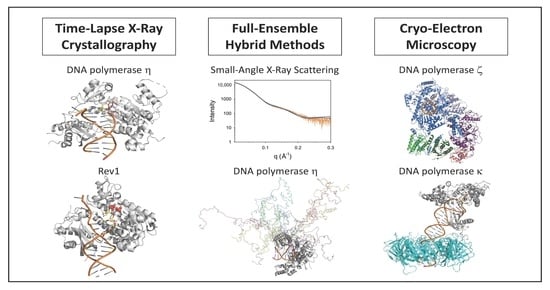
Recent Advances in Understanding the Structures of Translesion Synthesis DNA Polymerases
Abstract
:1. Introduction
2. Time-Lapse X-ray Crystallography
2.1. DNA Polymerase Eta
2.2. Rev1
3. Full-Ensemble Hybrid Methods
4. Cryo-Electron Microscopy
4.1. DNA Polymerase Zeta
4.2. DNA Polymerase Kappa and PCNA
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Friedberg, E.C.; Wagner, R.; Radman, M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 2002, 296, 1627–1630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, S.; Prakash, L. Translesion DNA synthesis in eukaryotes: A one- or two-polymerase affair. Genes Dev. 2002, 16, 1872–1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, A.R. Replication of damaged DNA. Cell Cycle 2003, 2, 300–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, S.; Johnson, R.E.; Prakash, L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu. Rev. Biochem. 2005, 74, 317–353. [Google Scholar] [CrossRef]
- Lehmann, A.R. Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett. 2005, 579, 873–876. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, A.R.; Niimi, A.; Ogi, T.; Brown, S.; Sabbioneda, S.; Wing, J.F.; Kannouche, P.L.; Green, C.M. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair 2007, 6, 891–899. [Google Scholar] [CrossRef]
- Waters, L.S.; Minesinger, B.K.; Wiltrout, M.E.; D’Souza, S.; Woodruff, R.V.; Walker, G.C. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 2009, 73, 134–154. [Google Scholar] [CrossRef] [Green Version]
- Washington, M.T.; Carlson, K.D.; Freudenthal, B.D.; Pryor, J.M. Variations on a theme: Eukaryotic Y-family DNA polymerases. Biochim. Biophys. Acta 2010, 1804, 1113–1123. [Google Scholar] [CrossRef] [Green Version]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Pryor, J.M.; Dieckman, L.M.; Boehm, E.M.; Washington, M.T. Eukaryotic Y-Family Polymerases: A Biochemical and Structural Perspective. In Nucleic Acid Polymerases; Murakami, K.S., Trakselis, M.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 30, pp. 85–108. [Google Scholar]
- Powers, K.T.; Washington, M.T. Eukaryotic translesion synthesis: Choosing the right tool for the job. DNA Repair 2018, 71, 127–134. [Google Scholar] [CrossRef]
- Marians, K.J. Lesion Bypass and the Reactivation of Stalled Replication Forks. Annu. Rev. Biochem. 2018, 87, 217–238. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.; Koonin, E.V.; Bruford, E.; Blanco, L.; Burtis, K.C.; Christman, M.F.; Copeland, W.C.; Friedberg, E.C.; Hanaoka, F.; Hinkle, D.C.; et al. Eukaryotic DNA polymerases: Proposal for a revised nomenclature. J. Biol. Chem. 2001, 276, 43487–43490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haracska, L.; Yu, S.L.; Johnson, R.E.; Prakash, L.; Prakash, S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 2000, 25, 458–461. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.D.; Washington, M.T. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase eta. Mol. Cell Biol. 2005, 25, 2169–2176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haracska, L.; Unk, I.; Johnson, R.E.; Johansson, E.; Burgers, P.M.; Prakash, S.; Prakash, L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001, 15, 945–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryor, J.M.; Washington, M.T. Pre-steady state kinetic studies show that an abasic site is a cognate lesion for the yeast Rev1 protein. DNA Repair 2011, 10, 1138–1144. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.E.; Prakash, S.; Prakash, L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science 1999, 283, 1001–1004. [Google Scholar] [CrossRef]
- Washington, M.T.; Johnson, R.E.; Prakash, S.; Prakash, L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc. Natl. Acad. Sci. USA 2000, 97, 3094–3099. [Google Scholar] [CrossRef]
