HLA Genotypes and Type 1 Diabetes and Its Relationship to Reported Race/Skin Color in Their Relatives: A Brazilian Multicenter Study
Abstract
:1. Introduction
2. Methods
2.1. Study Design and Sample
2.2. Data Collection
2.3. HLA Typing
2.4. Statistical Analysis
3. Results
3.1. Overview of Demographic Data and HLA Allele Frequencies of the Studied Population with Type 1 Diabetes
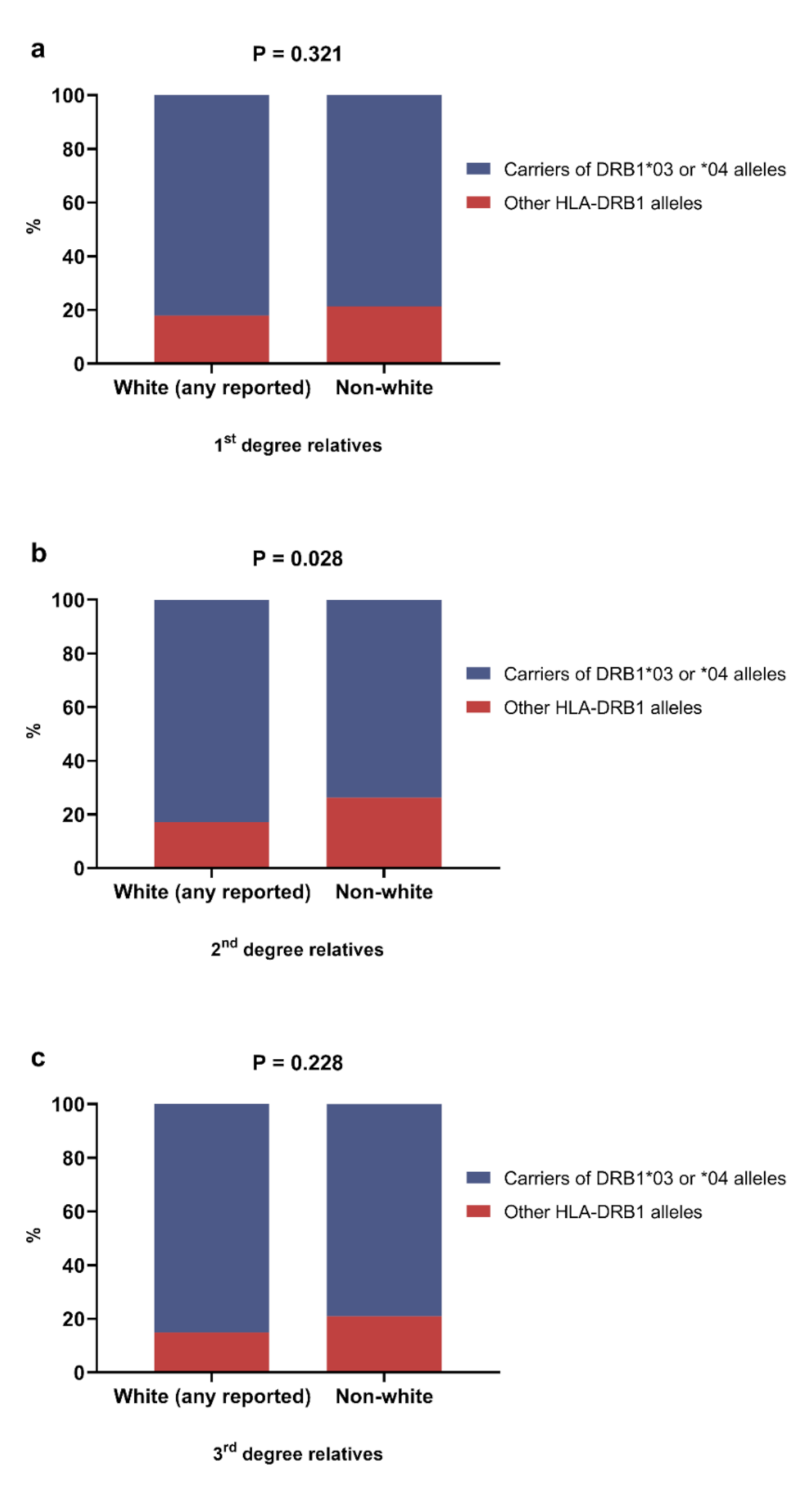
3.2. Overview of the Association between HLA Alleles and Reported Race/Skin Color of the Relatives
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noble, J.A. Immunogenetics of type 1 diabetes: A comprehensive review. J. Autoimmun. 2015, 64, 101–112. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019; pp. 32–56. [Google Scholar]
- Negrato, C.A.; Lauris, J.R.P.; Saggioro, I.B.; Corradini, M.C.M.; Borges, P.R.; Crês, M.C.; Junior, A.L.; Guedes, M.F.S.; Gomes, M.B. Increasing incidence of type 1 diabetes between 1986 and 2015 in Bauru, Brazil. Diabetes Res. Clin. Pract. 2017, 127, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Sharp, S.A.; Rich, S.S.; Wood, A.R.; Jones, S.E.; Beaumont, R.N.; Harrison, J.W.; Schneider, D.A.; Locke, J.M.; Tyrrell, J.; Weedon, M.N.; et al. Development and Standardization of an Improved Type 1 Diabetes Genetic Risk Score for Use in Newborn Screening and Incident Diagnosis. Diabetes Care 2019, 42, 200–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas-Villarraga, A.; Botello-Corzo, D.; Anaya, J.-M. HLA-Class II in Latin American patients with type 1 diabetes. Autoimmun. Rev. 2010, 9, 666–673. [Google Scholar] [CrossRef]
- Noble, J.A.; Johnson, J.; Lane, J.A.; Valdes, A.M. HLA Class II Genotyping of African American Type 1 Diabetic Patients Reveals Associations Unique to African Haplotypes. Diabetes 2013, 62, 3292–3299. [Google Scholar] [CrossRef] [Green Version]
