iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Isolation and Quantification
2.3. PCR and iPBS Marker Analyses
2.4. Data Scoring and Analysis
3. Results
3.1. Polymorphism Revealed by iPBS Primers
3.2. Genetic Diversity
3.3. Heterozygosity and Diversity of Varieties
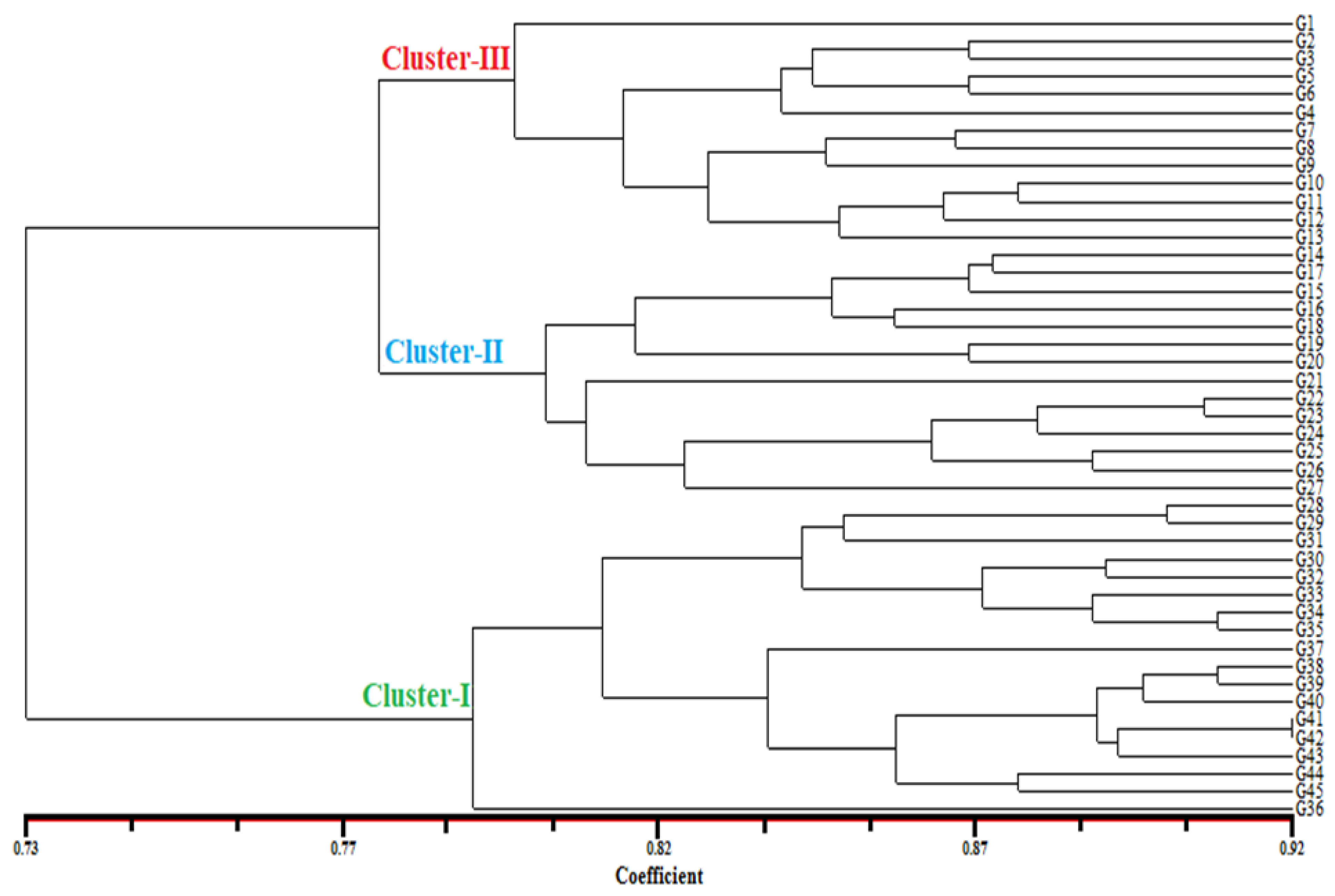
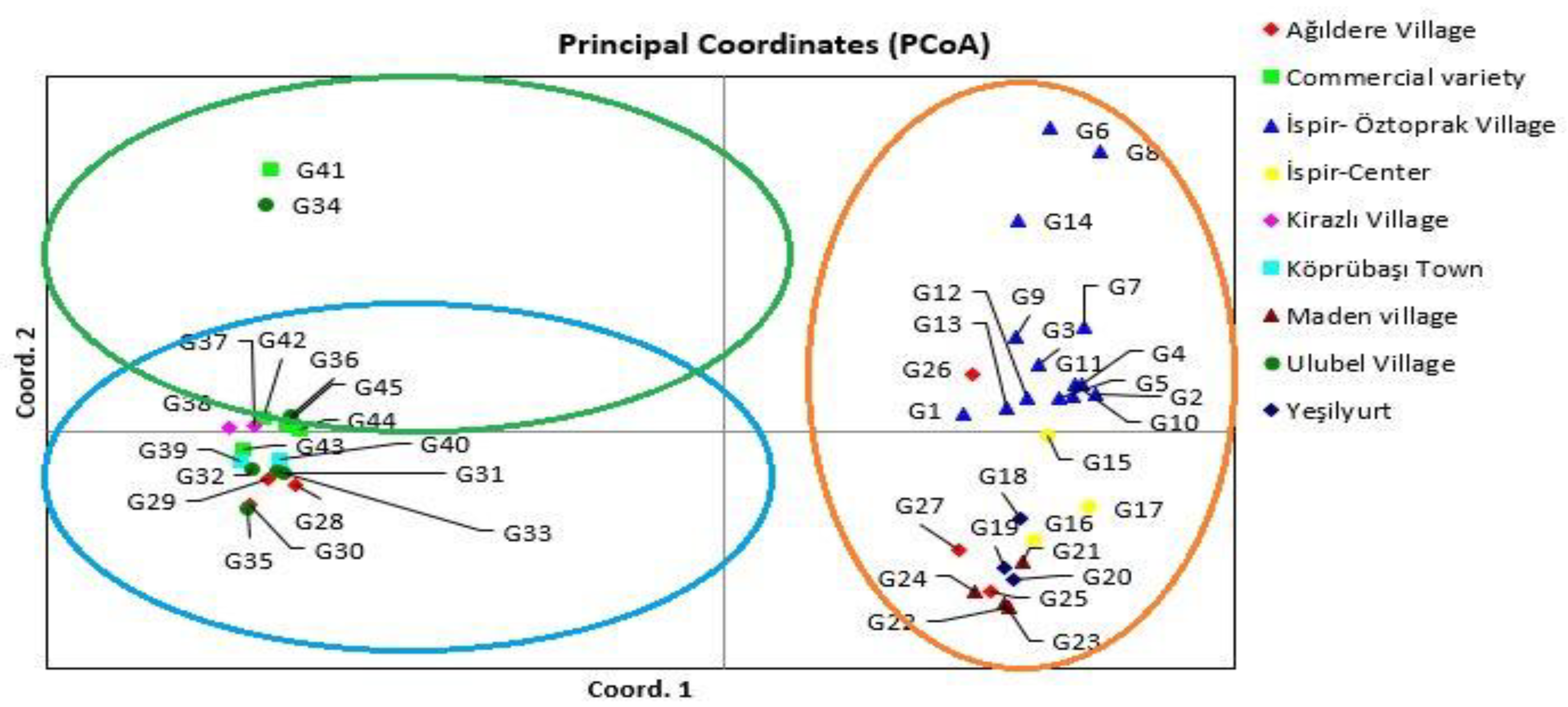
3.4. Principal Coordinate Analysis (PCoA) and Dendrogram Generated from 26 iPBS Markers
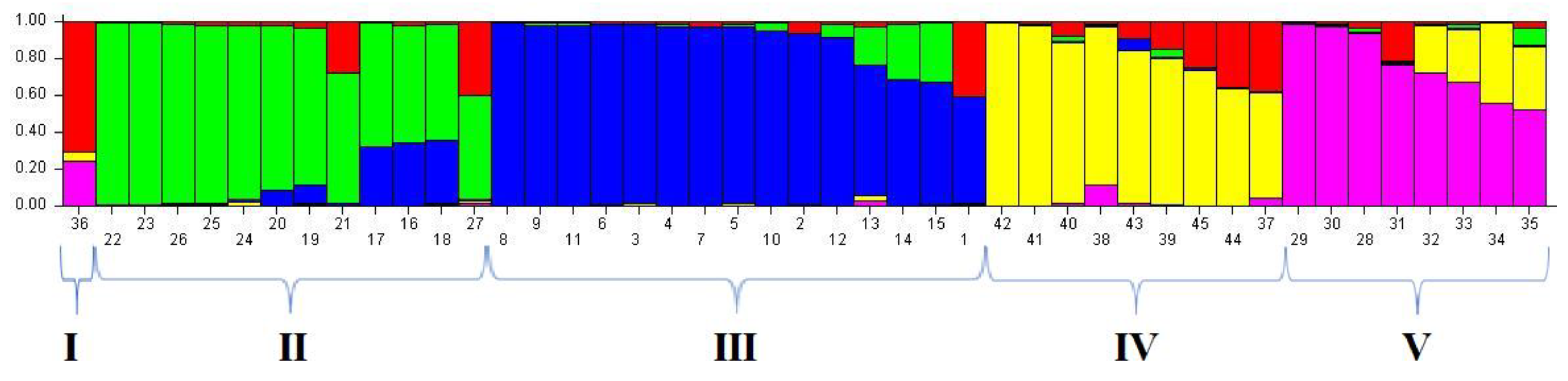
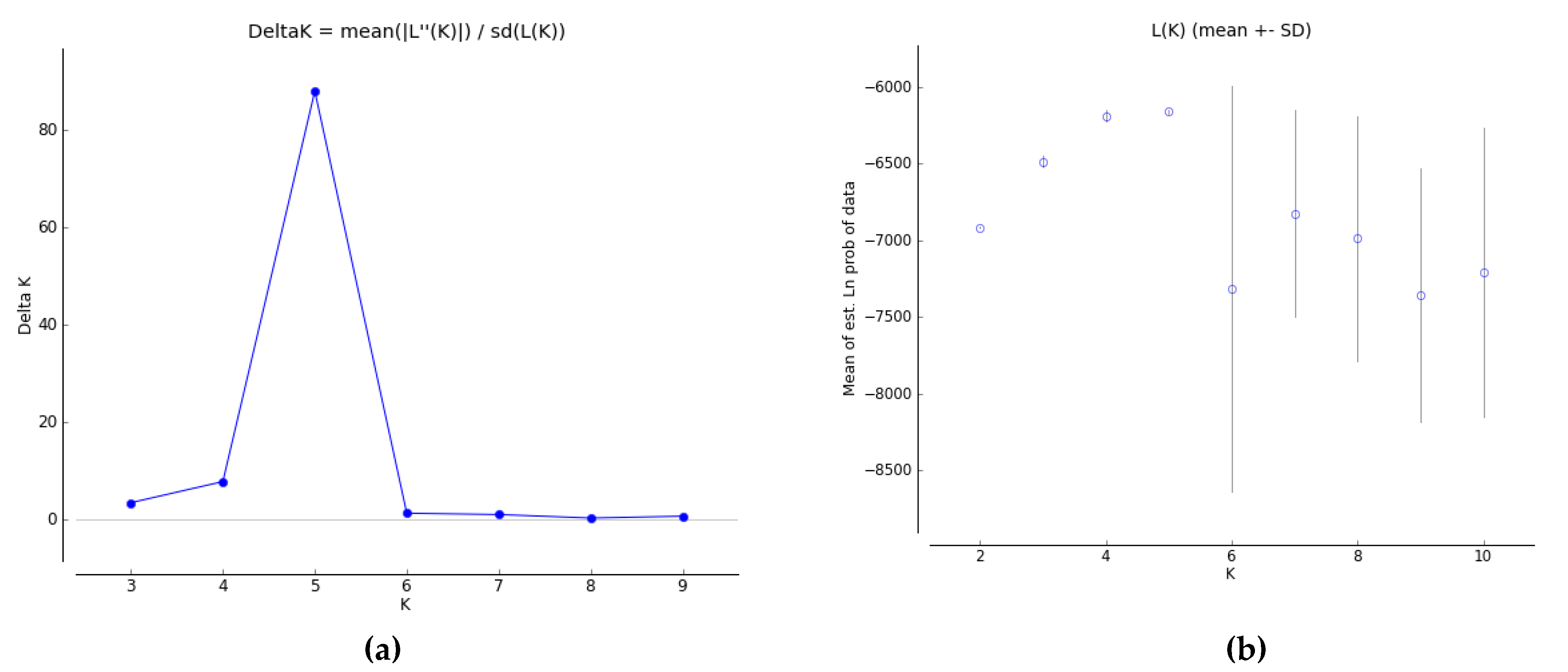
3.5. Population Genetic Structure Analysis for iPBS Markers
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savić, A.; Pipan, B.; Vasić, M.; Meglič, V. Genetic diversity of common bean (Phaseolus vulgaris L.) germplasm from Serbia, as revealed by single sequence repeats (SSR). Sci. Hortic. 2021, 288, 110405. [Google Scholar] [CrossRef]
- De Ron, A.M.; González, A.M.; Rodiño, A.P.; Santalla, M.; Godoy, L.; Papa, R. History of the common bean crop: Its evolution beyond its areas of origin and domestication. Arbor 2016, 192, a317. [Google Scholar] [CrossRef] [Green Version]
- Karık, Ü.; Nadeem, M.A.; Habyarimana, E.; Ercişli, S.; Yildiz, M.; Yılmaz, A.; Yang, S.H.; Chung, G.; Baloch, F.S. Exploring the Genetic Diversity and Population Structure of Turkish Laurel Germplasm by the iPBS-Retrotransposon Marker System. Agronomy 2019, 9, 647. [Google Scholar] [CrossRef] [Green Version]
- Hamouda, M.; Haider, A.S.; Elbehairy, E.; Elshanshory, A.R. Genetic variation among common beans cultivars (Phaseolus vulgaris L.) as revealed by morphological, protein and molecular markers. Egypt. J. Exp. Biol. 2020, 16, 129–139. [Google Scholar]
- Cabral, P.D.S.; de Souza, L.C.; da Costa, G.F.; Silva, F.H.L.; Soares, T. Investigation of the genetic diversity of common bean (Phaseolus vulgaris.) cultivars using molecular markers. Genet. Mol. Res. 2018, 17, 1–11. [Google Scholar] [CrossRef]
- Gioia, T.; Logozzo, G.; Marzario, S.; Spagnoletti Zeuli, P.; Gepts, P. Evolution of SSR diversity from wild types to US advanced cultivars in the Andean and Mesoamerican domestications of common bean (Phaseolus vulgaris). PLoS ONE 2019, 14, e0211342. [Google Scholar] [CrossRef] [Green Version]
- Ceylan, A.; Öcal, N.; Akbulut, M. Genetic diversity among the Turkish common bean cultivars (Phaseolus vulgaris L.) as assessed by SRAP, POGP and cpSSR markers. Biochem. Syst. Ecol. 2014, 54, 219–229. [Google Scholar] [CrossRef]
- Aydin, M.F.; Baloch, F.S. Exploring the genetic diversity and population structure of Turkish common bean germplasm by the iPBS-retrotransposons markers. Legum. Res. 2019, 42, 18–24. [Google Scholar] [CrossRef]
