Mutation of foxl1 Results in Reduced Cartilage Markers in a Zebrafish Model of Otosclerosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. Zebrafish Strains Used in This Study
2.3. Alcian Blue and Alizarin Red Staining
2.4. Whole Mount In Situ Hybridizations
2.5. Calcein Staining
2.6. TaqMan Real-Time Quantitative PCR
2.7. DXA Scanning
3. Results
3.1. foxl1 Mutant Generation
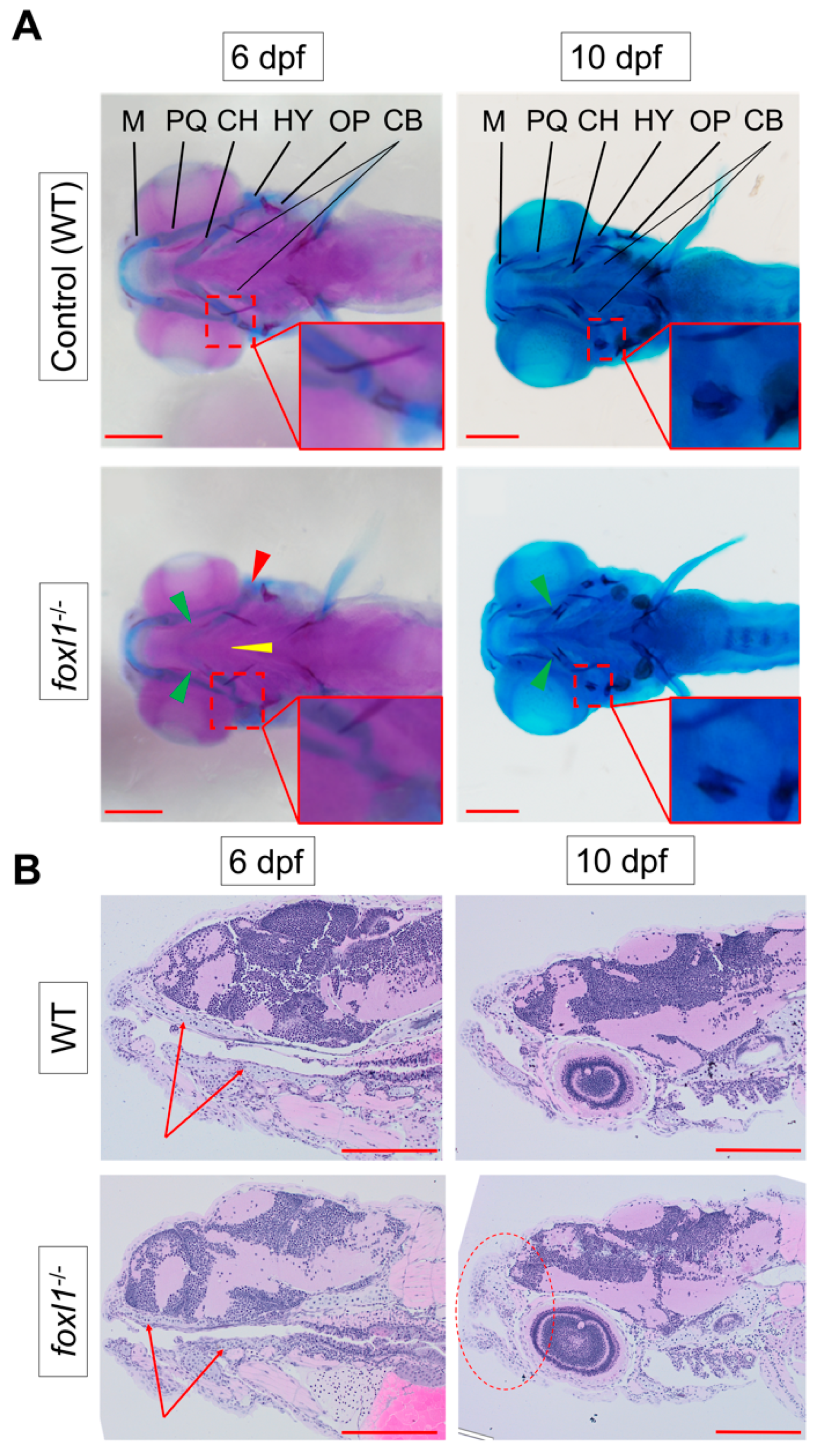
3.2. Craniofacial Cartilage Formation and Calcification in foxl1 Mutants
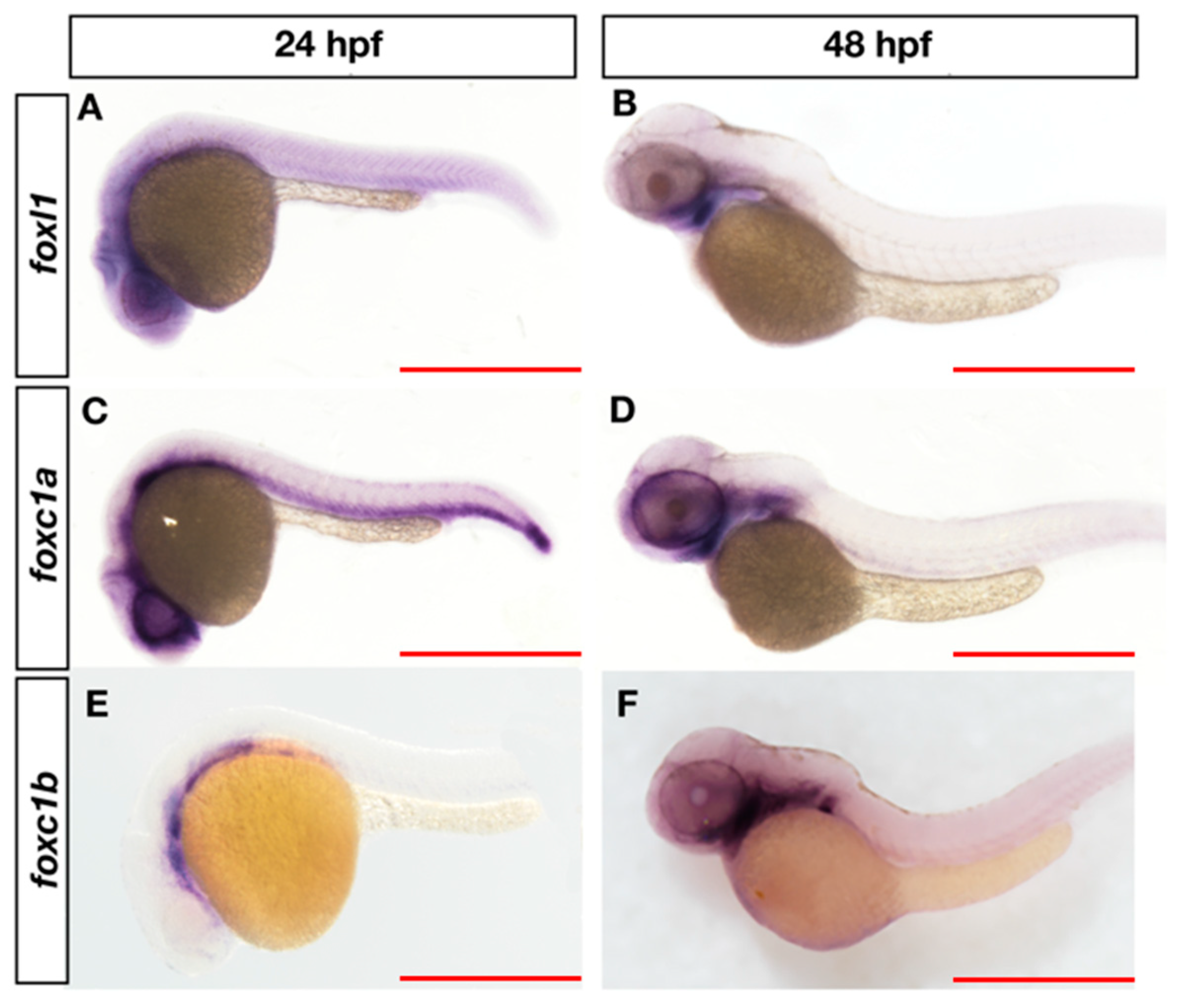
3.3. Overlapping Expression of foxl1, foxc1a, and foxc1b in Zebrafish
3.4. foxl1 Mutation in foxc1a and foxc1b Mutant Backgrounds
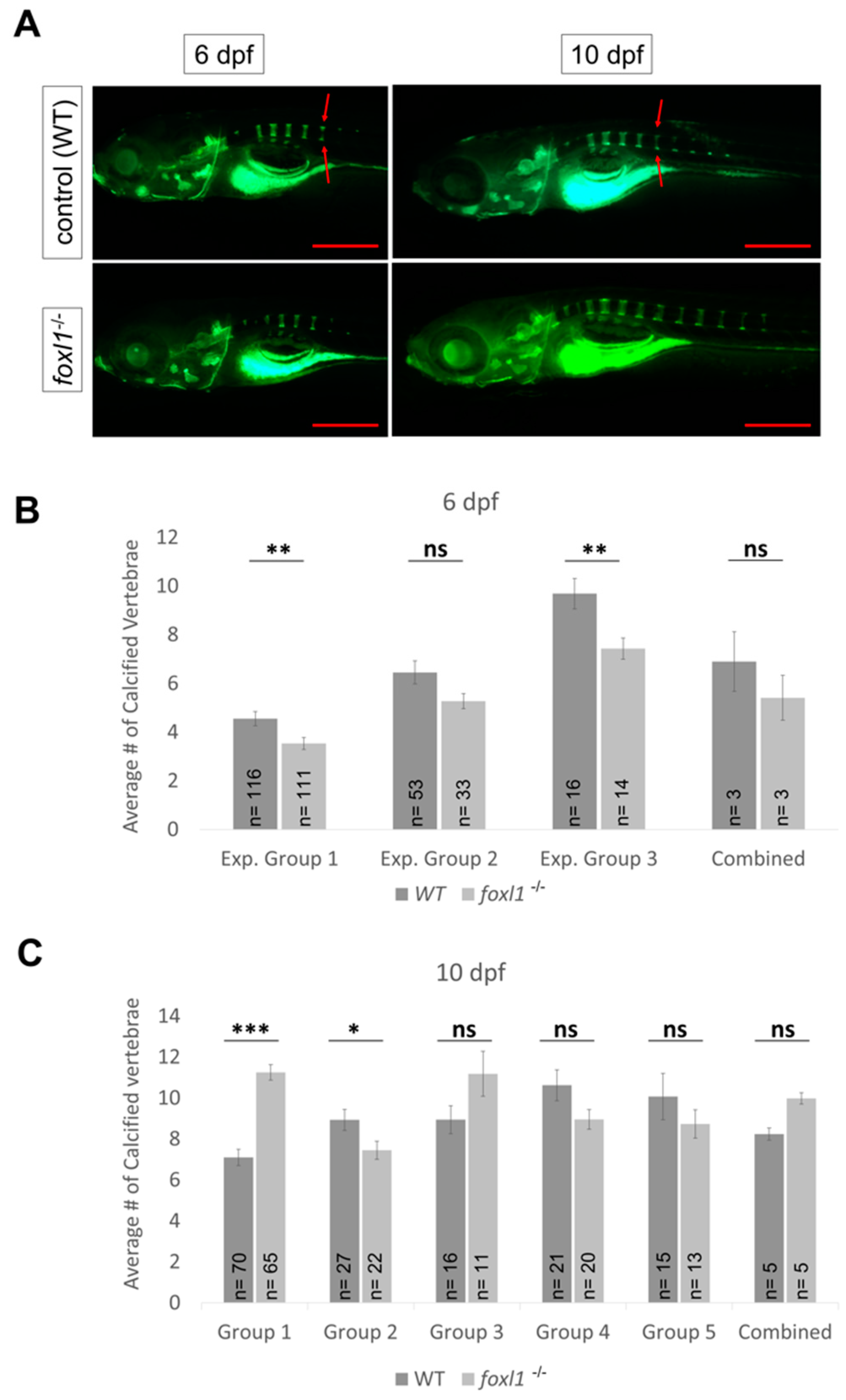
3.5. Axial Skeletal Calcification in Forkhead Mutants
3.6. Expression of Genes Required for Bone Formation in Forkhead Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berendsen, A.D.; Olsen, B.R. Bone Development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, A.; Kishida, M.G.; Kimmel, C.B.; Keynes, R.J. Building the Backbone: The Development and Evolution of Vertebral Patterning. Development 2015, 142, 1733–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javidan, Y.; Schilling, T.F. Development of Cartilage and Bone. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2004; Volume 76, pp. 415–436. ISBN 978-0-12-564171-5. [Google Scholar]
- Kozhemyakina, E.; Lassar, A.B.; Zelzer, E. A Pathway to Bone: Signaling Molecules and Transcription Factors Involved in Chondrocyte Development and Maturation. Development 2015, 142, 817–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dash, S.; Trainor, P.A. The Development, Patterning and Evolution of Neural Crest Cell Differentiation into Cartilage and Bone. Bone 2020, 137, 115409. [Google Scholar] [CrossRef]
- Winnier, G.E.; Hargett, L.; Hogan, B.L. The Winged Helix Transcription Factor MFH1 Is Required for Proliferation and Patterning of Paraxial Mesoderm in the Mouse Embryo. Genes Dev. 1997, 11, 926–940. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, V. Roles and Regulation of SOX Transcription Factors in Skeletogenesis. Curr. Top. Dev. Biol. 2019, 133, 171–193. [Google Scholar] [CrossRef]
