Expression of Modified Snowdrop Lectin (Galanthus nivalis Agglutinin) Protein Confers Aphids and Plutella xylostella Resistance in Arabidopsis and Cotton
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Synthesized ASGNA and Transgenic Plants
2.3. DNA Isolation
2.4. Determination of ASGNA Expression Levels in Transgenic Plants
2.5. Insect Bioassay
2.6. Transient GNA Expression in Cotton
3. Results
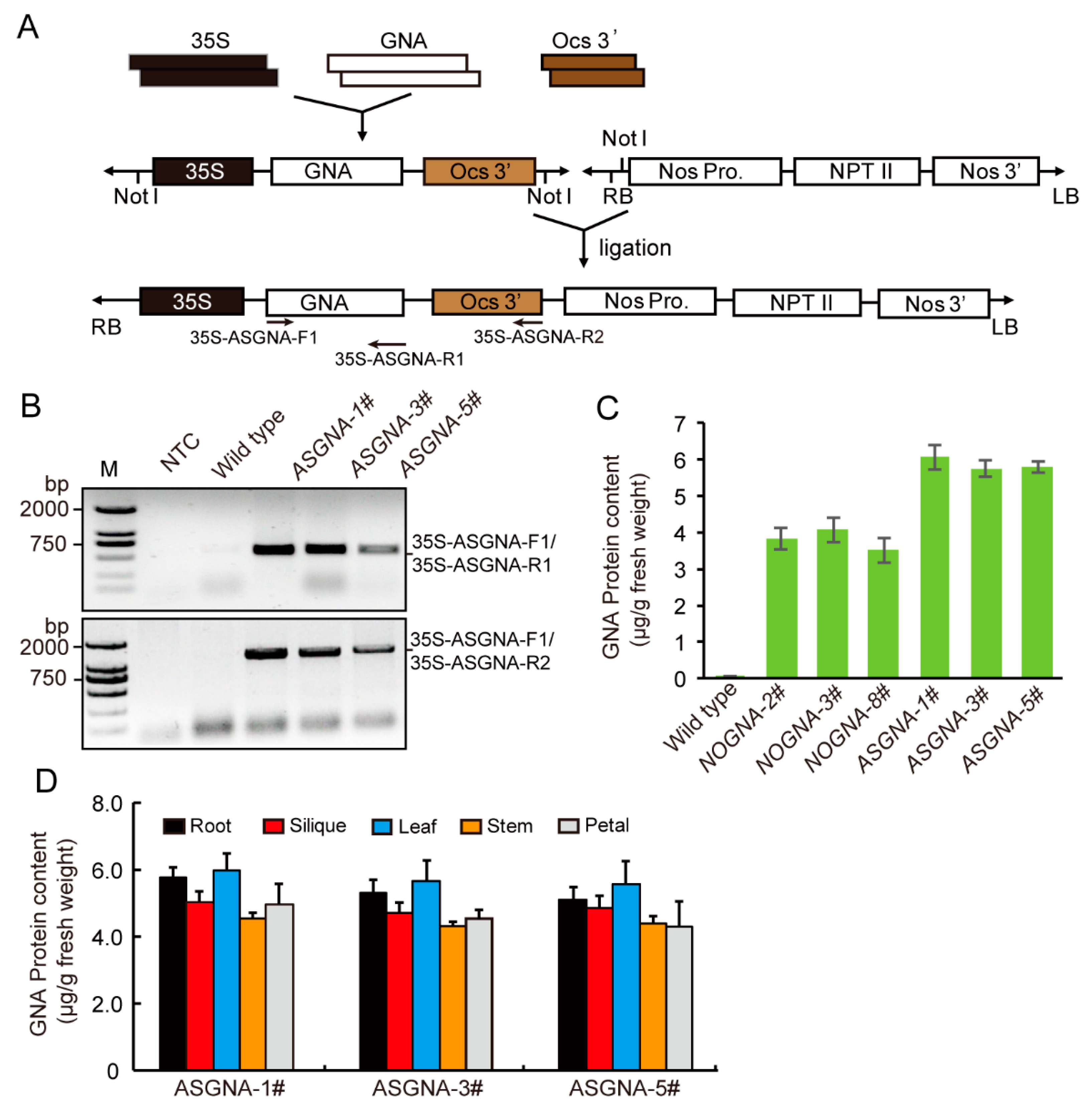
3.1. PCR Amplification of ASGNA Gene
3.2. Expression of ASGNA in Transgenic A. thaliana
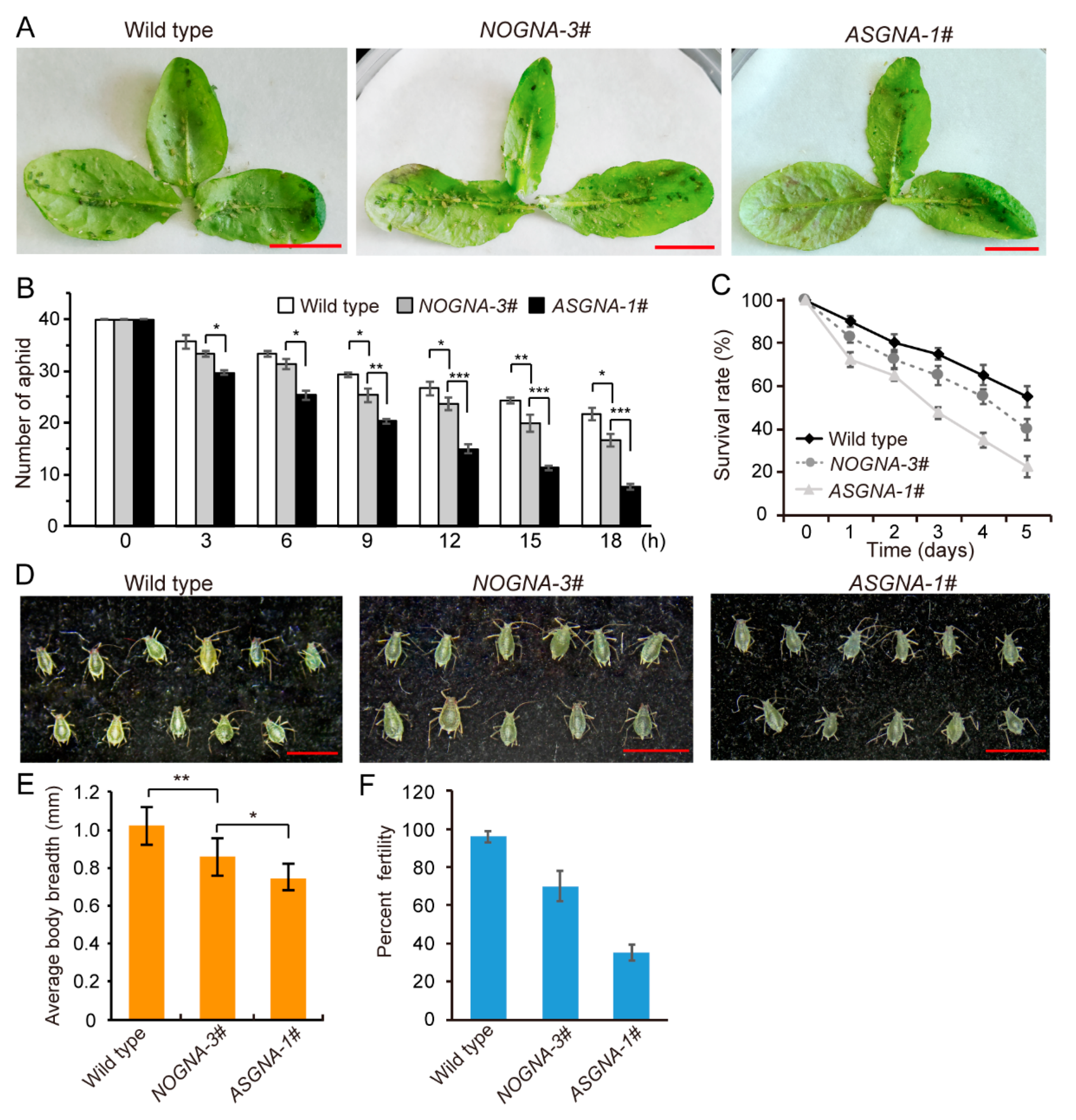
3.3. Effects of ASGNA Toxin on Aphids
3.4. Effects of ASGNA Toxin on P. xylostella
3.5. Transient Expression of ASGNA in Cotton Cotyledons Resulted in Enhanced Resistance to Aphids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, S.P.; Naranjo, S.E.; Wu, K.M. Biological control of cotton pests in China. Biol. Control 2014, 68, 6–14. [Google Scholar] [CrossRef]
- Wu, K.M.; Guo, Y.Y. The evolution of cotton pest management practices in China. Annu. Rev. Entomol. 2005, 50, 31–52. [Google Scholar] [CrossRef] [PubMed]
- Bestete, L.R.; Torres, J.B.; Silva, R.B.B.; Silva-Torres, C.S.A.; Bastos, C.S. Development of cotton pests exhibiting different feeding strategy on water-stressed and kaolin-treated cotton plants. J. Pest Sci. 2017, 90, 139–150. [Google Scholar] [CrossRef]
- Llandres, A.L.; Almohamad, R.; Brevault, T.; Renou, A.; Tereta, I.; Jean, J.; Goebel, F.R. Plant training for induced defense against insect pests: A promising tool for integrated pest management in cotton. Pest Manag. Sci. 2018, 74, 2004–2012. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chen, F.; Chen, C.; Chen, X.; Mao, Y. Transcriptome analysis of three cotton pests reveals features of gene expressions in the mesophyll feeder Apolygus lucorum. Sci. China Life Sci. 2017, 60, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.J.; Whitehouse, M.E.A.; Herron, G.A. The management of insect pests in Australian cotton: An evolving story. Annu. Rev. Entomol. 2018, 63, 215–237. [Google Scholar] [CrossRef]
- Wang, S.Y.; Qi, Y.F.; Desneux, N.; Shi, X.Y.; Biondi, A.; Gao, X.W. Sublethal and transgenerational effects of short-term and chronic exposures to the neonicotinoid nitenpyram on the cotton aphid Aphis. gossypii. J. Pest Sci. 2017, 90, 389–396. [Google Scholar] [CrossRef]