- Nemli, S.; Kianoosh, T.; Tanyolac, M.B. Genetic diversity and population structure of common bean (Phaseolus vulgaris L.) accessions through retrotransposon-based interprimer binding sites (iPBSs) markers. Turk. J. Agric. For. 2015, 39, 940–948. [Google Scholar] [CrossRef]
- Barut, M.; Nadeem, M.A.; Karaköy, T.; Baloch, F.S. DNA fingerprinting and genetic diversity analysis of world quinoa germplasm using iPBS-retrotransposon marker system. Turk. J. Agric. For. 2020, 44, 479–491. [Google Scholar] [CrossRef]
- Pinar, H.; Yahya, H.N.; Erċışlı, S.; Coskun, O.F.; Yaman, M.; Turgunbaev, K.; Uzun, A. Molecular Characterization of Barberry Genotypes from Turkey and Kyrgyzstan. Erwerbs-Obstbau 2021, 63, 403–407. [Google Scholar] [CrossRef]
- Uzun, A.; Yaman, M.; Pinar, H.; Gok, B.D.; Gazel, I. Leaf and fruit characteristics and genetic diversity of wild fruit cerasus prostrata genotypes collected from the Central Anatolia, Turkey. Acta Sci. Pol. Hortorum Cultus 2021, 20, 53–62. [Google Scholar] [CrossRef]
- Yaman, M. Determination of genetic diversity in european cranberrybush (Viburnum opulus L.) genotypes based on morphological, phytochemical and ISSR markers. Genet. Resour. Crop Evol. 2022, 69, 1889–1899. [Google Scholar] [CrossRef]
- Yildiz, E.; Pinar, H.; Uzun, A.; Yaman, M.; Sumbul, A.; Ercisli, S. Identification of genetic diversity among Juglans regia L. genotypes using molecular, morphological, and fatty acid data. Genet. Resour. Crop Evol. 2021, 68, 1425–1437. [Google Scholar] [CrossRef]
- Le Corre, V.; Kremer, A. Genetic variability at neutral markers, quantitative trait loci and trait in a subdivided population under selection. Genetics 2003, 164, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.R.P. Hybrid and Varietal Genetic Purity Testing Methods for Crop Improvement. Int. J. Appl. Biol. Pharm. Technol. 2014, 5, 197–199. [Google Scholar]
- Corrado, G.; Caramante, M.; Piffanelli, P.; Rao, R. Genetic diversity in Italian tomato landraces: Implications for the development of a core collection. Sci. Hortic. 2014, 168, 138–144. [Google Scholar] [CrossRef]
- Svetleva, D.; Pereira, G.; Carlier, J.; Cabrita, L.; Leitão, J.; Genchev, D. Molecular characterization of Phaseolus vulgaris L. genotypes included in Bulgarian collection by ISSR and AFLP™ analyses. Sci. Hortic. 2006, 109, 198–206. [Google Scholar] [CrossRef]
- Biswas, M.; Hassan, J.; Hossain, M. Assessment of genetic diversity in French bean (Phaseolus vulgaris L) based on RAPD marker. Afr. J. Biotechnol. 2010, 9, 5073–5077. [Google Scholar]