- Long, F.; Ornitz, D.M. Development of the Endochondral Skeleton. Cold Spring Harb. Perspect. Biol. 2013, 5, a008334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, Y.; Tanaka, N.; Abe, Y.; Tanaka, N. Roles of the Hedgehog Signaling Pathway in Epidermal and Hair Follicle Development, Homeostasis, and Cancer. J. Dev. Biol. 2017, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Ahlgren, S.C.; Bronner-Fraser, M. Inhibition of Sonic Hedgehog Signaling in Vivo Results in Craniofacial Neural Crest Cell Death. Curr. Biol. 1999, 9, 1304–1314. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, B.E.; Henner, A.; Stewart, S.; Stankunas, K. Shh Promotes Direct Interactions between Epidermal Cells and Osteoblast Progenitors to Shape Regenerated Zebrafish Bone. Development 2017, 144, 1165–1176. [Google Scholar] [CrossRef] [Green Version]
- Avaron, F.; Smith, A.; Akimenko, M.-A. Sonic Hedgehog Signaling in the Developing and Regenerating Fins of Zebrafish; Landes Bioscience: Austin, TX, USA, 2013. [Google Scholar]
- Carballo, G.B.; Honorato, J.R.; de Lopes, G.P.F.; Spohr, T.C.L. de S. e A Highlight on Sonic Hedgehog Pathway. Cell Commun. Signal. 2018, 16, 11. [Google Scholar] [CrossRef] [PubMed]
- Chung, U.; Schipani, E.; McMahon, A.P.; Kronenberg, H.M. Indian Hedgehog Couples Chondrogenesis to Osteogenesis in Endochondral Bone Development. J. Clin. Investig. 2001, 107, 295–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberhart, J.K.; Swartz, M.E.; Crump, J.G.; Kimmel, C.B. Early Hedgehog Signaling from Neural to Oral Epithelium Organizes Anterior Craniofacial Development. Development 2006, 133, 1069–1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, M. Networking of WNT, FGF, Notch, BMP, and Hedgehog Signaling Pathways during Carcinogenesis. Stem Cell Rev. Totowa 2007, 3, 30–38. [Google Scholar] [CrossRef]
- Rice, R.; Rice, D.P.C.; Thesleff, I. Foxc1 Integrates Fgf and Bmp Signalling Independently of Twist or Noggin during Calvarial Bone Development. Dev. Dyn. 2005, 233, 847–852. [Google Scholar] [CrossRef]
- Asharani, P.V.; Keupp, K.; Semler, O.; Wang, W.; Li, Y.; Thiele, H.; Yigit, G.; Pohl, E.; Becker, J.; Frommolt, P.; et al. Attenuated BMP1 Function Compromises Osteogenesis, Leading to Bone Fragility in Humans and Zebrafish. Am. J. Hum. Genet. 2012, 90, 661–674. [Google Scholar] [CrossRef] [Green Version]
- Katoh, M.; Katoh, M. Transcriptional Regulation of WNT2B Based on the Balance of Hedgehog, Notch, BMP and WNT Signals. Int. J. Oncol. 2009, 34, 1411–1415. [Google Scholar] [CrossRef] [Green Version]
- Wan, M.; Cao, X. BMP Signaling in Skeletal Development. Biochem. Biophys. Res. Commun. 2005, 328, 651–657. [Google Scholar] [CrossRef]
- Chrystal, P.W.; French, C.R.; Jean, F.; Havrylov, S.; van Baarle, S.; Peturson, A.-M.; Xu, P.; Crump, J.G.; Pilgrim, D.B.; Lehmann, O.J.; et al. The Axenfeld-Rieger Syndrome Gene FOXC1 Contributes to Left-Right Patterning. Genes 2021, 12, 170. [Google Scholar] [CrossRef]
