Discovery and Functional Characterization of Two Regulatory Variants Underlying Lupus Susceptibility at 2p13.1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Meta-Analysis and Conditional Analysis
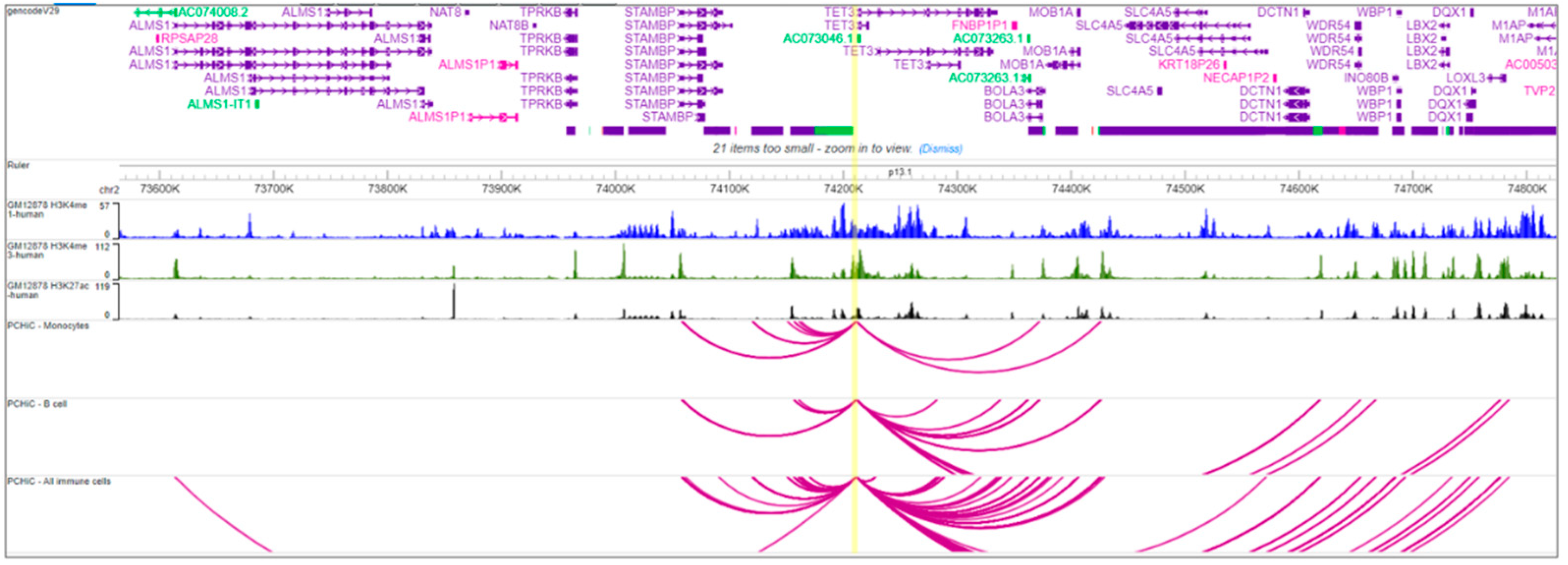
2.2. Bioinformatics, eQTL and sQTL Analysis, and 3D-Chromatin Interaction Analysis
2.3. Non-Coding RNA Interaction Analysis
2.4. Expression Level Analysis of DGUOK and DGUOK-AS1 in Four B-lymphocyte Lines
2.5. Luciferase Reporter Assay
2.6. Allele-Specific ChIP–qPCR
2.7. Western Blotting
3. Results
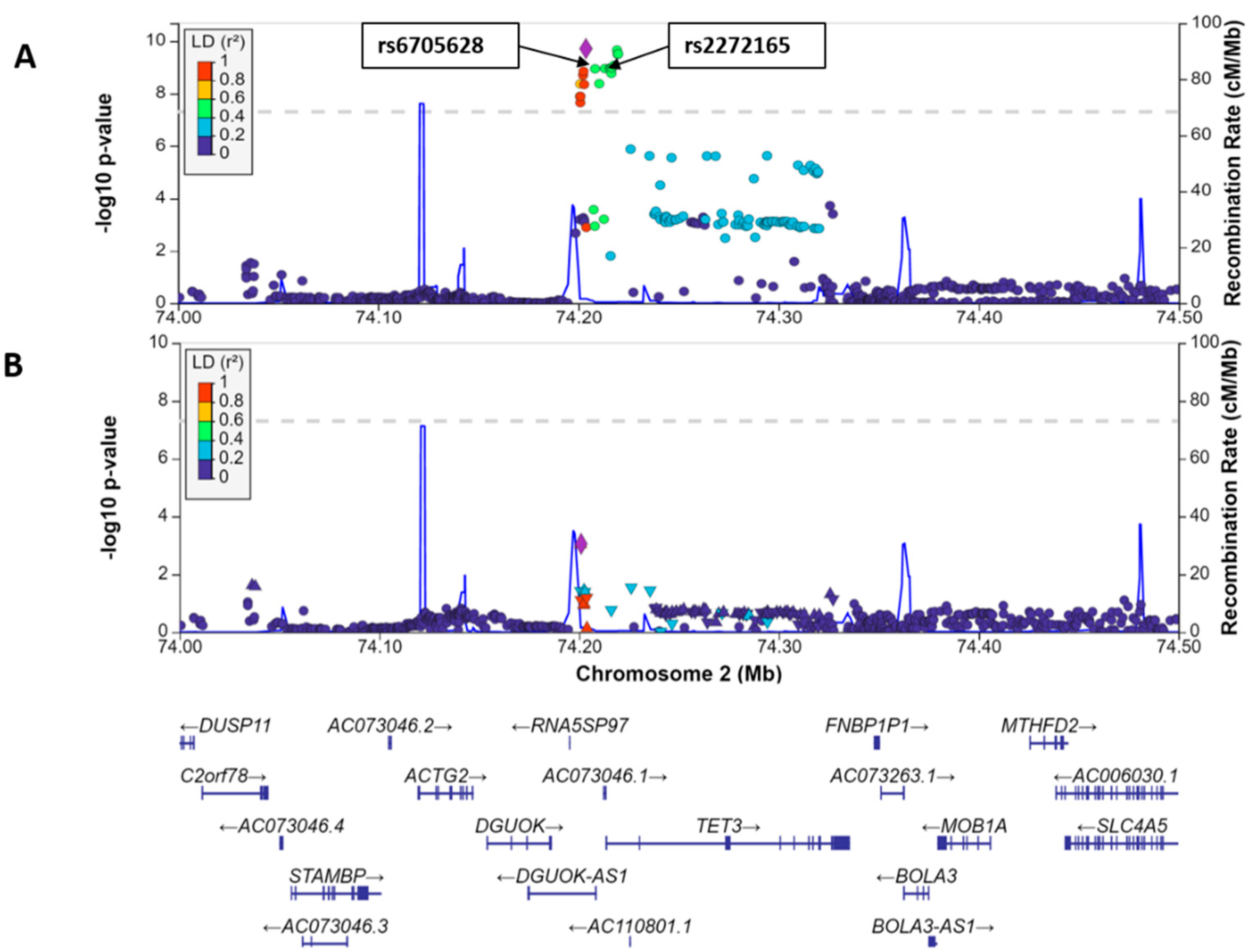
3.1. Meta-Analysis of Genome-Wide Association Studies
3.2. Bioinformatics of the Region
3.3. Non-Coding RNA Interactions
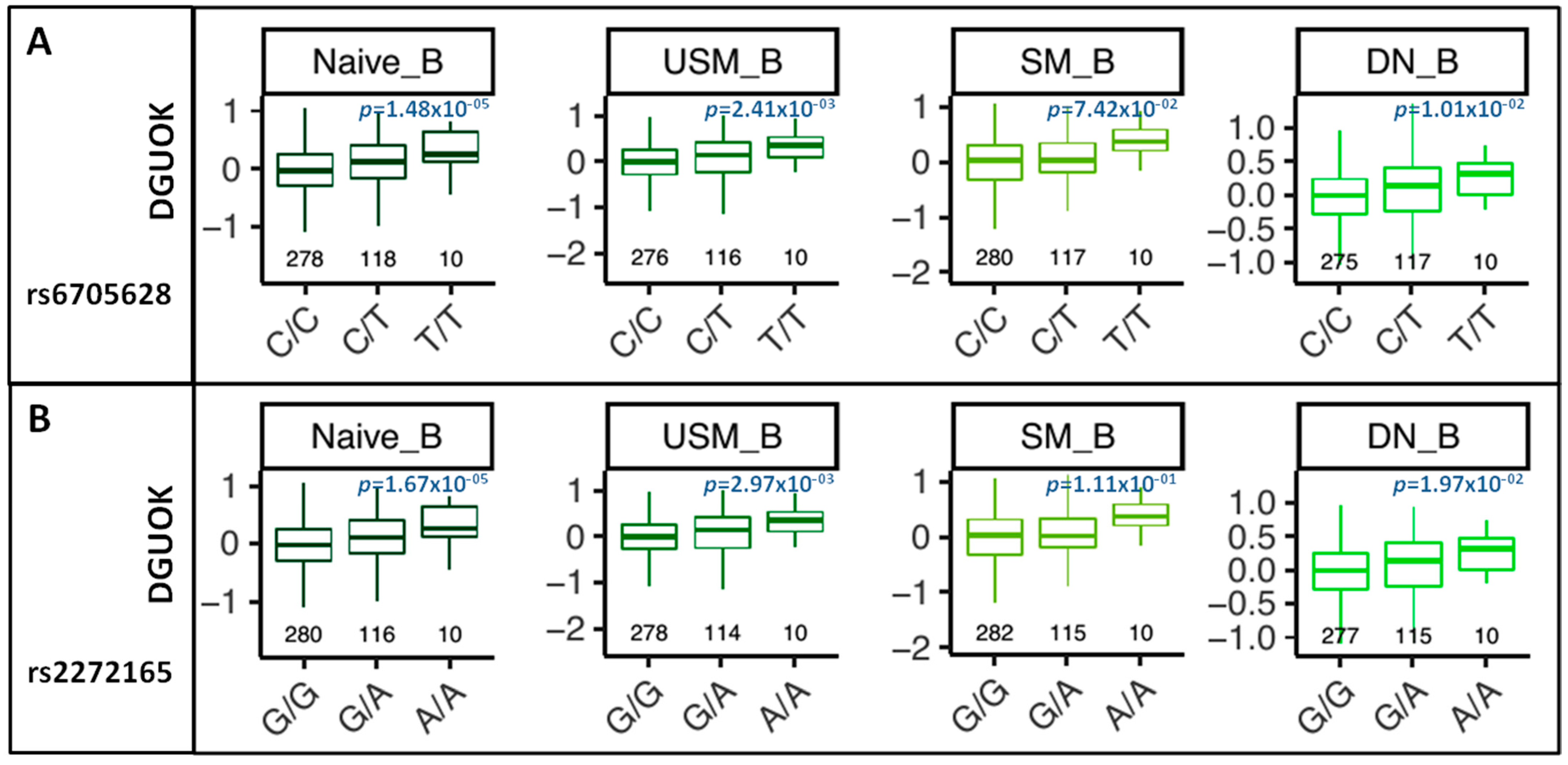
3.4. Relationship of DGUOK and DGUOK-AS1
3.5. eQTL and sQTL Analysis
3.6. Validation of Allele-Specific Regulatory Effects of SNPs
3.7. Differential Allele-Specific Binding of Regulatory Proteins to SNP Regions
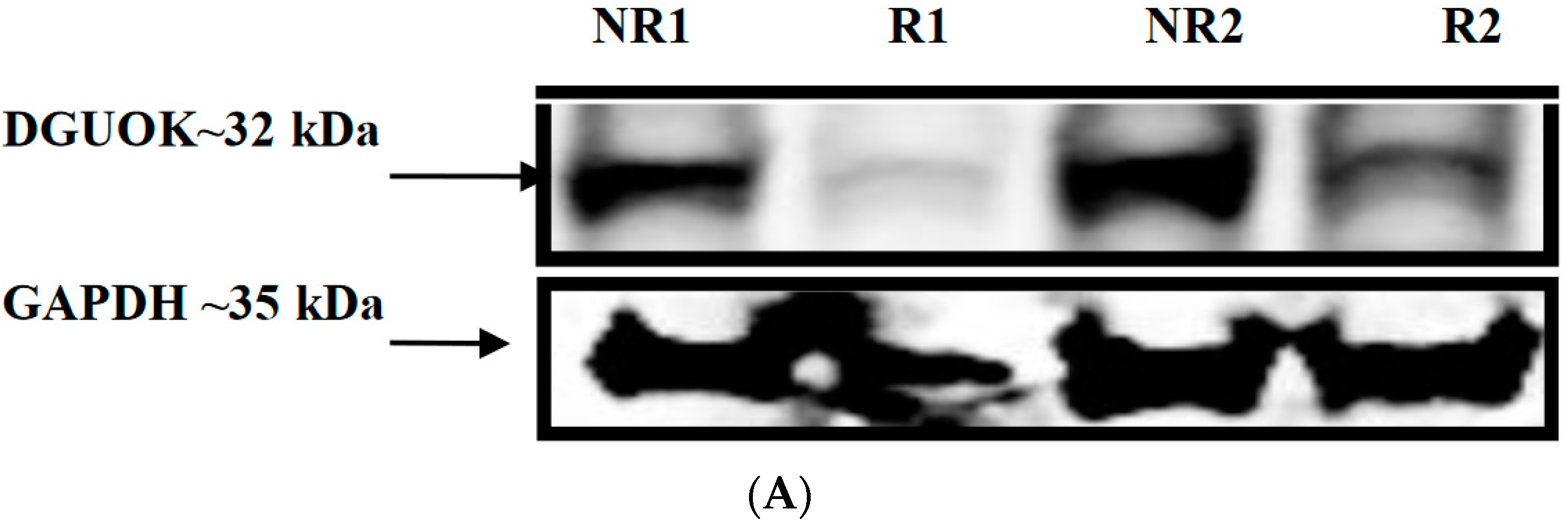
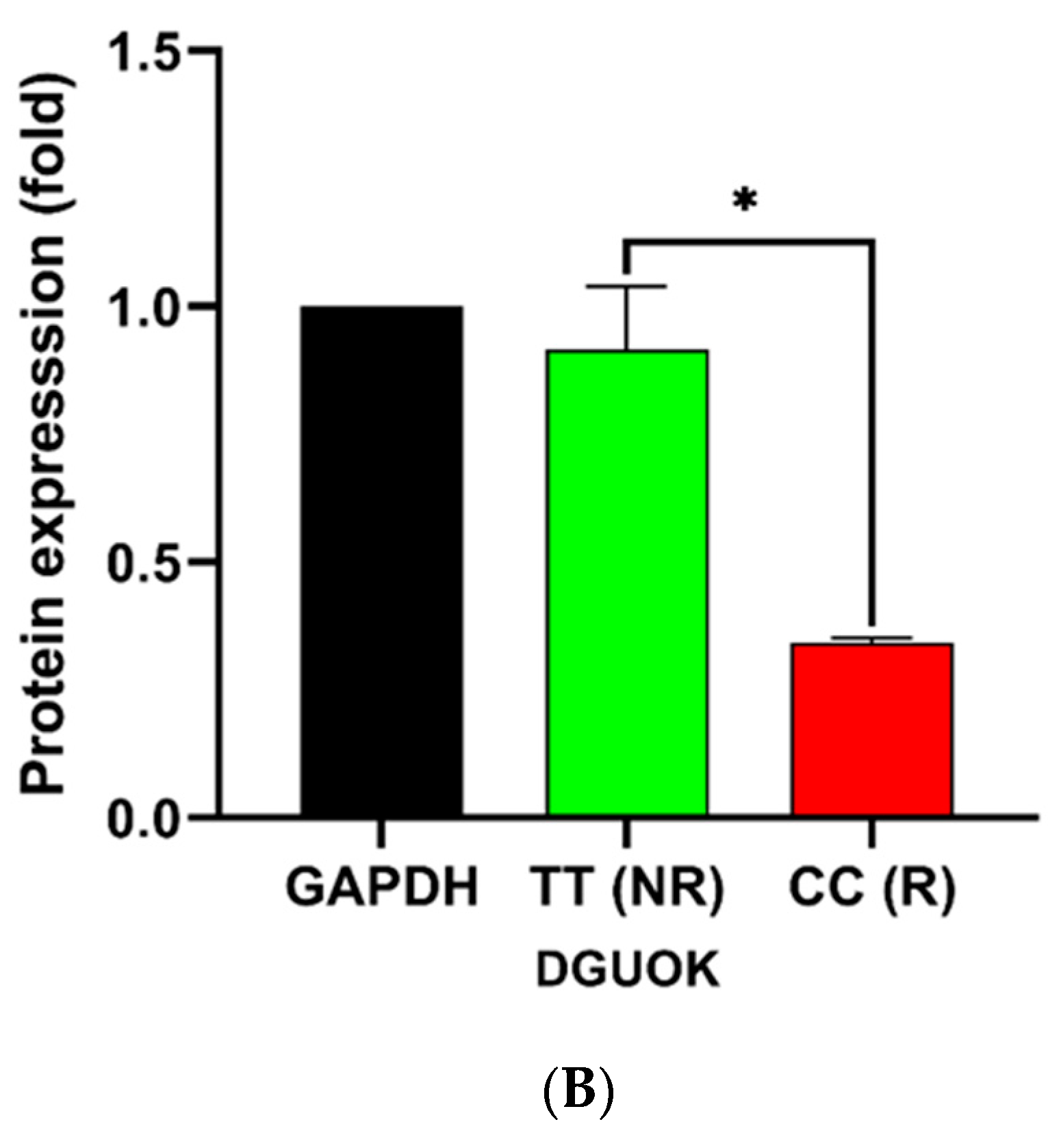
3.8. Western Blot Analysis of DGUOK
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Karrar, S.; Graham, D.S.C. Abnormal B Cell Development in Systemic Lupus Erythematosus: What the Genetics Tell Us. Arthritis Rheumatol. 2018, 70, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Morawski, P.A.; Bolland, S. Expanding the B Cell-Centric View of Systemic Lupus Erythematosus. Trends Immunol. 2017, 38, 373–382. [Google Scholar] [CrossRef] [Green Version]
- García-Carrasco, M.; Mendoza Pinto, C.; Solís Poblano, J.C.; Morales, I.E.; Cervera, R.; Anaya, J.M. Systemic lupus erythematosus. In Autoimmunity: From Bench to Bedside; Shoenfeld, Y., Anaya, J.M., Rojas-Villarraga, A., Levy, R.A., Cervera, R., Eds.; El Rosario University Press: Bogota, Colombia, 2013. [Google Scholar]