- Madakbaş, S.Y.; Sarıkamış, G.; Başak, H.; Karadavut, U.; Özmen, C.Y.; Daşçı, M.G.; Cayan, S. Genetic characterization of green bean (Phaseolus vulgaris L.) accessions from Turkey with SCAR and SSR markers. Biochem. Genet. 2016, 54, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Chavarro, M.C.; Blair, M.W. SNP marker diversity in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 2011, 123, 827–845. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.A.; Rangel, P.N.; Brondani, C.; Martins, W.S.; Melo, L.C.; Carneiro, M.S.; Borba, T.C.; Brondani, R.P. The characterization of a new set of EST-derived simple sequence repeat (SSR) markers as a resource for the genetic analysis of Phaseolus vulgaris. BMC Genet. 2011, 12, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parmar, S.C.; Thakur, N.; Nath, A.K. Assessment of Genetic Diversity among Kidney Bean (Phaseolus vulgaris L.) Cultivars using EST-Simple Sequence Repeat (SSR) Markers. Appl. Biol. Res. 2018, 20, 163–170. [Google Scholar] [CrossRef]
- Nadeem, M.A. Deciphering the genetic diversity and population structure of Turkish bread wheat germplasm using iPBS-retrotransposons markers. Mol. Biol. Rep. 2021, 48, 6739–6748. [Google Scholar] [CrossRef]
- Arystanbekkyzy, M.; Nadeem, M.A.; Aktas, H.; Yeken, M.Z.; Zencirci, N.; Nawaz, M.A.; Ali, F.; Haider, M.S.; Tunc, K.; Chung, G.; et al. Phylogenetic and taxonomic relationship of Turkish wild and cultivated emmer (Triticum turgidum ssp. dicoccoides) revealed by iPBS retrotransposons markers. Int. J. Agric. Biol. 2019, 21, 155–163. [Google Scholar]
- Öztürk, H.İ.; Dursun, A.; Hosseinpour, A.; Haliloğlu, K. Genetic diversity of pinto and fresh bean (Phaseolus vulgaris L.) germplasm collected from Erzincan province of Turkey by inter-primer binding site (iPBS) retrotransposon markers. Turk. J. Agric. For. 2020, 44, 417–427. [Google Scholar] [CrossRef]
- Zeinalzadehtabrizi, H.; Hosseinpour, A.; Aydin, M.; Haliloglu, K. A modified genomic DNA extraction method from leaves of sunflower for PCR based analyzes. J. Biodivers. Environ. Sci. 2015, 7, 222–225. [Google Scholar]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Hosseinpour, A.; Karahan, F.; İlhan, E.; İlçim, A.; Haliloğlu, K. Genetic structure and diversity of Adonis L. (Ranunculaceae) populations collected from Turkey by inter-primer binding site (iPBS) retrotransposon markers. Turk.J. Bot. 2019, 43, 585–596. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Anderson, J.A.; Churchill, G.; Autrique, J.; Tanksley, S.; Sorrells, M. Optimizing parental selection for genetic linkage maps. Genome 1993, 3, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C.; Yang, R.; Boyle, T.B.; Ye, Z.; Mao, J.X. POPGENE, the user-friendly shareware for population genetic analysis. Mol. Biol. Biotechnol. Centre 1997, 10, 295–301. Available online: https://www.scienceopen.com/document?vid=5db9fe1d-3632-465d-9e0a-5b2c7f417d08 (accessed on 20 November 2018).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Smouse, R.P.P.; Peakall, R. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar]