- Kim, S.H.; Cho, K.-W.; Choi, H.S.; Park, S.J.; Rhee, Y.; Jung, H.-S.; Lim, S.-K. The Forkhead Transcription Factor Foxc2 Stimulates Osteoblast Differentiation. Biochem. Biophys. Res. Commun. 2009, 386, 532–536. [Google Scholar] [CrossRef]
- Kume, T.; Jiang, H.; Topczewska, J.M.; Hogan, B.L.M. The Murine Winged Helix Transcription Factors, Foxc1 and Foxc2, Are Both Required for Cardiovascular Development and Somitogenesis. Genes Dev. 2001, 15, 2470–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Yu, H.V.; Tseng, K.-C.; Flath, M.; Fabian, P.; Segil, N.; Crump, J.G. Foxc1 Establishes Enhancer Accessibility for Craniofacial Cartilage Differentiation. eLife 2021, 10, e63595. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Balczerski, B.; Ciozda, A.; Louie, K.; Oralova, V.; Huysseune, A.; Crump, J.G. Fox Proteins Are Modular Competency Factors for Facial Cartilage and Tooth Specification. Development 2018, 145, dev.165498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, W.; Gao, H.; Fan, L.; Duan, D.; Wang, C.; Wang, K. Foxc2 Regulates Osteogenesis and Angiogenesis of Bone Marrow Mesenchymal Stem Cells. BMC Musculoskelet. Disord. 2013, 14, 199. [Google Scholar] [CrossRef] [Green Version]
- Dai, S.; Qu, L.; Li, J.; Chen, Y. Toward a Mechanistic Understanding of DNA Binding by Forkhead Transcription Factors and Its Perturbation by Pathogenic Mutations. Nucleic Acids Res. 2021, 49, 10235–10249. [Google Scholar] [CrossRef]
- Hannenhalli, S.; Kaestner, K.H. The Evolution of Fox Genes and Their Role in Development and Disease. Nat. Rev. Genet. 2009, 10, 233–240. [Google Scholar] [CrossRef] [Green Version]
- Nifuji, A.; Miura, N.; Kato, N.; Kellermann, O.; Noda, M. Bone Morphogenetic Protein Regulation of Forkhead/Winged Helix Transcription Factor Foxc2 (Mfh1) in a Murine Mesodermal Cell Line C1 and in Skeletal Precursor Cells. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2001, 16, 1765–1771. [Google Scholar] [CrossRef]
- Sun, J.; Ishii, M.; Ting, M.-C.; Maxson, R. Foxc1 Controls the Growth of the Murine Frontal Bone Rudiment by Direct Regulation of a Bmp Response Threshold of Msx2. Dev. Camb. Engl. 2013, 140, 1034–1044. [Google Scholar] [CrossRef] [Green Version]
- Mariotti, M.; Carnovali, M.; Banfi, G. Danio Rerio: The Janus of the Bone from Embryo to Scale. Clin. Cases Miner. Bone Metab. 2015, 12, 188–194. [Google Scholar] [CrossRef]