- Moulton, V.R.; Suarez-Fueyo, A.; Meidan, E.; Li, H.; Mizui, M.; Tsokos, G.C. Pathogenesis of Human Systemic Lupus Erythematosus: A Cellular Perspective. Trends Mol. Med. 2017, 23, 615–635. [Google Scholar] [CrossRef]
- Tsokos, G.C. Autoimmunity and organ damage in systemic lupus erythematosus. Nat. Immunol. 2020, 21, 605–614. [Google Scholar] [CrossRef]
- Izmirly, P.M.; Parton, H.; Wang, L.; McCune, W.J.; Lim, S.S.; Drenkard, C.; Ferucci, E.D.; Dall’Era, M.; Gordon, C.; Helmick, C.G.; et al. Prevalence of Systemic Lupus Erythematosus in the United States: Estimates From a Meta-Analysis of the Centers for Disease Control and Prevention National Lupus Registries. Arthritis Rheumatol. 2021, 73, 991–996. [Google Scholar] [CrossRef]
- Alarcon-Segovia, D.; Alarcón-Riquelme, M.E.; Cardiel, M.H.; Caeiro, F.; Massardo, L.; Villa, A.R.; Pons-Estel, B.A.; on behalf of the Grupo Latinoamericano de Estudio del Lupus Eritematoso (GLADEL). Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005, 52, 1138–1147. [Google Scholar] [PubMed]
- Drenkard, C.; Lim, S.S. Update on lupus epidemiology: Advancing health disparities research through the study of minority populations. Curr. Opin. Rheumatol. 2019, 31, 689–696. [Google Scholar] [CrossRef]
- Maningding, E.; Dall’Era, M.; Trupin, L.; Murphy, L.B.; Yazdany, J. Racial and Ethnic Differences in the Prevalence and Time to Onset of Manifestations of Systemic Lupus Erythematosus: The California Lupus Surveillance Project. Arthritis Care Res. 2020, 72, 622–629. [Google Scholar] [CrossRef]
- Deapen, D.; Escalante, A.; Weinrib, L.; Horwitz, D.; Bachman, B.; Roy-Burman, P.; Walker, A.; Mack, T.M. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992, 35, 311–318. [Google Scholar] [PubMed]
- Kuo, C.F.; Grainge, M.J.; Valdes, A.M.; See, L.; Luo, S.; Yu, K.; Zhang, W.; Doherty, M. Familial Aggregation of Systemic Lupus Erythematosus and Coaggregation of Autoimmune Diseases in Affected Families. JAMA Intern. Med. 2015, 175, 1518–1526. [Google Scholar] [CrossRef]