- Santos, D.C.; Porto, L.C.; Oliveira, R.V.; Secco, D.; Hanhoerderster, L.; Pizarro, M.H.; Barros, B.S.V.; Mello, L.G.N.; Muniz, L.H.; Silva, D.A.; et al. HLA class II genotyping of admixed Brazilian patients with type 1 diabetes according to self-reported color/race in a nationwide study. Sci. Rep. 2020, 10, 6628. [Google Scholar] [CrossRef]
- Halagan, M.; Oliveira, D.C.; Maiers, M.; Fabreti-Oliveira, R.A.; Moraes, M.E.H.; Visentainer, J.E.L.; Pereira, N.F.; Romero, M.; Cardoso, J.F.; Porto, L.C. The distribution of HLA haplotypes in the ethnic groups that make up the Brazilian Bone Marrow Volunteer Donor Registry (REDOME). Immunogenetics 2018, 70, 511–522. [Google Scholar] [CrossRef]
- Gomes, K.F.B.; Santos, A.S.; Semzezem, C.; Correia, M.R.; Brito, L.A.; Ruiz, M.O.; Fukui, R.T.; Matioli, S.R.; Passos-Bueno, M.R.; da Silva, M.E.R. The influence of population stratification on genetic markers associated with type 1 diabetes. Sci. Rep. 2017, 7, 43513. [Google Scholar] [CrossRef] [Green Version]
- Gomes, M.B.; Gabrielli, A.B.; Santos, D.C.; Pizarro, M.H.; Barros, B.S.V.; Negrato, C.A.; Dib, S.A.; Porto, L.C.; Silva, D.A. Self-reported color-race and genomic ancestry in an admixed population: A contribution of a nationwide survey in patients with type 1 diabetes in Brazil. Diabetes Res. Clin. Pract. 2018, 140, 245–252. [Google Scholar] [CrossRef]
- De Manta, F.S.N.; Pereira, R.; Vianna, R.; de Araújo, A.R.B.; Leite Góes Gitaí, D.; Aparecida da Silva, D.; de Vargas Wolfgramm, E.; da Mota Pontes, I.; Ivan Aguiar, J.; Ozório Moraes, M.; et al. Revisiting the Genetic Ancestry of Brazilians Using Autosomal AIM-Indels. PLoS ONE 2013, 8, e75145. [Google Scholar] [CrossRef]
- Pena, S.D.J.; Santos, F.R.; Tarazona-Santos, E. Genetic admixture in Brazil. Am. J. Med. Genet. Part C Semin. Med. Genet. 2020, 184, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Instituto Brasileiro de Geografia e Estatística (IBGE). Censo 2000. Available online: http://www.ibge.gov.br/censo (accessed on 10 May 2020).