- Washington, M.T.; Prakash, L.; Prakash, S. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase eta. Proc. Natl. Acad. Sci. USA 2003, 100, 12093–12098. [Google Scholar] [CrossRef] [Green Version]
- Washington, M.T.; Minko, I.G.; Johnson, R.E.; Wolfle, W.T.; Harris, T.M.; Lloyd, R.S.; Prakash, S.; Prakash, L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases iota and kappa. Mol. Cell. Biol. 2004, 24, 5687–5693. [Google Scholar] [CrossRef] [Green Version]
- Pence, M.G.; Blans, P.; Zink, C.N.; Hollis, T.; Fishbein, J.C.; Perrino, F.W. Lesion bypass of N2-ethylguanine by human DNA polymerase iota. J. Biol. Chem. 2009, 284, 1732–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.Y.; Angel, K.C.; Guengerich, F.P. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J. Biol. Chem. 2006, 281, 21062–21072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wu, X.; Guo, D.; Rechkoblit, O.; Wang, Z. Activities of human DNA polymerase kappa in response to the major benzo[a]pyrene DNA adduct: Error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair 2002, 1, 559–569. [Google Scholar] [CrossRef]
- Jha, V.; Ling, H. Structural basis of accurate replication beyond a bulky major benzo[a]pyrene adduct by human DNA polymerase kappa. DNA Repair 2017, 49, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Washington, M.T.; Minko, I.G.; Johnson, R.E.; Haracska, L.; Harris, T.M.; Lloyd, R.S.; Prakash, S.; Prakash, L. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase zeta. Mol. Cell. Biol. 2004, 24, 6900–6906. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.Y.; Guengerich, F.P. Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J. Biol. Chem. 2008, 283, 23645–23655. [Google Scholar] [CrossRef] [Green Version]
- Silverstein, T.D.; Jain, R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta. Structure 2010, 18, 1463–1470. [Google Scholar] [CrossRef] [Green Version]
- Patra, A.; Nagy, L.D.; Zhang, Q.; Su, Y.; Muller, L.; Guengerich, F.P.; Egli, M. Kinetics, structure, and mechanism of 8-Oxo-7,8-dihydro-2’-deoxyguanosine bypass by human DNA polymerase eta. J. Biol. Chem. 2014, 289, 16867–16882. [Google Scholar] [CrossRef] [Green Version]
- Silverstein, T.D.; Johnson, R.E.; Jain, R.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature 2010, 465, 1039–1043. [Google Scholar] [CrossRef]
- Biertümpfel, C.; Zhao, Y.; Kondo, Y.; Ramón-Maiques, S.; Gregory, M.; Lee, J.Y.; Masutani, C.; Lehmann, A.R.; Hanaoka, F.; Yang, W. Structure and mechanism of human DNA polymerase eta. Nature 2010, 465, 1044–1048. [Google Scholar] [CrossRef] [Green Version]
- Nair, D.T.; Johnson, R.E.; Prakash, S.; Prakash, L.; Aggarwal, A.K. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature 2004, 430, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.T.; Johnson, R.E.; Prakash, S.; Prakash, L.; Aggarwal, A.K. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 2005, 309, 2219–2222. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.T.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Protein-template-directed synthesis across an acrolein-derived DNA adduct by yeast Rev1 DNA polymerase. Structure 2008, 16, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.T.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. DNA synthesis across an abasic lesion by yeast REV1 DNA polymerase. J. Mol. Biol. 2011, 406, 18–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freudenthal, B.D.; Beard, W.A.; Wilson, S.H. New structural snapshots provide molecular insights into the mechanism of high-fidelity DNA synthesis. DNA Repair 2015, 32, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Freudenthal, B.D.; Beard, W.A.; Shock, D.D.; Wilson, S.H. Observing a DNA polymerase choose right from wrong. Cell 2013, 154, 157–168. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Zhao, Y.; Yamagata, Y.; Hua, Y.J.; Yang, W. Watching DNA polymerase eta make a phosphodiester bond. Nature 2012, 487, 196–201. [Google Scholar] [CrossRef] [Green Version]