- Yin, X.; Kim, K.; Suetsugu, H.; Bang, S.; Wen, L.; Koido, M.; Ha, E.; Liu, L.; Sakamoto, Y.; Jo, S. Meta-analysis of 208370 East Asians identifies 113 susceptibility loci for systemic lupus erythematosus. Ann. Rheum. Dis. 2020, 80, 632–640. [Google Scholar] [CrossRef]
- Ha, E.; Bae, S.C.; Kim, K. Recent advances in understanding the genetic basis of systemic lupus erythematosus. Semin. Immunopathol. 2021, 44, 29–46. [Google Scholar] [CrossRef]
- Cannon, M.E.; Mohlke, K.L. Deciphering the Emerging Complexities of Molecular Mechanisms at GWAS Loci. Am. J. Hum. Genet. 2018, 103, 637–653. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Tang, H.; Zhang, Y.; Tang, X.; Zhang, J.; Sun, L.; Yang, J.; Cui, Y.; Zhang, L.; Hirankarn, N.; et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am. J. Hum. Genet. 2013, 92, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Morris, D.L.; Sheng, Y.; Zhang, Y.; Wang, Y.; Zhu, Z.; Tombleson, P.; Chen, L.; Cunninghame Graham, D.S.; Bentham, J.; Roberts, A.L.; et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 2016, 48, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Molineros, J.E.; Looger, L.L.; Zhou, X.; Kim, K.; Okada, Y.; Ma, J.; Qi, Y.; Kim-Howard, X.; Motghare, P.; et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 2016, 48, 323–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julià, A.; López-Longo, F.J.; Venegas, J.J.P.; Bonàs-Guarch, S.; Olivé, À.; Andreu, J.L.; Aguirre-Zamorano, M.Á.; Vela, P.; Nolla, J.M. Genome-wide association study meta-analysis identifies five new loci for systemic lupus erythematosus. Arthritis Res. Ther. 2018, 20, 100. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Willer, C.J.; Li, Y.; Abecasis, G.R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010, 26, 2190–2191. [Google Scholar] [CrossRef]
