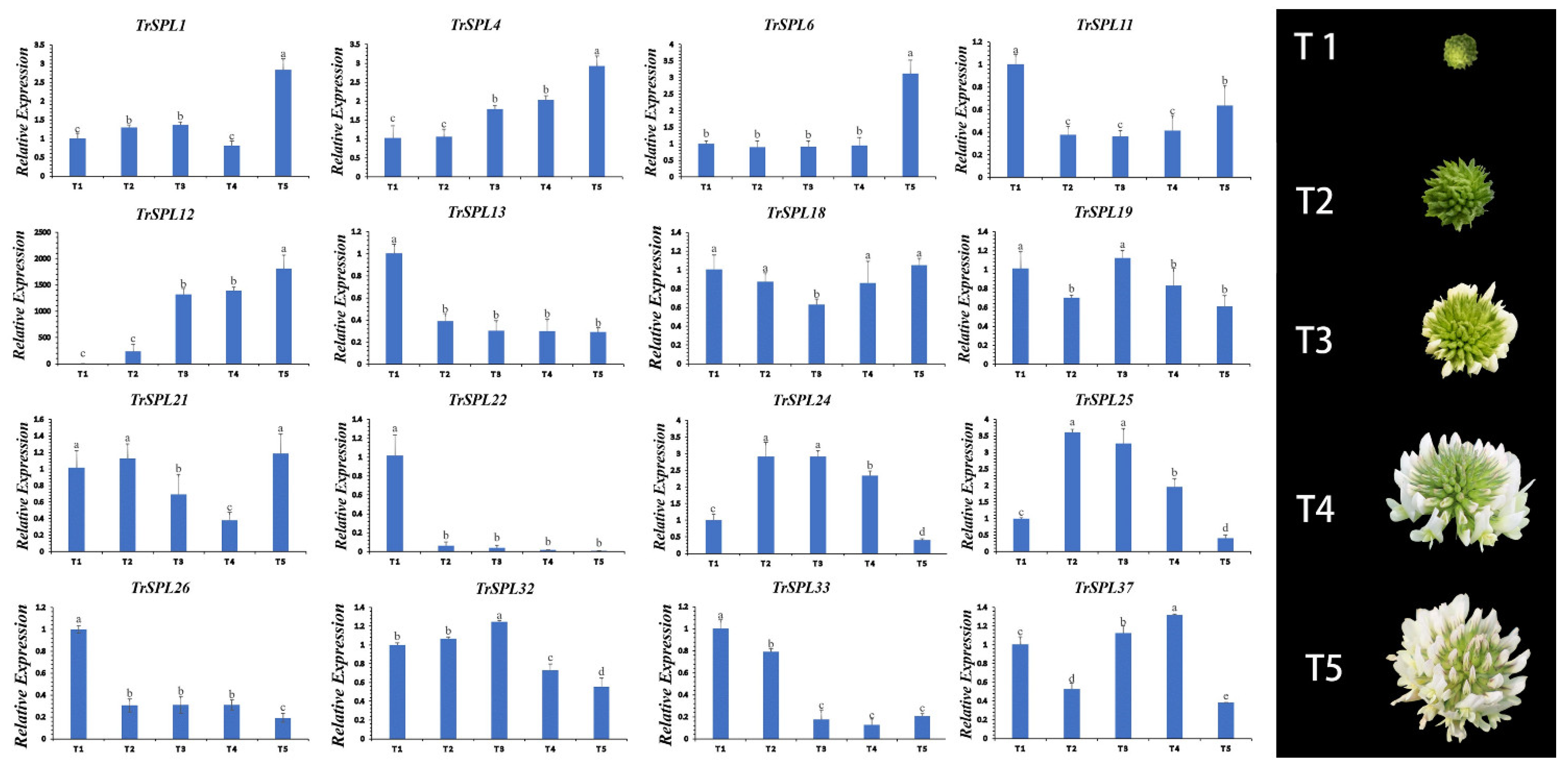
3.5. Expression Patterns of TrSPL Genes in Different Inflorescence Development Stage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SPL | Squa promoter binding protein–like |
| SBP | Squamosa promoter binding protein |
| CDS | Coding sequence |
| HMM | Hidden Markov model |
| MW | Molecular weight |
| PI | Isoelectric point |
| NLS | Nuclear localization signal |
References
- Daday, H. Gene frequencies in wild populations of Trifolium repens L. Heredity 1958, 12, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Coombe, D. Trifolium occidentale, a new species related to T. repens L. Watsonia 1961, 5, 68–87. [Google Scholar]
- Raffl, C.; Holderegger, R.; Parson, W.; Erschbamer, B. Patterns in genetic diversity of Trifolium pallescens populations do not reflect chronosequence on alpine glacier forelands. Heredity 2008, 100, 526–532. [Google Scholar] [CrossRef] [Green Version]
- Birte, B.; Bernadette, J.; Đura, K.; John, H. Legume Seed Production Meeting Market Requirements and Economic Impacts. Crit. Rev. Plant Sci. 2014, 34, 412–427. [Google Scholar] [CrossRef]
- Abberton, M.T.; Marshall, A.H. Progress in breeding perennial clovers for temperate agriculture. J. Agric. Sci. 2005, 143, 117–135. [Google Scholar] [CrossRef]
- Caradus, J.; Woodfield, D.; Stewart, A. Overview and vision for white clover. NZGA Res. Pract. Ser. 1995, 6, 1–6. [Google Scholar] [CrossRef]
- Gibson, P.; Cope, W. White clover. Clover Sci. Technol. 1985, 25, 471–490. [Google Scholar]
- Kilcher, M.R. Plant Development, Stage of Maturity and Nutrient Composition. J. Range Manag. 1981, 34, 363. [Google Scholar] [CrossRef] [Green Version]
- Fiorella, D.B.N.; Toshihiko, Y. Molecular Regulation of Flowering Time in Grasses. Agronomy 2017, 7, 17. [Google Scholar] [CrossRef]
- Christian, J. Flowering time regulation: Agrochemical control of flowering. Nat. Plants 2017, 3, 17045. [Google Scholar] [CrossRef]
- Pederson, G.A.; Brink, G.E. Seed Production of White Clover Cultivars and Naturalized Populations when Grown in a Pasture. Crop Sci. 2000, 40, 1109–1114. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, A.G.; Moraga, R.; Tausen, M.; Gupta, V.; Bilton, T.P.; Campbell, M.A.; Ashby, R.; Nagy, I.; Khan, A.; Larking, A.; et al. Breaking Free: The Genomics of Allopolyploidy-Facilitated Niche Expansion in White Clover. Plant Cell 2019, 31, 1466–1487. [Google Scholar] [CrossRef] [Green Version]
- Cardon, G.; Hohmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. MGG 1996, 250, 7–16. [Google Scholar]