- Tümer, Z.; Bach-Holm, D. Axenfeld–Rieger Syndrome and Spectrum of PITX2 and FOXC1 Mutations. Eur. J. Hum. Genet. 2009, 17, 1527–1539. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Wang, W.-Q.; Li, X.-Q.; Zhao, J.; Yang, K.; Feng, Y.; Guo, M.-M.; Liu, M.; Liu, X.; Wang, X.; et al. A Novel Variant in FOXC1 Associated with Atypical Axenfeld-Rieger Syndrome. BMC Med. Genom. 2021, 14, 277. [Google Scholar] [CrossRef] [PubMed]
- French, C.R. Mechanistic Insights into Axenfeld-Rieger Syndrome from Zebrafish Foxc1 and Pitx2 Mutants. Int. J. Mol. Sci. 2021, 22, 10001. [Google Scholar] [CrossRef] [PubMed]
- Abdelfatah, N.; Mostafa, A.A.; French, C.R.; Doucette, L.P.; Penney, C.; Lucas, M.B.; Griffin, A.; Booth, V.; Rowley, C.; Besaw, J.E.; et al. A Pathogenic Deletion in Forkhead Box L1 (FOXL1) Identifies the First Otosclerosis (OTSC) Gene. Hum. Genet. 2021, 141, 965–979. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-J.; Ahn, S.H.; Kim, H.-M.; Ikegawa, S.; Yang, T.-L.; Guo, Y.; Deng, H.-W.; Koh, J.-M.; Lee, S.H. Replication of Caucasian Loci Associated with Osteoporosis-Related Traits in East Asians. J. Bone Metab. 2016, 23, 233–242. [Google Scholar] [CrossRef] [Green Version]
- Kung, A.W.C. Novel Genetic Loci Associated with Osteoporosis. Bone 2010, 47, S376. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Zhang, L.; Papasian, C.J.; Deng, H.-W. Genome-Wide Association Studies for Osteoporosis: A 2013 Update. J. Bone Metab. 2014, 21, 99–116. [Google Scholar] [CrossRef] [Green Version]
- the Genetic Factors for Osteoporosis (GEFOS) Consortium; Rivadeneira, F.; Styrkársdottir, U.; Estrada, K.; Halldórsson, B.V.; Hsu, Y.-H.; Richards, J.B.; Zillikens, M.C.; Kavvoura, F.K.; Amin, N.; et al. Twenty Bone-Mineral-Density Loci Identified by Large-Scale Meta-Analysis of Genome-Wide Association Studies. Nat. Genet. 2009, 41, 1199–1206. [Google Scholar] [CrossRef] [Green Version]
- Aoki, R.; Shoshkes-Carmel, M.; Gao, N.; Shin, S.; May, C.L.; Golson, M.L.; Zahm, A.M.; Ray, M.; Wiser, C.L.; Wright, C.V.E.; et al. Foxl1-Expressing Mesenchymal Cells Constitute the Intestinal Stem Cell Niche. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Madison, B.B.; McKenna, L.B.; Dolson, D.; Epstein, D.J.; Kaestner, K.H. FoxF1 and FoxL1 Link Hedgehog Signaling and the Control of Epithelial Proliferation in the Developing Stomach and Intestine. J. Biol. Chem. 2009, 284, 5936–5944. [Google Scholar] [CrossRef] [Green Version]
- Umali, J.; Hawkey-Noble, A.; French, C.R. Loss of Foxc1 in Zebrafish Reduces Optic Nerve Size and Cell Number in the Retinal Ganglion Cell Layer. Vision Res. 2019, 156, 66–72. [Google Scholar] [CrossRef]