- Yang, J.; Ferreira, T.; Morris, A.P.; Medland, S.E.; Madden, P.A.F.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; Weedon, M.N.; Loos, R.J.; et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012, 44, 369–375. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendl, J.; Musil, M.; Štourač, J.; Zendulka, J.; Damborský, J.; Brezovský, J. PredictSNP2: A Unified Platform for Accurately Evaluating SNP Effects by Exploiting the Different Characteristics of Variants in Distinct Genomic Regions. PLoS Comput. Biol. 2016, 12, e1004962. [Google Scholar] [CrossRef]
- Ota, M.; Nagafuchi, Y.; Hatano, H.; Ishigaki, K.; Terao, C.; Takeshima, Y.; Yanaoka, H.; Kobayashi, S.; Okubo, M.; Shirai, H.; et al. Dynamic landscape of immune cell-specific gene regulation in immune-mediated diseases. Cell 2021, 184, 3006–3021.e17. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Purushotham, D.; Harrison, J.K.; Hsu, S.; Zhuo, X.; Fan, C.; Liu, S.; Xu, V.; Chen, S.; Xu, J.; et al. WashU Epigenome Browser update 2022. Nucleic Acids Res. 2022. [Google Scholar] [CrossRef]
- Javierre, B.M.; Burren, O.; Wilder, S.P.; Kreuzhuber, R.; Hill, S.; Sewitz, S.; Cairns, J.; Wingett, S.W.; Várnai, C.; Thiecke, M.J.; et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 2016, 167, 1369–1384.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Tastsoglou, S.; Skoufos, G.; Karavangeli, A.; Pierros, V.; Zacharopoulou, E.; Hatzigeorgiou, A.G. DIANA-LncBase v3: Indexing experimentally supported miRNA targets on non-coding transcripts. Nucleic Acids Res. 2019, 48, D101–D110. [Google Scholar] [CrossRef]
- Fukunaga, T.; Iwakiri, J.; Ono, Y.; Hamadae, M. LncRRIsearch: A Web Server for lncRNA-RNA Interaction Prediction Integrated With Tissue-Specific Expression and Subcellular Localization Data. Front. Genet. 2019, 10, 462. [Google Scholar] [CrossRef] [Green Version]