- Gomes, M.B.; Negrato, C.A. Adherence to insulin therapeutic regimens in patients with type 1 diabetes. A nationwide survey in Brazil. Diabetes Res. Clin. Pract. 2016, 120, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Hurley, C.K.; Kempenich, J.; Wadsworth, K.; Sauter, J.; Hofmann, J.A.; Schefzyk, D.; Schmidt, A.H.; Galarza, P.; Cardozo, M.B.R.; Dudkiewicz, M.; et al. Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0.0. HLA 2020, 95, 516–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.W.; Thompson, E.A. Performing the Exact Test of Hardy-Weinberg Proportion for Multiple Alleles. Biometrics 1992, 48, 361. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz-Villena, A.; Martínez-Laso, J.; Gómez-Casado, E.; Díaz-Campos, N.; Santos, P.; Martinho, A.; Breda-Coimbra, H. Relatedness among Basques, Portuguese, Spaniards, and Algerians studied by HLA allelic frequencies and haplotypes. Immunogenetics 1997, 47, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Romòn, I.; Montes, C.; Ligeiro, D.; Trindade, H.; Sanchez-Mazas, A.; Nunes, J.M.; Buhler, S. Mapping the HLA diversity of the Iberian Peninsula. Hum. Immunol. 2016, 77, 832–840. [Google Scholar] [CrossRef] [Green Version]
- Noble, J.A.; Erlich, H.A. Genetics of Type 1 Diabetes. Cold Spring Harb. Perspect. Med. 2012, 2, a007732. [Google Scholar] [CrossRef] [Green Version]
- Instituto Brasileiro de Geografia e Estatística (IBGE). Brasil: 500 Anos de Povoamento; IBGE: Rio de Janeiro, Brazil, 2007; ISBN 978-85-240-3940-9. [Google Scholar]
- Salzano, F.M.; Sans, M. Interethnic admixture and the evolution of Latin American populations. Genet. Mol. Biol. 2014, 37, 151–170. [Google Scholar] [CrossRef] [Green Version]
- Perreira, K.M.; Telles, E.E. The color of health: Skin color, ethnoracial classification, and discrimination in the health of Latin Americans. Soc. Sci. Med. 2014, 116, 241–250. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Lin, J.; Xie, Z.; Xiang, Y.; Zheng, P.; Huang, G.; Li, X.; Liao, Y.; Hagopian, W.A.; Wang, C.-Y.; et al. HLA Genetic Discrepancy Between Latent Autoimmune Diabetes in Adults and Type 1 Diabetes: LADA China Study No. 6. J. Clin. Endocrinol. Metab. 2016, 101, 1693–1700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikegami, H.; Kawabata, Y.; Noso, S.; Fujisawa, T.; Ogihara, T. Genetics of type 1 diabetes in Asian and Caucasian populations. Diabetes Res. Clin. Pract. 2007, 77, S116–S121. [Google Scholar] [CrossRef] [PubMed]
- Pena, S.D.J.; Di Pietro, G.; Fuchshuber-Moraes, M.; Genro, J.P.; Hutz, M.H.; de Souza Gomes Kehdy, F.; Kohlrausch, F.; Magno, L.A.V.; Montenegro, R.C.; Moraes, M.O.; et al. The Genomic Ancestry of Individuals from Different Geographical Regions of Brazil Is More Uniform Than Expected. PLoS ONE 2011, 6, e17063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryc, K.; Durand, E.Y.; Macpherson, J.M.; Reich, D.; Mountain, J.L. The Genetic Ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet. 2015, 96, 37–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiani, J.; Hajilooi, M.; Furst, D.; Rezaei, H.; Shahryari-Hesami, S.; Kowsarifard, S.; Zamani, A.; Solgi, G. HLA class II susceptibility pattern for type 1 diabetes (T1D) in an Iranian population. Int. J. Immunogenet. 2015, 42, 279–286. [Google Scholar] [CrossRef]