- Mhlaba, Z.B.; Mashilo, J.; Shimelis, H.; Assefa, A.B.; Modi, A.T. Progress in genetic analysis and breeding of tepary bean (Phaseolus acutifolius A. Gray), A review. Sci. Hortic. 2018, 237, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Todorovska, E. Retrotransposons and their role in plant—Genome evolution. Biotechnol. Biotechnol. Equip. 2007, 21, 294–305. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Elston, R. Linkage information content of polymorphic genetic markers. Hum. Hered. 1999, 49, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Baloch, F.S.; Alsaleh, A.; de Miera, L.E.S.; Hatipoğlu, R.; Çiftçi, V.; Karaköy, T.; Yıldız, M.; Özkan, H. DNA based iPBS-retrotransposon markers for investigating the population structure of pea (Pisum sativum) germplasm from Turkey. Biochem. Syst. Ecol. 2015, 61, 244–252. [Google Scholar] [CrossRef]
- Mehmood, A.; Jaskani, M.J.; Ahmad, S.; Ahma, R. Evaluation of genetic diversity in open pollinated guava by iPBS primers. Pak. J. Agric. Sci. 2013, 50, 591–597. [Google Scholar]
- Guo, D.L.; Guo, M.X.; Hou, X.G.; Zhang, G.H. Molecular diversity analysis of grape varieties based on iPBS markers. Biochem. Syst. Ecol. 2014, 52, 27–32. [Google Scholar] [CrossRef]
- Shimira, F.; Boyaci, H.F.; Çilesiz, Y.; Nadeem, M.A.; Baloch, F.S.; Taşkin, H. Exploring the genetic diversity and population structure of scarlet eggplant germplasm from Rwanda through iPBS-retrotransposon markers. Mol. Biol. Rep. 2021, 48, 6323–6333. [Google Scholar] [CrossRef]
- Ellstrand, N.C.; Elam, D.R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Evol. Syst. 1993, 24, 217–242. [Google Scholar] [CrossRef]
- Mohammadi, S.A.; Prasanna, B. Analysis of genetic diversity in crop plants—salient statistical tools and considerations. Crop Sci. 2003, 43, 1235–1248. [Google Scholar] [CrossRef] [Green Version]
- Karagoz, H.; Cakmakci, R.; Hosseinpour, A.; Ozkan, G.; Haliloglu, K. Analysis of genetic variation and population structure among of oregano (Origanum acutidens L.) accessions revealed by agro-morphological traits, oil constituents and retrotransposon-based inter-primer binding sites (iPBS) markers. Genet. Resour. Crop Evol. 2020, 67, 1367–1384. [Google Scholar] [CrossRef]
- Singh, V.J.; Bhowmick, P.K.; Vinod, K.K.; Krishnan, S.G.; Nandakumar, S.; Kumar, A.; Kumar, M.; Shekhawat, S.; Dixit, B.K.; Malik, A.; et al. Population Structure of a Worldwide Collection of Tropical Japonica Rice Indicates Limited Geographic Differentiation and Shows Promising Genetic Variability Associated with New Plant Type. Genes 2022, 13, 484. [Google Scholar] [CrossRef]