- Osorio, D.; Yu, X.; Serpedin, E.; Cai, J.J. Single-cell RNA sequencing of a European and an African lymphoblastoid cell line. Sci. Data 2019, 6, 112. [Google Scholar] [CrossRef] [Green Version]
- SoRelle, E.D.; Dai, J.; Bonglack, E.N.; Heckenberg, E.M.; Zhou, J.Y.; Giamberardino, S.N.; Bailey, J.A.; Gregory, S.G.; Chan, C.; Luftig, M.A. Single-cell RNA-seq reveals transcriptomic heterogeneity mediated by host-pathogen dynamics in lymphoblastoid cell lines. eLife 2021, 10, e62586. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liang, Y.; Ma, T.; Yang, Q. Identification of DGUOK-AS1 as a Prognostic Factor in Breast Cancer by Bioinformatics Analysis. Front. Oncol. 2020, 10, 1092. [Google Scholar] [CrossRef]
- Liang, Y.; Ye, F.; Wang, Y.; Li, Y.; Li, Y.; Song, X.; Luo, D.; Long, L.; Han, D.; Liu, Y.; et al. DGUOK-AS1 acts as a tumor promoter through regulating miR-204-5p/IL-11 axis in breast cancer. Mol. Ther. Nucleic Acids 2021, 26, 1079–1091. [Google Scholar] [CrossRef]
- Vanarsa, K.; Henderson, J.; Soomro, S.; Qin, L.; Zhang, T.; Jordan, N.; Putterman, C.; Blanco, I.; Saxena, R.; Mohan, C. Upregulation of Proinflammatory Bradykinin Peptides in Systemic Lupus Erythematosus and Rheumatoid Arthritis. J. Immunol. 2020, 205, 369–376. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [Green Version]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef]
- Bost, C.; Arleevskaya, M.I.; Brooks, W.H.; Plaza, S.; Guery, J.-C.; Renaudineau, Y. Long non-coding RNA Xist contribution in systemic lupus erythematosus and rheumatoid arthritis. Clin. Immunol. 2022, 236, 108937. [Google Scholar] [CrossRef]
- Garton, J.; Barron, M.D.; Ratliff, M.L.; Webb, C.F. New Frontiers: ARID3a in SLE. Cells 2019, 8, 1136. [Google Scholar] [CrossRef] [Green Version]
- Ha, E.; Bae, S.-C.; Kim, K. Large-scale meta-analysis across East Asian and European populations updated genetic architecture and variant-driven biology of rheumatoid arthritis, identifying 11 novel susceptibility loci. Ann. Rheum. Dis. 2021, 80, 558–565. [Google Scholar] [CrossRef]