- Yamaguchi, A.; Wu, M.-F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The MicroRNA-Regulated SBP-Box Transcription Factor SPL3 Is a Direct Upstream Activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Bong Soo, P.; Hui-Zhu, M.; Jun Sung, S.; Naohiko, O.; Ying, L.; Niu, Y.; Nur Fatimah Binte, M.; Chung-Hao, H.; Nam-Hai, C. Regulation of flowering time by SPL10/MED25 module in Arabidopsis. New Phytol. 2019, 224, 493–504. [Google Scholar] [CrossRef]
- Unte, U.S.; Sorensen, A.-M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Francesca, G.; Fabio, F. SPL transcription factors prevent inflorescence reversion in rice. Mol. Plant 2021, 14, 1041–1043. [Google Scholar] [CrossRef]
- Lei, W.; Qifa, Z. Boosting Rice Yield by Fine-Tuning SPL Gene Expression. Trends Plant Sci. 2017, 22, 643–646. [Google Scholar] [CrossRef]
- Ting, Z.; Yue, L.; Liting, M.; Xiaoying, W.; Dazhong, Z.; Yucui, H.; Qin, D.; Lingjian, M. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef]
- Guangyan, F.; Jiating, H.; Zhongfu, Y.; Qiuxu, L.; Yang, S.; Xiaoheng, X.; Gang, N.; Linkai, H.; Wei, L.; Xinquan, Z. Genome-wide identification, phylogenetic analysis, and expression analysis of the SPL gene family in orchardgrass (Dactylis glomerata L.). Genomics 2021, 113, 2413–2425. [Google Scholar] [CrossRef]
- Wang, H.F.; Lu, Z.C.; Xu, Y.T.; Kong, L.C.; Shi, J.J.; Liu, Y.F.; Fu, C.X.; Wang, X.S.; Wang, Z.Y.; Zhou, C.E.; et al. Genome-wide characterization of SPL family in Medicago truncatula reveals the novel roles of miR156/SPL module in spiky pod development. BMC Genom. 2019, 20, 14. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Jia, T.; Hou, J.; Iqbal, M.Z.; Zhang, Y.; Cheng, B.; Feng, H.; Li, Z.; Liu, L.; Zhou, J.; Feng, G.; et al. Overexpression of the white clover TrSAMDC1 gene enhanced salt and drought resistance in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 165, 147–160. [Google Scholar] [CrossRef]
- Preston, J.C.; Hileman, L.C. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 2015, 8, 677–688. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Zheng, F.; Wang, J.; Zhang, C.; Xiao, F.; Ye, J.; Li, C.; Ye, Z.; Zhang, J. miR156a-targeted SBP-Box transcription factor SlSPL13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnol. J. 2020, 18, 1670–1682. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.X.; Zhang, Z.L.; Zheng, J.J.; Lu, X.L.; Zhang, X.Q.; Xue, D.W. Genome-wide identification, phylogenetic and expression analysis of SBP-box gene family in barley (Hordeum vulgare L.). Plant Growth Regul. 2020, 90, 137–149. [Google Scholar] [CrossRef]
- Li, J.; Gao, X.Y.; Sang, S.Y.; Liu, C.N. Genome-wide identification, phylogeny, and expression analysis of the SBP-box gene family in Euphorbiaceae. BMC Genom. 2019, 20, 912–915. [Google Scholar] [CrossRef]