- Weaver, T.M.; Cortez, L.M.; Khoang, T.H.; Washington, M.T.; Agarwal, P.K.; Freudenthal, B.D. Visualizing Rev1 catalyze protein-template DNA synthesis. Proc. Natl. Acad. Sci. USA 2020, 117, 25494–25504. [Google Scholar] [CrossRef]
- Perera, L.; Freudenthal, B.D.; Beard, W.A.; Pedersen, L.G.; Wilson, S.H. Revealing the role of the product metal in DNA polymerase β catalysis. Nucleic Acids Res. 2017, 45, 2736–2745. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Konigsberg, W.H. Two metal-ion catalysis: Inhibition of DNA polymerase activity by a third divalent metal ion. Front. Mol. Biosci. 2022, 9, 824794. [Google Scholar] [CrossRef]
- Malaby, A.W.; Chakravarthy, S.; Irving, T.C.; Kathuria, S.V.; Bilsel, O.; Lambright, D.G. Methods for analysis of size-exclusion chromatography-small-angle X-ray scattering and reconstruction of protein scattering. J. Appl. Crystallogr. 2015, 48, 1102–1113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambo, R.P.; Tainer, J.A. Bridging the solution divide: Comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr. Opin. Struct. Biol. 2010, 20, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boldon, L.; Laliberte, F.; Liu, L. Review of the fundamental theories behind small angle X-ray scattering, molecular dynamics simulations, and relevant integrated application. Nano Rev. 2015, 6, 25661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allison, J.R. Using simulation to interpret experimental data in terms of protein conformational ensembles. Curr. Opin. Struct. Biol. 2017, 43, 79–87. [Google Scholar] [CrossRef]
- Kikhney, A.G.; Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015, 589, 2570–2577. [Google Scholar] [CrossRef] [Green Version]
- Hub, J.S. Interpreting solution X-ray scattering data using molecular simulations. Curr. Opin. Struct. Biol. 2018, 49, 18–26. [Google Scholar] [CrossRef]
- Rambo, R.P.; Tainer, J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 2013, 496, 477–481. [Google Scholar] [CrossRef]
- Powers, K.T.; Gildenberg, M.S.; Washington, M.T. Modeling conformationally flexible proteins with X-ray scattering and molecular simulations. Comput. Struct. Biotechnol. J. 2019, 17, 570–578. [Google Scholar] [CrossRef]
- Pelikan, M.; Hura, G.L.; Hammel, M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen. Physiol. Biophys. 2009, 28, 174–189. [Google Scholar] [CrossRef]
- Powers, K.T.; Elcock, A.H.; Washington, M.T. The C-terminal region of translesion synthesis DNA polymerase η is partially unstructured and has high conformational flexibility. Nucleic Acids Res. 2018, 46, 2107–2120. [Google Scholar] [CrossRef] [Green Version]
- Bienko, M.; Green, C.M.; Crosetto, N.; Rudolf, F.; Zapart, G.; Coull, B.; Kannouche, P.; Wider, G.; Peter, M.; Lehmann, A.R.; et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 2005, 310, 1821–1824. [Google Scholar] [CrossRef] [PubMed]
- Ripley, B.M.; Reusch, D.T.; Washington, M.T. Yeast DNA polymerase η possesses two PIP-like motifs that bind PCNA and Rad6-Rad18 with different specificities. DNA Repair 2020, 95, 102968. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, H.; Hanafusa, T.; Ohashi, E.; Vaziri, C. Separate roles of structured and unstructured regions of Y-family DNA polymerases. Adv. Protein Chem. Struct. Biol. 2009, 78, 99–146. [Google Scholar] [PubMed] [Green Version]
- Johnson, R.E.; Washington, M.T.; Haracska, L.; Prakash, S.; Prakash, L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature 2000, 406, 1015–1019. [Google Scholar] [CrossRef]