- Ramos, P.S.; Shaftman, S.R.; Ward, R.C.; Langefeld, C.D. Genes Associated with SLE Are Targets of Recent Positive Selection. Autoimmune Dis. 2014, 2014, 203435. [Google Scholar] [CrossRef] [Green Version]
- Freisinger, P.; Fütterer, N.; Lankes, E.; Gempel, K.; Berger, T.M.; Spalinger, J.; Hoerbe, A.; Schwantes, C.; Lindner, M.; Santer, R.; et al. Hepatocerebral Mitochondrial DNA Depletion Syndrome Caused by Deoxyguanosine Kinase (DGUOK) Mutations. Arch. Neurol. 2006, 63, 1129–1134. [Google Scholar] [CrossRef]
- Chen, P.-M.; Tsokos, G.C. Mitochondria in the Pathogenesis of Systemic Lupus Erythematosus. Curr. Rheumatol. Rep. 2022, 24, 88–95. [Google Scholar] [CrossRef]
- Jacob, C.O.; Eisenstein, M.; Dinauer, M.C.; Ming, W.; Liu, Q.; John, S.; Quismorio, F.P.; Reiff, A.; Myones, B.L.; Kaufman, K.M.; et al. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc. Natl. Acad. Sci. USA 2012, 109, E59–E67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsson, L.M.; Johansson, Å.C.; Gullstrand, B.; Jönsen, A.; Saevarsdottir, S.; Rönnblom, L.; Leonard, D.; Wetterö, J.; Sjöwall, C.; Svenungsson, E.; et al. A single nucleotide polymorphism in the NCF1 gene leading to reduced oxidative burst is associated with systemic lupus erythematosus. Ann. Rheum. Dis. 2017, 76, 1607–1613. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-T.; Lin, C.-S.; Chen, W.-S.; Liao, H.-T.; Tsai, C.-Y.; Wei, Y.-H. Leukocyte Mitochondrial DNA Alteration in Systemic Lupus Erythematosus and Its Relevance to the Susceptibility to Lupus Nephritis. Int. J. Mol. Sci. 2012, 13, 8853–8868. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Ma, X.; Zhao, Y.-F.; Zhang, C. MiR-1-3p facilitates Th17 differentiation associating with multiple sclerosis via targeting ETS1. Eur. Rev. Med. Pharm. Sci. 2020, 24, 6881–6892. [Google Scholar]