- Park, Y.; Eisenbarth, G.S. Genetic susceptibility factors of Type 1 diabetes in Asians. Diabetes. Metab. Res. Rev. 2001, 17, 2–11. [Google Scholar] [CrossRef]
- Harrison, J.W.; Tallapragada, D.S.P.; Baptist, A.; Sharp, S.A.; Bhaskar, S.; Jog, K.S.; Patel, K.A.; Weedon, M.N.; Chandak, G.R.; Yajnik, C.S.; et al. Type 1 diabetes genetic risk score is discriminative of diabetes in non-Europeans: Evidence from a study in India. Sci. Rep. 2020, 10, 9450. [Google Scholar] [CrossRef]
- Thomson, G.; Valdes, A.M.; Noble, J.A.; Kockum, I.; Grote, M.N.; Najman, J.; Erlich, H.A.; Cucca, F.; Pugliese, A.; Steenkiste, A.; et al. Relative predispositional effects of HLA class II DRB1-DQB1 haplotypes and genotypes on type 1 diabetes: A meta-analysis. Tissue Antigens 2007, 70, 110–127. [Google Scholar] [CrossRef]
- Sanchez-Mazas, A.; Fernandez-Viña, M.; Middleton, D.; Hollenbach, J.A.; Buhler, S.; Di, D.; Rajalingam, R.; Dugoujon, J.-M.; Mack, S.J.; Thorsby, E. Immunogenetics as a tool in anthropological studies. Immunology 2011, 133, 143–164. [Google Scholar] [CrossRef]
- Sanchez-Mazas, A.; Lemaître, J.-F.; Currat, M. Distinct evolutionary strategies of human leucocyte antigen loci in pathogen-rich environments. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 830–839. [Google Scholar] [CrossRef] [Green Version]
- Boquett, J.A.; Bisso-Machado, R.; Zagonel-Oliveira, M.; Schüler-Faccini, L.; Fagundes, N.J.R. HLA diversity in Brazil. HLA 2020, 95, 3–14. [Google Scholar] [CrossRef] [PubMed]


| Variables | Mean ± SD | n | % |
|---|---|---|---|
| Gender | |||
| Male | 317 | 47.89 | |
| Female | 345 | 52.11 | |
| Age, years | 29.32 ± 12.52 | ||
| Age of diabetes diagnosis, years | 14.5 ± 9.2 | ||
| Self-reported race/skin color (IBGE classification) | |||
| White (Branca) | 355 | 53.63 | |
| Black (Preta) | 43 | 6.50 | |
| Brown (Parda) | 255 | 38.52 | |
| Yellow (Amarela) | 8 | 1.21 | |
| Indigenous (Indígena) | 1 | 0.15 | |
| Birthplace (Brazilian geographic macro region) | |||
| Southeast | 367 | 55.44 | |
| South | 53 | 8.01 | |
| Northeast | 214 | 32.33 | |
| Midwest | 2 | 0.30 | |
| North | 26 | 3.93 |
| HLA Haplotypes | n | % |
|---|---|---|
| HLA-DRB1* | ||
| DRB1*01 | 110 | 8.31 |
| DRB1*03 | 396 | 29.91 |
| DRB1*04 | 400 | 30.21 |
| DRB1*07 | 109 | 8.23 |
| DRB1*08 | 45 | 3.40 |
| DRB1*09 | 37 | 2.79 |
| DRB1*10 | 13 | 0.98 |
| DRB1*11 | 52 | 3.93 |
| DRB1*12 | 9 | 0.68 |
| DRB1*13 | 73 | 5.51 |
| DRB1*14 | 15 | 1.13 |
| DRB1*15 | 35 | 2.64 |