- Whitesell, T.R.; Chrystal, P.W.; Ryu, J.-R.; Munsie, N.; Grosse, A.; French, C.R.; Workentine, M.L.; Li, R.; Zhu, L.J.; Waskiewicz, A.; et al. Foxc1 Is Required for Embryonic Head Vascular Smooth Muscle Differentiation in Zebrafish. Dev. Biol. 2019, 453, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Mackay, E.W.; Apschner, A.; Schulte-Merker, S. A Bone to Pick with Zebrafish. BoneKEy Rep. 2013, 2, 445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witten, P.E.; Harris, M.P.; Huysseune, A.; Winkler, C. Chapter 13—Small Teleost Fish Provide New Insights into Human Skeletal Diseases. In Methods in Cell Biology; Detrich, H.W., Westerfield, M., Zon, L.I., Eds.; The Zebrafish; Academic Press: London, UK, 2017; Volume 138, pp. 321–346. [Google Scholar]
- Nikaido, M.; Tada, M.; Saji, T.; Ueno, N. Conservation of BMP Signaling in Zebrafish Mesoderm Patterning. Mech. Dev. 1997, 61, 75–88. [Google Scholar] [CrossRef]
- Wada, N.; Javidan, Y.; Nelson, S.; Carney, T.J.; Kelsh, R.N.; Schilling, T.F. Hedgehog Signaling Is Required for Cranial Neural Crest Morphogenesis and Chondrogenesis at the Midline in the Zebrafish Skull. Development 2005, 132, 3977–3988. [Google Scholar] [CrossRef] [Green Version]
- Raterman, S.T.; Metz, J.R.; Wagener, F.A.D.T.G.; Von den Hoff, J.W. Zebrafish Models of Craniofacial Malformations: Interactions of Environmental Factors. Front. Cell Dev. Biol. 2020, 8, 1346. [Google Scholar] [CrossRef]
- Mork, L.; Crump, G. Zebrafish Craniofacial Development: A Window into Early Patterning. Curr. Top. Dev. Biol. 2015, 115, 235–269. [Google Scholar] [CrossRef] [Green Version]
- Bergen, D.J.M.; Kague, E.; Hammond, C.L. Zebrafish as an Emerging Model for Osteoporosis: A Primary Testing Platform for Screening New Osteo-Active Compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef] [Green Version]
- Anthwal, N.; Joshi, L.; Tucker, A.S. Evolution of the Mammalian Middle Ear and Jaw: Adaptations and Novel Structures. J. Anat. 2013, 222, 147–160. [Google Scholar] [CrossRef]
- Takechi, M.; Kuratani, S. History of Studies on Mammalian Middle Ear Evolution: A Comparative Morphological and Developmental Biology Perspective. J. Exp. Zoolog. B Mol. Dev. Evol. 2010, 314B, 417–433. [Google Scholar] [CrossRef]
- Ankamreddy, H.; Bok, J.; Groves, A.K. Uncovering the Secreted Signals and Transcription Factors Regulating the Development of Mammalian Middle Ear Ossicles. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2020, 249, 1410–1424. [Google Scholar] [CrossRef]