- Zargar, S.M.; Farhat, S.; Mahajan, R.; Bhakhri, A.; Sharma, A. Unraveling the efficiency of RAPD and SSR markers in diversity analysis and population structure estimation in common bean. Saudi J. Biol. Sci. 2016, 23, 139–149. [Google Scholar] [CrossRef] [Green Version]
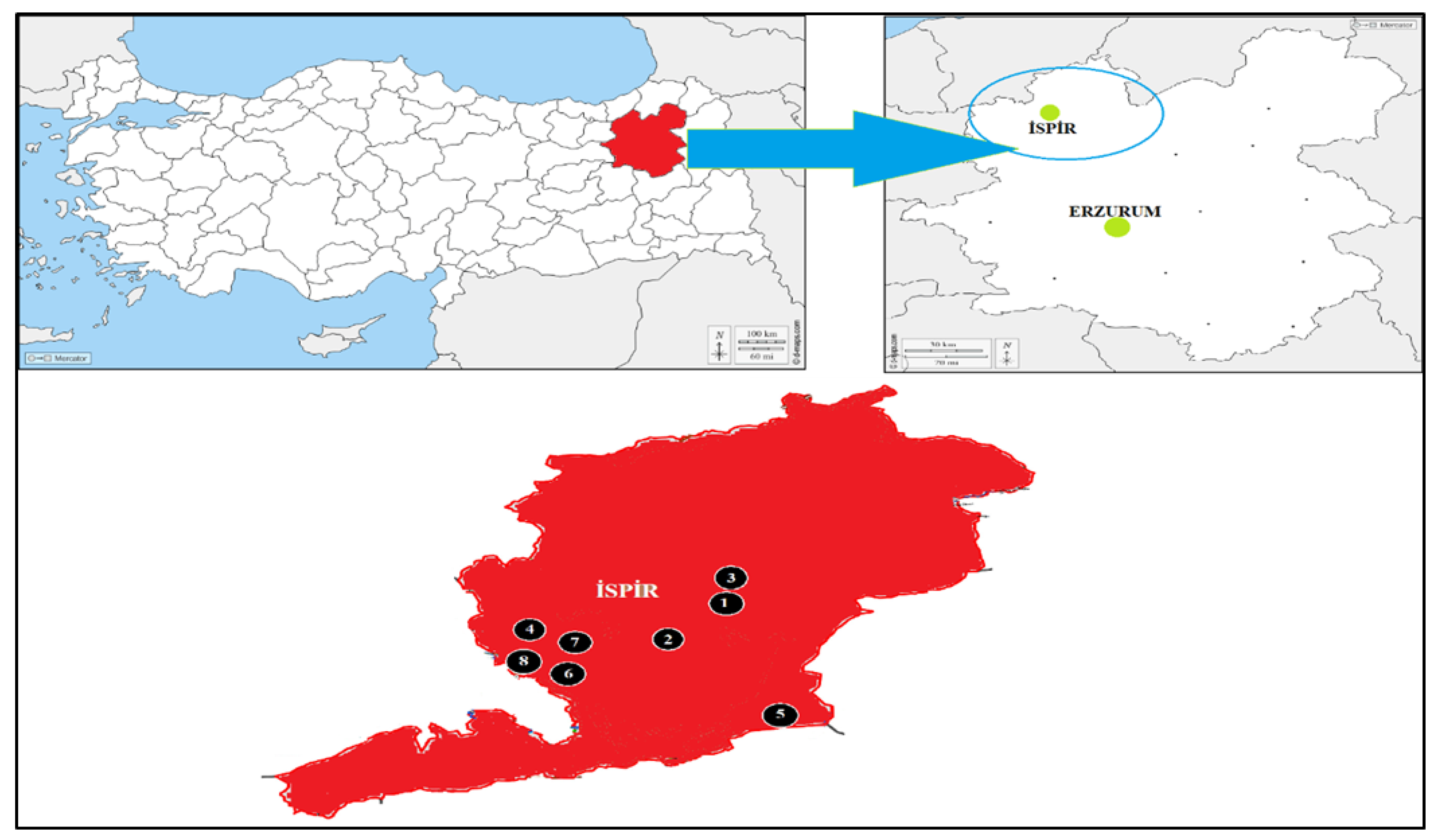
| Variety | Collected Location | Latitude | Longitude | Altitude (m) |
|---|---|---|---|---|
| G1 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G2 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G3 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G4 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G5 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G6 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G7 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G8 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G9 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G10 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G11 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G12 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G13 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G14 | Ispir- Öztoprak village | 40.518 | 41.052 | 1431 |
| G15 | Ispir-center | 40.485 | 41.002 | 1264 |
| G16 | Ispir-center | 40.468 | 40.983 | 1168 |
| G17 | Ispir-center | 40.468 | 40.983 | 1168 |
| G18 | Yeşilyurt | 40.518 | 41.069 | 1549 |
| G19 | Yeşilyurt | 40.518 | 41.069 | 1549 |
| G20 | Yeşilyurt | 40.518 | 41.069 | 1549 |
| G21 | Maden village | 40.435 | 40.851 | 1226 |
| G22 | Maden village | 40.435 | 40.851 | 1226 |
| G23 | Maden village | 40.435 | 40.851 | 1226 |
| G24 | Maden village | 40.435 | 40.851 | 1226 |
| G25 | Ağıldere village | 40.401 | 40.834 | 1470 |
| G26 | Ağıldere village | 40.401 | 40.834 | 1470 |
| G27 | Ağıldere village | 40.401 | 40.834 | 1470 |
| G28 | Ağıldere village | 40.401 | 40.834 | 1470 |
| G29 | Ağıldere village | 40.401 | 40.834 | 1470 |
| G30 | Ağıldere village | 40.401 | 40.834 | 1470 |
| G31 | Ulubel village | 40.418 | 40.868 | 1424 |
| G32 | Ulubel village | 40.418 | 40.868 | 1424 |
| G33 | Ulubel village | 40.418 | 40.868 | 1424 |
| G34 | Ulubel village | 40.418 | 40.868 | 1424 |
| G35 | Ulubel village | 40.418 | 40.868 | 1424 |
| G36 | Ulubel village | 40.418 | 40.868 | 1424 |
| G37 | Kirazlı village | 40.436 | 40.887 | 1220 |
| G38 | Kirazlı village | 40.436 | 40.887 | 1220 |
| G39 | Köprübaşı town | 40.434 | 40.819 | 1286 |
| G40 | Köprübaşı town | 40.434 | 40.819 | 1286 |
| G41 | Aras-98 | Commercial cultivars | ||
| G42 | Elkoca-05 | |||
| G43 | Göynük-98 | |||
| G44 | Karacaşehir-90 | |||
| G45 | Yakutiye-98 | |||
| Marker | Primers Sequences (5′→3′) | Marker | Primers Sequences (5′→3′) |
|---|---|---|---|
| iPBS-2074 | GCTCTGATACCA | iPBS-2377 | ACGAAGGGACCA |
| iPBS-2077 | CTCACGATGCCA | iPBS-2378 | GGTCCTCATCCA |
| iPBS-2078 | GCGGAGTCGCCA | iPBS-2380 | CAACCTGATCCA |