- Wang, B.; Wang, D.; Yan, T.; Yuan, H. MiR-138-5p promotes TNF-α-induced apoptosis in human intervertebral disc degeneration by targeting SIRT1 through PTEN/PI3K/Akt signaling. Exp. Cell Res. 2016, 345, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Alkhateeb, D.M.; Altamemi, A. Circulating Microrna (148b and 150) as Potential Biomarker in Iga Nephropathy and Lupus Nephritis. Ann. Rom. Soc. Cell Biol. 2021, 25, 6294–6309. [Google Scholar]
- Xiao, H.; Wei, N.; Su, M.; Xiong, Z. Down-regulation of serum miR-151a-3p is associated with renal tissue activity in class IV lupus nephritis. Clin. Exp. Rheumatol. 2019, 37, 67–72. [Google Scholar] [PubMed]
- Emmi, G.; Bagni, G.; Lastraioli, E.; di Patti, F.; Bettiol, A.; Fiorillo, C.; Becatti, M.; Silvestri, E.; Urban, M.L.; Emmi, L.; et al. A unique circulating miRNA profile highlights thrombo-inflammation in Behçet’s syndrome. Ann. Rheum. Dis. 2022, 81, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Gao, W.; Xue, L.; Wang, J.; Zhao, L. The lncRNA SLCO4A1-AS1/miR-876-3p/RBBP6 axis regulates cell proliferation and apoptosis in acute lymphocytic leukemia via the JNK signaling pathway. Int. J. Lab. Hematol. 2021, 43, 1050–1061. [Google Scholar] [CrossRef]


| rs6705628 (T/C) | rs2272165 (A/G) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnicity | Cohort | PMID | # Cases | # Controls | FU(A1) | FA(A1) | OR (95% CI) | p-Value | FU(A1) | FA(A1) | OR (95% CI) | p-Value |
| ASN | Meta-analysis | 4144 | 11,014 | 0.19 | 0.16 | 0.79 (0.72–0.87) | 5.56 × 10–10 | 0.19 | 0.16 | 0.79 (0.71–0.86) | 3.48 × 10−10 | |
| HC: Morris et al., 2016 | 27399966 | 1659 | 3398 | 0.18 | 0.15 | 0.72 (0.63–0.81) | 3.23 × 10−8 | 0.18 | 0.15 | 0.71 (0.63–0.80) | 1.86 × 10−8 | |
| Korean: Sun et al., 2016 | 26808113 | 1710 | 6836 | 0.20 | 0.17 | 0.86 (0.77–0.961) | 6.42 × 10−3 | 0.20 | 0.17 | 0.86 (0.77–0.96) | 7.63 × 10−3 | |