- Baranovskiy, A.G.; Lada, A.G.; Siebler, H.M.; Zhang, Y.; Pavlov, Y.I.; Tahirov, T.H. DNA polymerase delta and zeta switch by sharing accessory subunits of DNA polymerase delta. J. Biol. Chem. 2012, 287, 17281–17287. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.E.; Prakash, L.; Prakash, S. Pol31 and Pol32 subunits of yeast DNA polymerase delta are also essential subunits of DNA polymerase zeta. Proc. Natl. Acad. Sci. USA 2012, 109, 12455–12460. [Google Scholar] [CrossRef] [Green Version]
- Makarova, A.V.; Stodola, J.L.; Burgers, P.M. A four-subunit DNA polymerase zeta complex containing Pol delta accessory subunits is essential for PCNA-mediated mutagenesis. Nucleic Acids Res. 2012, 40, 11618–11626. [Google Scholar] [CrossRef] [Green Version]
- Hara, K.; Hashimoto, H.; Murakumo, Y.; Kobayashi, S.; Kogame, T.; Unzai, S.; Akashi, S.; Takeda, S.; Shimizu, T.; Sato, M. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J. Biol. Chem. 2010, 285, 12299–12307. [Google Scholar] [CrossRef] [Green Version]
- Malik, R.; Kopylov, M.; Gomez-Llorente, Y.; Jain, R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Ubarretxena-Belandia, I.; Aggarwal, A.K. Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis. Nat. Struct. Mol. Biol 2020, 27, 913–924. [Google Scholar] [CrossRef]
- Jain, R.; Rice, W.J.; Malik, R.; Johnson, R.E.; Prakash, L.; Prakash, S.; Ubarretxena-Belandia, I.; Aggarwal, A.K. Cryo-EM structure and dynamics of eukaryotic DNA polymerase delta holoenzyme. Nat. Struct. Mol. Biol. 2019, 26, 955–962. [Google Scholar] [CrossRef]
- Washington, M.T.; Johnson, R.E.; Prakash, L.; Prakash, S. Human DINB1-encoded DNA polymerase kappa is a promiscuous extender of mispaired primer termini. Proc. Natl. Acad. Sci. USA 2002, 99, 1910–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haracska, L.; Prakash, L.; Prakash, S. Role of human DNA polymerase kappa as an extender in translesion synthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 16000–16005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lone, S.; Townson, S.A.; Uljon, S.N.; Johnson, R.E.; Brahma, A.; Nair, D.T.; Prakash, S.; Prakash, L.; Aggarwal, A.K. Human DNA polymerase kappa encircles DNA: Implications for mismatch extension and lesion bypass. Mol. Cell 2007, 25, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Irimia, A.; Eoff, R.L.; Guengerich, F.P.; Egli, M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase kappa opposite the 7,8-dihydro-8-oxo-2′-deoxyguanosine adduct. J. Biol. Chem. 2009, 284, 22467–22480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jha, V.; Bian, C.; Xing, G.; Ling, H. Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase kappa. Nucleic Acids Res. 2016, 44, 4957–4967. [Google Scholar] [CrossRef] [Green Version]
- Lancey, C.; Tehseen, M.; Bakshi, S.; Percival, M.; Takahashi, M.; Sobhy, M.A.; Raducanu, V.S.; Blair, K.; Muskett, F.W.; Ragan, T.J.; et al. Cryo-EM structure of human Pol kappa bound to DNA and mono-ubiquitylated PCNA. Nat. Commun. 2021, 12, 6095. [Google Scholar] [CrossRef]
- Yuan, Z.; Georgescu, R.; Schauer, G.D.; O’Donnell, M.E.; Li, H. Structure of the polymerase epsilon holoenzyme and atomic model of the leading strand replisome. Nat. Commun. 2020, 11, 3156. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, J.A.; Frevert, Z.; Washington, M.T. Recent Advances in Understanding the Structures of Translesion Synthesis DNA Polymerases. Genes 2022, 13, 915. https://doi.org/10.3390/genes13050915
Ling JA, Frevert Z, Washington MT. Recent Advances in Understanding the Structures of Translesion Synthesis DNA Polymerases. Genes. 2022; 13(5):915. https://doi.org/10.3390/genes13050915
Chicago/Turabian StyleLing, Justin A., Zach Frevert, and M. Todd Washington. 2022. "Recent Advances in Understanding the Structures of Translesion Synthesis DNA Polymerases" Genes 13, no. 5: 915. https://doi.org/10.3390/genes13050915
APA StyleLing, J. A., Frevert, Z., & Washington, M. T. (2022). Recent Advances in Understanding the Structures of Translesion Synthesis DNA Polymerases. Genes, 13(5), 915. https://doi.org/10.3390/genes13050915