- Wu, K.M.; Lu, Y.H.; Feng, H.Q.; Jiang, Y.Y.; Zhao, J.Z. Suppression of cotton bollworm in multiple crops in china in areas with Bt toxin-containing cotton. Science 2008, 321, 1676–1678. [Google Scholar] [CrossRef] [Green Version]
- Lacey, L.A.; Georgis, R. Entomopathogenic nematodes for control of insect pests above and below ground with comments on commercial production. J. Nematol. 2012, 44, 218–225. [Google Scholar]
- Savci, S. An agricultural pollutant: Chemical fertilizer. Int. J. Environ. Sci. Dev. 2012, 3, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.H.; Wu, K.M.; Jiang, Y.Y.; Xia, B.; Li, P.; Feng, H.Q.; Wyckhuys, K.A.G.; Guo, Y.Y. Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science 2010, 328, 1151–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kathage, J.; Qaim, M. Economic impacts and impact dynamics of Bt (Bacillus thuringiensis) cotton in India. Proc. Natl. Acad. Sci. USA 2012, 109, 11652–11656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, K.S.; Ravi, K.C.; Suresh, P.J.; Sumerford, D.; Head, G.P. Field resistance to the Bacillus thuringiensis protein Cry1Ac expressed in Bollgard hybrid cotton in pink bollworm, Pectinophora gossypiella (Saunders), populations in India. Pest Manag. Sci. 2016, 72, 738–746. [Google Scholar] [CrossRef]
- Katara, J.L.; Kaur, S.; Kumari, G.K.; Singh, N.K. Prevalence of cry2-type genes in Bacillus thuringiensis isolates recovered from diverse habitats in India and isolation of a novel cry2Af2 gene toxic to Helicoverpa armigera (cotton boll worm). Can. J. Microbiol. 2016, 62, 1003–1012. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.J.; Luo, J.Y.; Van der Werf, W.; Ma, Y.; Xia, J.Y. Effect of pyramiding Bt and CpTI genes on resistance of cotton to Helicoverpa armigera (Lepidoptera:Noctuidae) under laboratory and field conditions. J. Econ. Entomol. 2011, 104, 673–684. [Google Scholar] [CrossRef] [Green Version]
- Din, S.U.; Azam, S.; Rao, A.Q.; Shad, M.; Ahmed, M.; Gul, A.; Latif, A.; Ali, M.A.; Husnain, T.; Shahid, A.A. Development of broad-spectrum and sustainable resistance in cotton against major insects through the combination of Bt and plant lectin genes. Plant Cell Rep. 2021, 40, 707–721. [Google Scholar] [CrossRef]
- Fourie, D.; van den Berg, J.; du Plessis, H. Efficacy of Bacillus thuringiensis sprays and cotton cultivars expressing Cry proteins in the control of Earias biplaga (Walker) (Lepidoptera: Noctuidae). Afr. Entomol. 2017, 25, 335–340. [Google Scholar] [CrossRef]