| iPBS-2079 | AGGTGGGCGCCA | iPBS-2381 | GTCCATCTTCCA |
| iPBS-2080 | CAGACGGCGCCA | iPBS-2383 | GCATGGCCTCCA |
| iPBS-2095 | GCTCGGATACCA | iPBS-2384 | GTAATGGGTCCA |
| iPBS-2231 | ACTTGGATGCTGATACCA | iPBS-2385 | CCATTGGGTCCA |
| iPBS-2270 | ACCTGGCGTGCCA | iPBS-2386 | CTGATCAACCCA |
| iPBS-2271 | GGCTCGGATGCCA | iPBS-2389 | ACATCCTTCCCA |
| iPBS-2274 | ATGGTGGGCGCCA | iPBS-2390 | GCAACAACCCCA |
| iPBS-2276 | ACCTCTGATACCA | iPBS-2391 | ATCTGTCAGCCA |
| iPBS-2278 | GCTCATGATACCA | iPBS-2392 | TAGATGGTGCCA |
| iPBS-2298 | AGAAGAGCTCTGATACCA | iPBS-2402 | TCTAAGCTCTTGATACCA |
| Marker | Number of Alleles | Major Allele Frequency | PIC * | Marker | Number of Alleles | Major Allele Frequency | PIC * |
|---|---|---|---|---|---|---|---|
| iPBS-2074 | 40 | 0.651 | 0.430 | iPBS-2377 | 45 | 0.715 | 0.309 |
| iPBS-2077 | 23 | 0.653 | 0.387 | iPBS-2378 | 64 | 0.805 | 0.241 |
| iPBS-2078 | 71 | 0.682 | 0.323 | iPBS-2380 | 51 | 0.678 | 0.336 |
| iPBS-2079 | 35 | 0.810 | 0.226 | iPBS-2381 | 57 | 0.687 | 0.359 |
| iPBS-2080 | 43 | 0.756 | 0.316 | iPBS-2383 | 23 | 0.528 | 0.495 |
| iPBS-2095 | 64 | 0.691 | 0.352 | iPBS-2384 | 56 | 0.761 | 0.252 |
| iPBS-2231 | 52 | 0.655 | 0.398 | iPBS-2385 | 63 | 0.728 | 0.313 |
| iPBS-2270 | 25 | 0.877 | 0.153 | iPBS-2386 | 64 | 0.612 | 0.397 |
| iPBS-2271 | 36 | 0.674 | 0.311 | iPBS-2389 | 65 | 0.587 | 0.396 |
| iPBS-2274 | 80 | 0.743 | 0.342 | iPBS-2390 | 62 | 0.654 | 0.431 |
| iPBS-2276 | 42 | 0.732 | 0.329 | iPBS-2391 | 53 | 0.668 | 0.341 |
| iPBS-2278 | 57 | 0.700 | 0.338 | iPBS-2392 | 47 | 0.654 | 0.379 |
| iPBS-2298 | 72 | 0.888 | 0.151 | iPBS-2402 | 60 | 0.776 | 0.292 |
| Mean | 52 | 0.706 | 0.331 | ||||
| Variety | ne * | h ** | I * | Variety | ne * | h ** | I * |
|---|---|---|---|---|---|---|---|
| G1 | 1.491 | 0.329 | 0.511 | G24 | 1.530 | 0.347 | 0.531 |
| G2 | 1.538 | 0.350 | 0.534 | G25 | 1.586 | 0.369 | 0.556 |
| G3 | 1.540 | 0.351 | 0.535 | G26 | 1.550 | 0.355 | 0.540 |
| G4 | 1.601 | 0.376 | 0.563 | G27 | 1.470 | 0.320 | 0.500 |
| G5 | 1.521 | 0.343 | 0.526 | G28 | 1.658 | 0.397 | 0.586 |
| G6 | 1.568 | 0.362 | 0.548 | G29 | 1.696 | 0.410 | 0.601 |
| G7 | 1.609 | 0.379 | 0.566 | G30 | 1.642 | 0.391 | 0.580 |
| G8 | 1.604 | 0.377 | 0.564 | G31 | 1.688 | 0.408 | 0.598 |
| G9 | 1.593 | 0.372 | 0.560 | G32 | 1.588 | 0.370 | 0.557 |
| G10 | 1.591 | 0.372 | 0.559 | G33 | 1.586 | 0.369 | 0.556 |
| G11 | 1.576 | 0.365 | 0.552 | G34 | 1.524 | 0.344 | 0.528 |
| G12 | 1.589 | 0.371 | 0.558 | G35 | 1.476 | 0.322 | 0.503 |
| G13 | 1.549 | 0.354 | 0.539 | G36 | 1.720 | 0.419 | 0.609 |
| G14 | 1.568 | 0.362 | 0.548 | G37 | 1.648 | 0.393 | 0.582 |
| G15 | 1.562 | 0.360 | 0.546 | G38 | 1.520 | 0.342 | 0.526 |
| G16 | 1.538 | 0.350 | 0.535 | G39 | 1.567 | 0.362 | 0.548 |