| HC: Sun et al., 2016 | 26808113 | 490 | 493 | 0.22 | 0.20 | 0.85 (0.67–1.07) | 1.49 × 10−1 | 0.22 | 0.20 | 0.84 (0.67–1.06) | 1.40 × 10−1 | |
| Malaysian: Sun et al., 2016 | 26808113 | 285 | 287 | 0.17 | 0.11 | 0.66 (0.46–0.93) | 1.36 × 10−2 | 0.17 | 0.11 | 0.67 (0.47–0.94) | 1.72 × 10−2 | |
| EUR | Meta-analysis | 6108 | 10,590 | 0.01 | --- | 0.78 (0.55–1.0) | 1.88 × 10−2 | 0.01 | --- | 0.78 (0.55–1.0) | 1.90 × 10−2 | |
| EUR: Bentham et al., 2015 | 26502338 | 5201 | 9066 | 0.01 | --- | 0.84 (0.65–1.08) | 1.68 × 10−1 | 0.01 | --- | 0.84 (0.65–1.08) | 1.69 × 10−1 | |
| SPN: Julia et al., 2018 | 29848360 | 907 | 1524 | 0.01 | --- | 0.57 (0.31–1.04) | 7.21 × 10−2 | 0.01 | --- | 0.57 (0.31–1.04) | 7.19 × 10−2 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fazel-Najafabadi, M.; Rallabandi, H.-R.; Singh, M.K.; Maiti, G.P.; Morris, J.; Looger, L.L.; Nath, S.K. Discovery and Functional Characterization of Two Regulatory Variants Underlying Lupus Susceptibility at 2p13.1. Genes 2022, 13, 1016. https://doi.org/10.3390/genes13061016
Fazel-Najafabadi M, Rallabandi H-R, Singh MK, Maiti GP, Morris J, Looger LL, Nath SK. Discovery and Functional Characterization of Two Regulatory Variants Underlying Lupus Susceptibility at 2p13.1. Genes. 2022; 13(6):1016. https://doi.org/10.3390/genes13061016
Chicago/Turabian StyleFazel-Najafabadi, Mehdi, Harikrishna-Reddy Rallabandi, Manish K. Singh, Guru P. Maiti, Jacqueline Morris, Loren L. Looger, and Swapan K. Nath. 2022. "Discovery and Functional Characterization of Two Regulatory Variants Underlying Lupus Susceptibility at 2p13.1" Genes 13, no. 6: 1016. https://doi.org/10.3390/genes13061016
APA StyleFazel-Najafabadi, M., Rallabandi, H.-R., Singh, M. K., Maiti, G. P., Morris, J., Looger, L. L., & Nath, S. K. (2022). Discovery and Functional Characterization of Two Regulatory Variants Underlying Lupus Susceptibility at 2p13.1. Genes, 13(6), 1016. https://doi.org/10.3390/genes13061016