- Chen, Z.H.; Wei, K.; Chen, L.J.; Wu, Z.J.; Luo, J.Y.; Cui, J.J. Effects of the consecutive cultivation and periodic residue incorporation of Bacillus thuringiensis (Bt) cotton on soil microbe-mediated enzymatic properties. Agric. Ecosyst. Environ. 2017, 239, 154–160. [Google Scholar] [CrossRef]
- Carriere, Y.; Ellers-Kirk, C.; Sisterson, M.; Antilla, L.; Whitlow, M.; Dennehy, T.J.; Tabashnik, B.E. Long-term regional suppression of pink bollworm by Bacillus thuringiensis cotton. Proc. Natl. Acad. Sci. USA 2003, 100, 1519–1523. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.Z.; Cao, J.; Li, Y.; Collins, H.L.; Roush, R.T.; Earle, E.D.; Shelton, A.M. Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat. Biotechnol. 2003, 21, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Cui, J.; Meng, J.; Hu, W.; Luo, J.; Zheng, Y. Effects of transgenic Bt+CpTI cotton on the growth and reproduction of earthworm Eisenia foetida. Front. Biosci. 2009, 14, 4008–4014. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.S.; Shi, X.; Li, J.; Wu, T.Y.; Ren, Q.Q.; Zhang, Z.H.; Wang, M.Y.; Shang, X.X.; Liu, Y.; Xiao, S.H. Effects of root exudates of bivalent transgenic cotton (Bt+CpTI) plants on antioxidant proteins and growth of conventional cotton (Xinluhan 33). J. Environ. Biol. 2016, 37, 13–19. [Google Scholar]
- Wang, P.; Zhuo, X.R.; Tang, L.; Liu, X.S.; Wang, Y.F.; Wang, G.X.; Yu, X.Q.; Wang, J.L. C-type lectin interacting with β-integrin enhances hemocytic encapsulation in the cotton bollworm, Helicoverpa armigera. Insect Biochem. Mol. Biol. 2017, 86, 29–40. [Google Scholar] [CrossRef]
- Vanti, G.L.; Katageri, I.S.; Inamdar, S.R.; Hiremathada, V.; Swamy, B.M. Potent insect gut binding lectin from Sclerotium rolfsii impart resistance to sucking and chewing type insects in cotton. J. Biotechnol. 2018, 278, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.F.; Yu, C.M.; Tang, S.W.; Guo, S.D.; Zhang, D.; Wang, Y.Z.; Zhu, A.G.; Zhu, S.Y.; Xiong, H.P. Transgenic of ramie with synthetic CryIA+CpTI gene by Agrobacterium tumefaciens-mediated. Acta Agron. Sin. 2010, 36, 788–793. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Shi, J.; Yu, Z.; Pan, A.; Tang, X.; Ming, F. Impact of transgenic Cry1Ac + CpTI cotton on diversity and dynamics of rhizosphere bacterial community of different root environments. Sci. Total. Environ. 2018, 637–638, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Cui, H.Z.; Xia, L.; Wu, D.L.; Ni, W.; Zhang, Z.L.; Zhang, B.L.; Xu, Y.J. Development of bivalent insect-resistant transgenic cotton plants. Sci. Agric. Sin. 1999, 32, 1–7. [Google Scholar]