- Riese, M.; Hoehmann, S.; Saedler, H.; Muenster, T.; Huijser, P. Comparative analysis of the SBP-box gene families in P-patens and seed plants. Gene 2007, 401, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Frame, J.; Charlton, J.; Laidlaw, A.S. Temperate Forage Legumes; Cab International: London, UK, 1998. [Google Scholar]
- Huijser, P.; Klein, J.; Lönnig, W.; Meijer, H.; Saedler, H.; Sommer, H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992, 11, 1239–1249. [Google Scholar] [CrossRef]
- Mandel, M.A.; Cindy, G.-B.; Beth, S.; Martin, F.Y. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef]
- Li, M.Y.; He, Q.; Zhang, Y.; Sun, B.; Luo, Y.; Zhang, Y.; Chen, Q.; Wang, Y.; Zhang, F.; Zhang, Y.T.; et al. New insights into the evolution of the SBP-box family and expression analysis of genes in the growth and development of Brassica juncea. Biotechnol. Biotechnol. Equip. 2020, 34, 810–824. [Google Scholar] [CrossRef]
- Zhou, L.; Quan, S.W.; Ma, L.; Xu, H.; Yang, J.P.; Niu, J.X. Molecular characterization of SBP-box gene family during floral induction in walnut (Juglans regia L.). Tree Genet. Genomes 2019, 16, 12. [Google Scholar] [CrossRef]
- Cheng, H.T.; Hao, M.Y.; Wang, W.X.; Mei, D.S.; Tong, C.B.; Wang, H.; Liu, J.; Fu, L.; Hu, Q. Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol. 2016, 16, 196. [Google Scholar] [CrossRef] [Green Version]
- Schneeberger, K.; Ossowski, S.; Ott, F.; Klein, J.D.; Wang, X.; Lanz, C.; Smith, L.M.; Cao, J.; Fitz, J.; Warthmann, N. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA 2011, 108, 10249–10254. [Google Scholar] [CrossRef] [Green Version]
- Takuji, S.; International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 2005, 436, 793–800. [Google Scholar] [CrossRef]
- Xuewei, L.; Ling, K.; Jing, Z.; Yinpeng, X.; Liping, W.; Yan, Y.; Na, W.; Jidi, X.; Cuiying, L.; Wen, W.; et al. Improved hybrid de novo genome assembly of domesticated apple (Malus × domestica). GigaScience 2016, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Kang, L.; Qian, L.; Zheng, M.; Chen, L.; Chen, H.; Yang, L.; You, L.; Yang, B.; Yan, M.; Gu, Y. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat. Genet. 2021, 53, 1392–1402. [Google Scholar] [CrossRef]
- Jia-Ming, S.; Zhilin, G.; Jianlin, H.; Chaocheng, G.; Zhiquan, Y.; Shuo, W.; Dongxu, L.; Bo, W.; Shaoping, L.; Run, Z.; et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef]
- Warren, M.W.; Nicholas, W.E.; Helal, A.A.; Isabelle, M.V.; Hussain, S.W. Experimental evidence for the ancestry of allotetraploid Trifolium repens and creation of synthetic forms with value for plant breeding. BMC Plant Biol. 2012, 12, 55. [Google Scholar] [CrossRef] [Green Version]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [Green Version]