| DRB1*16 | 30 | 2.27 |
| HLA-DQA1* | ||
| DQA1*01 | 266 | 20.12 |
| DQA1*02 | 95 | 7.18 |
| DQA1*03 | 452 | 34.19 |
| DQA1*04 | 49 | 3.70 |
| DQA1*05 | 458 | 34.64 |
| DQA1*06 | 2 | 0.15 |
| HLA-DQB1* | ||
| DQB1*02 | 523 | 39.50 |
| DQB1*03 | 492 | 37.31 |
| DQB1*04 | 46 | 3.48 |
| DQB1*05 | 167 | 12.61 |
| DQB1*06 | 94 | 7.10 |
| Variables | adjusted OR | 95% CI | p Value |
|---|---|---|---|
| HLA-DRB1*01 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 1.35 | 0.83–2.21 | 0.219 |
| All relatives reported as black | 0.65 | 0.08–5.28 | 0.687 |
| Any relative reported as indigenous | 0.63 | 0.21–1.84 | 0.405 |
| Any relative reported as yellow | – | – | – |
| Any relative born in Europe | 0.84 | 0.50–1.43 | 0.536 |
| HLA-DRB1*03 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 1.53 | 1.04–2.24 | 0.028 * |
| All relatives reported as black | 1.29 | 0.34–4.89 | 0.703 |
| Any relative reported as indigenous | 1.07 | 0.55–2.08 | 0.831 |
| Any relative reported as yellow | 1.03 | 0.25–4.19 | 0.960 |
| Any relative born in Europe | 0.85 | 0.58–1.25 | 0.427 |
| HLA-DRB1*04 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 1.37 | 0.93–2.01 | 0.105 |
| All relatives reported as black | 0.43 | 0.10–1.78 | 0.250 |
| Any relative reported as indigenous | 1.12 | 0.57–2.20 | 0.721 |
| Any relative reported as yellow | 0.29 | 0.05–1.47 | 0.136 |
| Any relative born in Europe | 0.85 | 0.57–1.25 | 0.411 |
| HLA-DRB1*07 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 0.36 | 0.19–0.67 | 0.001 * |
| All relatives reported as black | 0.59 | 0.07–4.86 | 0.631 |
| Any relative reported as indigenous | 0.97 | 0.41–2.29 | 0.947 |
| Any relative reported as yellow | 1.59 | 0.31–8.08 | 0.571 |
| Any relative born in Europe | 1.51 | 0.90–2.54 | 0.117 |
| HLA-DRB1*08 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 0.55 | 0.22–1.39 | 0.211 |
| All relatives reported as black | 1.31 | 0.16–10.86 | 0.797 |
| Any relative reported as indigenous | – | – | – |
| Any relative reported as yellow | 3.51 | 0.68–18.10 | 0.132 |
| Any relative born in Europe | 0.39 | 0.13–1.16 | 0.093 |
| HLA-DRB1*09 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 0.16 | 0.03–0.72 | 0.017 * |
| All relatives reported as black | 1.94 | 0.23–16.24 | 0.537 |
| Any relative reported as indigenous | 0.80 | 0.18–3.50 | 0.768 |
| Any relative reported as yellow | 9.35 | 2.10–41.55 | 0.003 * |
| Any relative born in Europe | 1.35 | 0.55–3.28 | 0.506 |
| HLA-DRB1*11 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 0.34 | 0.13–0.86 | 0.023 * |
| All relatives reported as black | 1.44 | 0.17–11.89 | 0.733 |
| Any relative reported as indigenous | 1.55 | 0.56–4.22 | 0.391 |
| Any relative reported as yellow | – | – | – |
| Any relative born in Europe | 1.56 | 0.78–3.14 | 0.206 |
| HLA-DRB1*13 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 1.14 | 0.62–2.07 | 0.664 |