- Yao, Y.S.; Han, P.; Niu, C.Y.; Dong, Y.; Gao, X.W.; Cui, J.J.; Desneux, N. Transgenic Bt cotton does not disrupt the top-down forces regulating the cotton aphid in central china. PLoS ONE 2016, 11, e0166771. [Google Scholar] [CrossRef] [Green Version]
- Cao, C.-W.; Zhang, J.; Gao, X.-W.; Liang, P.; Guo, H.-L. Overexpression of carboxylesterase gene associated with organophosphorous insecticide resistance in cotton aphids, Aphis gossypii (Glover). Pestic. Biochem. Physiol. 2008, 90, 175–180. [Google Scholar] [CrossRef]
- Gong, Y.-H.; Yu, X.-R.; Shang, Q.-L.; Shi, X.-Y.; Gao, X.-W. Oral delivery mediated RNA interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton Aphid, Aphis gossypii Glover. PLoS ONE 2014, 9, e102823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, S.K.; Mishra, M.; Singh, H.; Ranjan, A.; Chandrashekar, K.; Verma, P.C.; Singh, P.K.; Tuli, R. Interaction of Allium sativum leaf agglutinin with midgut brush border membrane vesicles proteins and its stability in Helicoverpa armigera. Proteomics 2010, 10, 4431–4440. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Singh, P.K. Receptors of garlic ( Allium sativum ) lectins and their role in insecticidal action. Protein J. 2012, 31, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Zhu-Salzman, K.; Shade, R.E.; Koiwa, H.; Salzman, R.A.; Narasimhan, M.; Bressan, R.A.; Hasegawa, P.M.; Murdock, L.L. Carbohydrate binding and resistance to proteolysis control insecticidal activity of Griffonia simplicifolia lectin II. Proc. Natl. Acad. Sci. USA 1998, 95, 15123–15128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.Q.; Zhao, C.Y.; Zhou, Y.; Tian, Y.C. Aphid-resistant transgenic tobacco plants expressing modified gna gene. Acta Bot. Sin. 2001, 43, 592–597. [Google Scholar]