- Apschner, A.; Schulte-Merker, S.; Witten, P.E. Chapter 10—Not All Bones Are Created Equal—Using Zebrafish and Other Teleost Species in Osteogenesis Research. In Methods in Cell Biology; Detrich, H.W., Westerfield, M., Zon, L.I., Eds.; The Zebrafish: Disease Models and Chemical Screens; Academic Press: London, UK, 2011; Volume 105, pp. 239–255. [Google Scholar]
- Chang, Z.; Chen, P.-Y.; Chuang, Y.-J.; Akhtar, R. Zebrafish as a Model to Study Bone Maturation: Nanoscale Structural and Mechanical Characterization of Age-Related Changes in the Zebrafish Vertebral Column. J. Mech. Behav. Biomed. Mater. 2018, 84, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Du, S.J.; Frenkel, V.; Kindschi, G.; Zohar, Y. Visualizing Normal and Defective Bone Development in Zebrafish Embryos Using the Fluorescent Chromophore Calcein. Dev. Biol. 2001, 238, 239–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Lin, H.; Lan, F.; Wu, Y.; Yang, Z.; Zhang, J. Application of Bone Transgenic Zebrafish in Anti-Osteoporosis Chemical Screening. Anim. Models Exp. Med. 2018, 1, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kwon, R.Y.; Watson, C.J.; Karasik, D. Using Zebrafish to Study Skeletal Genomics. Bone 2019, 126, 37–50. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.M.; Kimmel, C. A Two-Color Acid-Free Cartilage and Bone Stain for Zebrafish Larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High-Resolution in Situ Hybridization to Whole-Mount Zebrafish Embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Peterson, S.M.; Freeman, J.L. RNA Isolation from Embryonic Zebrafish and CDNA Synthesis for Gene Expression Analysis. J. Vis. Exp. JoVE 2009, 30, 1470. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An Improvement of the 2ˆ (–Delta Delta CT) Method for Quantitative Real-Time Polymerase Chain Reaction Data Analysis. Biostat. Bioinforma. Biomath. 2013, 3, 71–85. [Google Scholar]
- Atan, D.; Atan, T.; Özcan, K.M.; Ensari, S.; Dere, H. Relation of Otosclerosis and Osteoporosis: A Bone Mineral Density Study. Auris. Nasus. Larynx 2016, 43, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Markou, K.; Goudakos, J. An Overview of the Etiology of Otosclerosis. Eur. Arch. Otorhinolaryngol. 2008, 266, 25. [Google Scholar] [CrossRef] [PubMed]
- Niedermeyer, H.P.; Arnold, W. Etiopathogenesis of Otosclerosis. ORL 2002, 64, 114–119. [Google Scholar] [CrossRef]
- Schrauwen, I.; Khalfallah, A.; Ealy, M.; Fransen, E.; Claes, C.; Huber, A.; Murillo, L.R.; Masmoudi, S.; Smith, R.J.H.; Van Camp, G. COL1A1 Association and Otosclerosis: A Meta-Analysis. Am. J. Med. Genet. A. 2012, 158A, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Niedermeyer, H.P.; Becker, E.T.; Arnold, W. Expression of Collagens in the Otosclerotic Bone. Adv. Otorhinolaryngol. 2007, 65, 45–49. [Google Scholar] [CrossRef] [Green Version]