| G17 | 1.538 | 0.350 | 0.534 | G40 | 1.528 | 0.345 | 0.529 |
| G18 | 1.570 | 0.363 | 0.549 | G41 | 1.564 | 0.361 | 0.546 |
| G19 | 1.470 | 0.320 | 0.500 | G42 | 1.562 | 0.360 | 0.546 |
| G20 | 1.526 | 0.345 | 0.529 | G43 | 1.586 | 0.370 | 0.556 |
| G21 | 1.540 | 0.351 | 0.535 | G44 | 1.556 | 0.358 | 0.543 |
| G22 | 1.514 | 0.340 | 0.523 | G45 | 1.505 | 0.335 | 0.518 |
| G23 | 1.521 | 0.342 | 0.526 | Mean | 1.566 | 0.361 | 0.546 |
| Population | n | na | ne | I | He | uHe | PPL (%) |
|---|---|---|---|---|---|---|---|
| Av | 6 | 0.908 | 1.305 | 0.253 | 0.173 | 0.208 | 43.40 |
| Iov | 14 | 1.098 | 1.270 | 0.254 | 0.165 | 0.178 | 24.72 |
| Ic | 3 | 0.519 | 1.166 | 0.132 | 0.092 | 0.138 | 53.58 |
| Kv | 2 | 0.389 | 1.132 | 0.092 | 0.066 | 0.132 | 20.75 |
| Kt | 2 | 0.336 | 1.104 | 0.072 | 0.052 | 0.104 | 13.21 |
| Mv | 4 | 0.613 | 1.182 | 0.158 | 0.107 | 0.143 | 10.38 |
| Uv | 6 | 0.781 | 1.218 | 0.195 | 0.130 | 0.156 | 26.98 |
| Yy | 3 | 0.560 | 1.190 | 0.151 | 0.106 | 0.158 | 35.66 |
| Com | 5 | 0.574 | 1.165 | 0.142 | 0.096 | 0.120 | 23.77 |
| Mean | 0.642 | 1.192 | 0.161 | 0.110 | 0.149 | 28.05 |
| Av | Com | Iov | Ic | Kv | Kt | Mv | Uv | Yy | |
|---|---|---|---|---|---|---|---|---|---|
| Av | 0.000 | ||||||||
| Com | 0.125 | 0.000 | |||||||
| Iov | 0.124 | 0.179 | 0.000 | ||||||
| Ic | 0.137 | 0.215 | 0.081 | 0.000 | |||||
| Kv | 0.128 | 0.072 | 0.209 | 0.232 | 0.000 | ||||
| Kt | 0.129 | 0.071 | 0.207 | 0.222 | 0.071 | 0.000 | |||
| Mv | 0.099 | 0.202 | 0.114 | 0.109 | 0.215 | 0.211 | 0.000 | ||
| Uv | 0.068 | 0.085 | 0.177 | 0.202 | 0.081 | 0.108 | 0.184 | 0.000 | |
| Yy | 0.119 | 0.207 | 0.104 | 0.086 | 0.229 | 0.212 | 0.087 | 0.197 | 0.000 |
| Axis | 1 | 2 | 3 |
|---|---|---|---|
| % | 32.34 | 6.35 | 5.23 |
| Cum % | 32.34 | 38.69 | 43.92 |
| Scheme | Degree of Freedom (DF) | Sum of Squares (SS) | Variance Component | % Of Total Variance | p-Value |
|---|---|---|---|---|---|
| Among Population | 8 | 1150.70 | 21.439 | 33% | 0.332 |
| Within Population | 36 | 1554.89 | 43.192 | 67% | 0.001 |
| Total | 44 | 2705.60 | 64.631 | 100% |
| Subpopulation | Subpopulation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Varieties | I | II | III | IV | V | Varieties | I | II | III | IV | V |
| G1 | 0.401 | 0.005 | 0.579 | 0.004 | 0.012 | G24 | 0.017 | 0.946 | 0.009 | 0.025 | 0.003 |
| G2 | 0.059 | 0.005 | 0.923 | 0.008 | 0.006 | G25 | 0.021 | 0.960 | 0.002 | 0.004 | 0.014 |
| G3 | 0.009 | 0.002 | 0.972 | 0.013 | 0.004 | G26 | 0.012 | 0.968 | 0.004 | 0.005 | 0.011 |
| G4 | 0.014 | 0.012 | 0.970 | 0.003 | 0.001 | G27 | 0.399 | 0.560 | 0.010 | 0.011 | 0.019 |
| G5 | 0.011 | 0.011 | 0.961 | 0.011 | 0.006 | G28 | 0.033 | 0.018 | 0.004 | 0.007 | 0.938 |
| G6 | 0.008 | 0.003 | 0.975 | 0.011 | 0.003 | G29 | 0.004 | 0.002 | 0.003 | 0.002 | 0.989 |