| All relatives reported as black | 1.03 | 0.12–8.48 | 0.973 |
| Any relative reported as indigenous | 1.28 | 0.47–3.46 | 0.615 |
| Any relative reported as yellow | – | – | – |
| Any relative born in Europe | 0.89 | 0.48–1.66 | 0.733 |
| HLA-DRB1*15 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 0.81 | 0.32–2.03 | 0.661 |
| All relatives reported as black | 2.44 | 0.29–20.54 | 0.409 |
| Any relative reported as indigenous | 1.61 | 0.46–5.69 | 0.452 |
| Any relative reported as yellow | – | – | – |
| Any relative born in Europe | 1.15 | 0.48–2.74 | 0.744 |
| HLA-DRB1*16 (Dependent variable) | |||
| Data from relatives up to 3rd degree (Predictors) | |||
| All relatives reported as white | 1.01 | 0.39–2.59 | 0.983 |
| All relatives reported as black | 3.01 | 0.35–25.53 | 0.311 |
| Any relative reported as indigenous | 1.29 | 0.28–5.82 | 0.734 |
| Any relative reported as yellow | 3.44 | 0.39–29.65 | 0.260 |
| Any relative born in Europe | 1.13 | 0.44–2.88 | 0.785 |
| HLA-DRB1*/-DRB1* Genotype | n | % |
|---|---|---|
| 03:01/03:01 | 16 | 9.76% |
| 03:01/04:02 | 15 | 9.15% |
| 03:01/04:04 | 13 | 7.93% |
| 03:01/04:05 | 8 | 4.88% |
| 03:01/04:01 | 6 | 3.66% |
| 01:01/03:01 | 5 | 3.05% |
| 03:01/13:02 | 5 | 3.05% |
| 04:01/04:05 | 5 | 3.05% |
| 03:01/07:01 | 4 | 2.44% |
| 01:01/04:01 | 3 | 1.83% |
| 01:01/04:05 | 3 | 1.83% |
| 01:02/03:01 | 3 | 1.83% |
| 03:01/08:01 | 3 | 1.83% |
| 03:01/13:01 | 3 | 1.83% |
| 03:01/16:01 | 3 | 1.83% |
| 04:05/04:05 | 3 | 1.83% |
| 01:01/04:04 | 2 | 1.22% |
| 01:02/04:01 | 2 | 1.22% |
| 03:01/04:03 | 2 | 1.22% |
| 03:01/09:01 | 2 | 1.22% |
| 04:01/04:02 | 2 | 1.22% |
| 04:01/13:02 | 2 | 1.22% |
| 04:02/04:02 | 2 | 1.22% |
| 04:02/04:05 | 2 | 1.22% |
| 04:05/10:01 | 2 | 1.22% |
| Other | 48 | <1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, M.B.; Porto, L.C.; Silva, D.A.; Negrato, C.A.; Pavin, E.J.; Montenegro Junior, R.; Dib, S.A.; Felício, J.S.; Santos, D.C.; Muniz, L.H.; et al. HLA Genotypes and Type 1 Diabetes and Its Relationship to Reported Race/Skin Color in Their Relatives: A Brazilian Multicenter Study. Genes 2022, 13, 972. https://doi.org/10.3390/genes13060972
Gomes MB, Porto LC, Silva DA, Negrato CA, Pavin EJ, Montenegro Junior R, Dib SA, Felício JS, Santos DC, Muniz LH, et al. HLA Genotypes and Type 1 Diabetes and Its Relationship to Reported Race/Skin Color in Their Relatives: A Brazilian Multicenter Study. Genes. 2022; 13(6):972. https://doi.org/10.3390/genes13060972
Chicago/Turabian StyleGomes, Marília B., Luís C. Porto, Dayse A. Silva, Carlos A. Negrato, Elizabeth João Pavin, Renan Montenegro Junior, Sergio A. Dib, João S. Felício, Deborah C. Santos, Luiza H. Muniz, and et al. 2022. "HLA Genotypes and Type 1 Diabetes and Its Relationship to Reported Race/Skin Color in Their Relatives: A Brazilian Multicenter Study" Genes 13, no. 6: 972. https://doi.org/10.3390/genes13060972