- Green, J.; Taylor, J.J.; Hindes, A.; Johnson, S.L.; Goldsmith, M.I. A Gain of Function Mutation Causing Skeletal Overgrowth in the Rapunzel Mutant. Dev. Biol. 2009, 334, 224–234. [Google Scholar] [CrossRef] [Green Version]
- Kitazawa, T.; Fujisawa, K.; Narboux-Nême, N.; Arima, Y.; Kawamura, Y.; Inoue, T.; Wada, Y.; Kohro, T.; Aburatani, H.; Kodama, T.; et al. Distinct Effects of Hoxa2 Overexpression in Cranial Neural Crest Populations Reveal That the Mammalian Hyomandibular-Ceratohyal Boundary Maps within the Styloid Process. Dev. Biol. 2015, 402, 162–174. [Google Scholar] [CrossRef] [Green Version]
- Nakada, C.; Satoh, S.; Tabata, Y.; Arai, K.; Watanabe, S. Transcriptional Repressor Foxl1 Regulates Central Nervous System Development by Suppressing Shh Expression in Zebra Fish. Mol. Cell. Biol. 2006, 26, 7246–7257. [Google Scholar] [CrossRef] [Green Version]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.-L.; et al. Genetic Compensation Triggered by Mutant MRNA Degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Stainier, D.Y.R. Genetic Compensation: A Phenomenon in Search of Mechanisms. PLoS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef] [Green Version]
- Salanga, C.M.; Salanga, M.C. Genotype to Phenotype: CRISPR Gene Editing Reveals Genetic Compensation as a Mechanism for Phenotypic Disjunction of Morphants and Mutants. Int. J. Mol. Sci. 2021, 22, 3472. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.E.; Mikulec, A.A.; Mikulec, K.H.; Merchant, S.N.; McKenna, M.J. Association between Osteoporosis and Otosclerosis in Women. J. Laryngol. Otol. 2004, 118, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, I.; Zaret, K.S.; Santisteban, P. The Forkhead Factor FoxE1 Binds to the Thyroperoxidase Promoter during Thyroid Cell Differentiation and Modifies Compacted Chromatin Structure. Mol. Cell. Biol. 2007, 27, 7302–7314. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, T.; Muthurajan, U.M.; Luger, K.; Tulin, A.V.; Zaret, K.S. Nucleosome-Binding Affinity as a Primary Determinant of the Nuclear Mobility of the Pioneer Transcription Factor FoxA. Genes Dev. 2009, 23, 804–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifi, M.; Walter, M.a. Axenfeld-Rieger Syndrome. Clin. Genet. 2018, 93, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Delanghe, J.R.; Langlois, M.R. Hemopexin: A Review of Biological Aspects and the Role in Laboratory Medicine. Clin. Chim. Acta 2001, 312, 13–23. [Google Scholar] [CrossRef]