- Hua, Z.; Weilong, K.; Ziyun, G.; Xinyi, F.; Xiaoxiao, D.; Chang, L.; Yangsheng, L. Evolutionary Analyses Reveal Diverged Patterns of SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family in Oryza Genus. Front. Plant Sci. 2019, 10, 565. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, S.S.; Chen, F.; Liu, B.J.; Wu, L.; Li, F.; Zhang, J.Q.; Bao, M.Z.; Liu, G.F. Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genom. 2018, 19, 193. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.D.; Sun, L.D.; Zhou, Y.Z.; Yang, W.R.; Cheng, T.R.; Wang, J.; Zhang, Q.X. Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genom. 2015, 290, 1701–1715. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.H.; Hohmann, S.; Nettesheim, K.; Saedler, H.; Huijser, P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. Cell Mol. Biol. 1997, 12, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Meenu, S.P.; Shisong, M.; Tessa, M.B.-S.; Kirk, C.; Peter, H.; Savithramma, P.D.-K. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef] [Green Version]
- Hongmin, H.; Qin, Y.; Xiping, W.; Hui, X. A SBP-Box Gene VpSBP5 from Chinese Wild Vitis Species Responds to Erysiphe necator and Defense Signaling Molecules. Plant Mol. Biol. Rep. 2013, 31, 1261–1270. [Google Scholar] [CrossRef]
- Nomoto, M.; Skelly, M.J.; Itaya, T.; Mori, T.; Suzuki, T.; Matsushita, T.; Tokizawa, M.; Kuwata, K.; Mori, H.; Yamamoto, Y.Y. Suppression of MYC transcription activators by the immune cofactor NPR1 fine-tunes plant immune responses. Cell Rep. 2021, 37, 110125. [Google Scholar] [CrossRef]
- Anna, S.; Simone, A.; Karen, H.; Bikram Datt, P.; Wolf-Rüdiger, S.; Isabel, B. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [Green Version]
- Hu-De, M.; Li-Juan, Y.; Zhan-Jie, L.; Yan, Y.; Ran, H.; Hui, L.; Meng, M. Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sergei, A.F.; Mitra, A.; Valerie, N.F.; Molly, M. Identification of transcription factors from NF-Y, NAC, and SPL families responding to osmotic stress in multiple tomato varieties. Plant Sci. 2018, 274, 441–450. [Google Scholar] [CrossRef]
- Jianwen, W.; Youju, Y.; Meng, X.; Liguo, F.; Li-an, X. Roles of the SPL gene family and miR156 in the salt stress responses of tamarisk (Tamarix chinensis). BMC Plant Biol. 2019, 19, 370. [Google Scholar] [CrossRef] [Green Version]


| Name | PI | MW (Da) | Length (aa) | Subcellular Localization |
|---|---|---|---|---|
| TrSPL1 | 6.12 | 86,752.27 | 773 | nucleus |
| TrSPL2 | 6.88 | 67,153.17 | 591 | nucleus |
| TrSPL3 | 8.18 | 109,672.44 | 989 | plasma membrane |
| TrSPL4 | 5.75 | 111,233.87 | 1010 | endomembrane system |
| TrSPL5 | 5.72 | 111,303.94 | 1014 | endomembrane system |
| TrSPL6 | 7.01 | 116,204.16 | 1053 | nucleus |