| G7 | 0.024 | 0.002 | 0.969 | 0.002 | 0.002 | G30 | 0.009 | 0.004 | 0.005 | 0.004 | 0.979 |
| G8 | 0.002 | 0.003 | 0.993 | 0.001 | 0.001 | G31 | 0.214 | 0.003 | 0.005 | 0.010 | 0.767 |
| G9 | 0.007 | 0.009 | 0.980 | 0.003 | 0.002 | G32 | 0.010 | 0.004 | 0.003 | 0.257 | 0.727 |
| G10 | 0.007 | 0.041 | 0.946 | 0.003 | 0.003 | G33 | 0.011 | 0.024 | 0.006 | 0.286 | 0.674 |
| G11 | 0.005 | 0.010 | 0.979 | 0.003 | 0.003 | G34 | 0.002 | 0.002 | 0.002 | 0.432 | 0.561 |
| G12 | 0.014 | 0.070 | 0.909 | 0.004 | 0.003 | G35 | 0.030 | 0.095 | 0.006 | 0.342 | 0.528 |
| G13 | 0.025 | 0.205 | 0.709 | 0.031 | 0.030 | G36 | 0.702 | 0.002 | 0.003 | 0.046 | 0.246 |
| G14 | 0.013 | 0.298 | 0.682 | 0.003 | 0.004 | G37 | 0.378 | 0.004 | 0.002 | 0.572 | 0.043 |
| G15 | 0.007 | 0.320 | 0.665 | 0.004 | 0.004 | G38 | 0.009 | 0.006 | 0.009 | 0.857 | 0.118 |
| G16 | 0.017 | 0.640 | 0.336 | 0.005 | 0.002 | G39 | 0.150 | 0.041 | 0.005 | 0.792 | 0.012 |
| G17 | 0.003 | 0.670 | 0.323 | 0.002 | 0.002 | G40 | 0.078 | 0.028 | 0.007 | 0.870 | 0.017 |
| G18 | 0.014 | 0.625 | 0.344 | 0.007 | 0.009 | G41 | 0.009 | 0.004 | 0.003 | 0.984 | 0.002 |
| G19 | 0.031 | 0.849 | 0.100 | 0.009 | 0.012 | G42 | 0.003 | 0.001 | 0.002 | 0.992 | 0.002 |
| G20 | 0.020 | 0.893 | 0.081 | 0.004 | 0.003 | G43 | 0.088 | 0.004 | 0.064 | 0.823 | 0.022 |
| G21 | 0.278 | 0.701 | 0.015 | 0.003 | 0.003 | G44 | 0.355 | 0.006 | 0.003 | 0.631 | 0.005 |
| G22 | 0.003 | 0.988 | 0.003 | 0.002 | 0.004 | G45 | 0.246 | 0.003 | 0.013 | 0.735 | 0.002 |
| G23 | 0.005 | 0.984 | 0.004 | 0.002 | 0.004 | ||||||
| Subpopulation (K) | Expected Heterozygosity (He) | FST |
|---|---|---|
| 1 | 0.3210 | 0.0002 |
| 2 | 0.1858 | 0.4371 |
| 3 | 0.1947 | 0.4061 |
| 4 | 0.1567 | 0.6372 |
| 5 | 0.1907 | 0.5440 |
| Mean | 0.2103 | 0.4049 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haliloğlu, K.; Türkoğlu, A.; Öztürk, H.I.; Özkan, G.; Elkoca, E.; Poczai, P. iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye. Genes 2022, 13, 1147. https://doi.org/10.3390/genes13071147
Haliloğlu K, Türkoğlu A, Öztürk HI, Özkan G, Elkoca E, Poczai P. iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye. Genes. 2022; 13(7):1147. https://doi.org/10.3390/genes13071147
Chicago/Turabian StyleHaliloğlu, Kamil, Aras Türkoğlu, Halil Ibrahim Öztürk, Güller Özkan, Erdal Elkoca, and Peter Poczai. 2022. "iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye" Genes 13, no. 7: 1147. https://doi.org/10.3390/genes13071147
APA StyleHaliloğlu, K., Türkoğlu, A., Öztürk, H. I., Özkan, G., Elkoca, E., & Poczai, P. (2022). iPBS-Retrotransposon Markers in the Analysis of Genetic Diversity among Common Bean (Phaseolus vulgaris L.) Germplasm from Türkiye. Genes, 13(7), 1147. https://doi.org/10.3390/genes13071147