- Tolosano, E.; Altruda, F. Hemopexin: Structure, Function, and Regulation. DNA Cell Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, I.; Kobayashi-Sun, J.; Hirakawa, Y.; Ouchi, M.; Yasuda, K.; Kamei, H.; Fukuhara, S.; Yamaguchi, M. Dual Role of Jam3b in Early Hematopoietic and Vascular Development. Development 2020, 147, dev181040. [Google Scholar] [CrossRef]
- Cesari, M.; Pahor, M.; Lauretani, F.; Penninx, B.W.H.J.; Bartali, B.; Russo, R.; Cherubini, A.; Woodman, R.; Bandinelli, S.; Guralnik, J.M.; et al. Bone Density and Hemoglobin Levels in Older Persons: Results from the InCHIANTI Study. Osteoporos. Int. J. Establ. Result Coop. Eur. Found. Osteoporos. Natl. Osteoporos. Found. USA 2005, 16, 691–699. [Google Scholar] [CrossRef] [Green Version]
- Chuang, M.-H.; Chuang, T.-L.; Koo, M.; Wang, Y.-F. Low Hemoglobin Is Associated with Low Bone Mineral Density and High Risk of Bone Fracture in Male Adults: A Retrospective Medical Record Review Study. Am. J. Mens Health 2019, 13, 1557988319850378. [Google Scholar] [CrossRef]
- Steer, K.; Stavnichuk, M.; Morris, M.; Komarova, S.V. Bone Health in Patients with Hematopoietic Disorders of Bone Marrow Origin: Systematic Review and Meta-Analysis. J. Bone Miner. Res. 2017, 32, 731–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hawkey-Noble, A.; Pater, J.A.; Kollipara, R.; Fitzgerald, M.; Maekawa, A.S.; Kovacs, C.S.; Young, T.-L.; French, C.R. Mutation of foxl1 Results in Reduced Cartilage Markers in a Zebrafish Model of Otosclerosis. Genes 2022, 13, 1107. https://doi.org/10.3390/genes13071107
Hawkey-Noble A, Pater JA, Kollipara R, Fitzgerald M, Maekawa AS, Kovacs CS, Young T-L, French CR. Mutation of foxl1 Results in Reduced Cartilage Markers in a Zebrafish Model of Otosclerosis. Genes. 2022; 13(7):1107. https://doi.org/10.3390/genes13071107
Chicago/Turabian StyleHawkey-Noble, Alexia, Justin A. Pater, Roshni Kollipara, Meriel Fitzgerald, Alexandre S. Maekawa, Christopher S. Kovacs, Terry-Lynn Young, and Curtis R. French. 2022. "Mutation of foxl1 Results in Reduced Cartilage Markers in a Zebrafish Model of Otosclerosis" Genes 13, no. 7: 1107. https://doi.org/10.3390/genes13071107
APA StyleHawkey-Noble, A., Pater, J. A., Kollipara, R., Fitzgerald, M., Maekawa, A. S., Kovacs, C. S., Young, T. -L., & French, C. R. (2022). Mutation of foxl1 Results in Reduced Cartilage Markers in a Zebrafish Model of Otosclerosis. Genes, 13(7), 1107. https://doi.org/10.3390/genes13071107