| TrSPL7 | 8.51 | 115,211.08 | 1044 | endomembrane system |
| TrSPL8 | 5.8 | 111,853.78 | 1004 | nucleus |
| TrSPL9 | 5.86 | 110,745.6 | 995 | nucleus |
| TrSPL10 | 8.92 | 37,297.64 | 335 | nucleus |
| TrSPL11 | 9.14 | 40,922.71 | 363 | nucleus |
| TrSPL12 | 6.9 | 14,269.05 | 124 | nucleus |
| TrSPL13 | 9.3 | 20,662.94 | 182 | nucleus |
| TrSPL14 | 9.41 | 20,574.88 | 182 | nucleus |
| TrSPL15 | 5.98 | 57,844.27 | 527 | nucleus |
| TrSPL16 | 6.46 | 55,524.04 | 509 | nucleus |
| TrSPL17 | 6.31 | 55,948.26 | 500 | nucleus |
| TrSPL18 | 8.97 | 30,286.98 | 267 | chloroplast |
| TrSPL19 | 8.88 | 48,472.58 | 439 | nucleus |
| TrSPL20 | 8.73 | 44,554.3 | 406 | nucleus |
| TrSPL21 | 8.17 | 45,859.11 | 408 | nucleus |
| TrSPL22 | 8.67 | 34,247.9 | 313 | nucleus |
| TrSPL23 | 8.67 | 34,313.96 | 312 | nucleus |
| TrSPL24 | 6.1 | 17,499.07 | 149 | nucleus |
| TrSPL25 | 6.1 | 17,472.04 | 149 | nucleus |
| TrSPL26 | 7.6 | 41,040.79 | 367 | nucleus |
| TrSPL27 | 7.61 | 40,962.63 | 367 | nucleus |
| TrSPL28 | 8.86 | 42,684.84 | 383 | nucleus |
| TrSPL29 | 8.62 | 42,817.9 | 385 | nucleus |
| TrSPL30 | 6.62 | 42,612.13 | 382 | nucleus |
| TrSPL31 | 6.78 | 42,977.52 | 387 | nucleus |
| TrSPL32 | 9.2 | 37,707.11 | 339 | chloroplast |
| TrSPL33 | 9.41 | 37,953.47 | 340 | nucleus |
| TrSPL34 | 6.99 | 37,614.66 | 343 | plasma membrane |
| TrSPL35 | 8.73 | 36,551.32 | 335 | nucleus |
| TrSPL36 | 7.04 | 16,415.99 | 142 | nucleus |
| TrSPL37 | 7.04 | 16,430.02 | 142 | nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Nie, G.; Yang, Z.; Ma, S.; Fan, J.; Hu, R.; Wu, F.; Zhang, X. Genome-Wide Identification, Characterization, and Expression Profiling Analysis of SPL Gene Family during the Inflorescence Development in Trifolium repens. Genes 2022, 13, 900. https://doi.org/10.3390/genes13050900
Ma J, Nie G, Yang Z, Ma S, Fan J, Hu R, Wu F, Zhang X. Genome-Wide Identification, Characterization, and Expression Profiling Analysis of SPL Gene Family during the Inflorescence Development in Trifolium repens. Genes. 2022; 13(5):900. https://doi.org/10.3390/genes13050900
Chicago/Turabian StyleMa, Jieyu, Gang Nie, Zhongfu Yang, Sainan Ma, Jinwan Fan, Ruchang Hu, Feifei Wu, and Xinquan Zhang. 2022. "Genome-Wide Identification, Characterization, and Expression Profiling Analysis of SPL Gene Family during the Inflorescence Development in Trifolium repens" Genes 13, no. 5: 900. https://doi.org/10.3390/genes13050900
APA StyleMa, J., Nie, G., Yang, Z., Ma, S., Fan, J., Hu, R., Wu, F., & Zhang, X. (2022). Genome-Wide Identification, Characterization, and Expression Profiling Analysis of SPL Gene Family during the Inflorescence Development in Trifolium repens. Genes, 13(5), 900. https://doi.org/10.3390/genes13050900