- Mi, X.X.; Liu, X.; Yan, H.L.; Liang, L.N.; Zhou, X.Y.; Yang, J.W.; Si, H.J.; Zhang, N. Expression of the Galanthus nivalis agglutinin (GNA) gene in transgenic potato plants confers resistance to aphids. Comptes Rendus Biol. 2017, 340, 7–12. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.; Wang, J.; Guan, F.; Zhang, J.; Yu, S.; Liu, S.; Xue, Y.; Li, L.; Wu, S.; Wang, X.; et al. Dominant point mutation in a tetraspanin gene associated with field-evolved resistance of cotton bollworm to transgenic Bt cotton. Proc. Natl. Acad. Sci. USA 2018, 115, 11760–11765. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, I.J.; Hughes, R.C.; Monsigny, M.; Osawa, T.; Sharon, N. What should be called a lectin? Nature 1980, 285, 66. [Google Scholar] [CrossRef]
- Sharon, N.; Lis, H. Lectins as cell recognition molecules. Science 1989, 246, 227–234. [Google Scholar] [CrossRef]
- Sharon, N. Lectins: Carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 2007, 282, 2753–2764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damme, E.; Allen, A.; Peumans, W. Isolation and characterization of a lectin with exclusive specificity towards mannose from snowdrop (Galanthus nivalis) bulbs. FEBS Lett. 1987, 215, 140–144. [Google Scholar] [CrossRef] [Green Version]
- Lupan, I.; Valimareanu, S.; Coste, A.; Popescu, O. Molecular cloning of agglutinin gene from Galanthus nivalis for Lettuce transformation. Rom. Biotechnol. Lett. 2010, 15, 69–77. [Google Scholar]
- Rao, K.V.; Rathore, K.S.; Hodges, T.K.; Fu, X.; Stoger, E.; Sudhakar, D.; Williams, S.; Christou, P.; Bharathi, M.; Bown, D.P.; et al. Expression of snowdrop lectin (GNA) in transgenic rice plants confers resistance to rice brown planthopper. Plant J. 1998, 15, 469–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.M.; Li, J.; Zhu, J.-Q.; Wang, X.; Wang, C.S.; Liu, S.S.; Chen, X.X.; Li, S. Transgenic plants expressing the AaIT/GNA fusion protein show increased resistance and toxicity to both chewing and sucking pests. Insect Sci. 2015, 23, 265–276. [Google Scholar] [CrossRef]
- Li, Y.; Romeis, J. Impact of snowdrop lectin (Galanthus nivalis agglutinin; GNA) on adults of the green lacewing, Chrysoperla carnea. J. Insect Physiol. 2009, 55, 135–142. [Google Scholar] [CrossRef]
- Peumans, W.J.; Smeets, K.; Van Nerum, K.; Van Leuven, F.; Van Damme, E.J. Lectin and alliinase are the predominant proteins in nectar from leek (Allium porrum L.) flowers. Planta 1997, 201, 298–302. [Google Scholar] [CrossRef]
- Down, R.; Ford, L.; Woodhouse, S.; Raemaekers, R.; Leitch, B.; Gatehouse, J.; Gatehouse, A. Snowdrop lectin (GNA) has no acute toxic effects on a beneficial insect predator, the 2-spot ladybird (Adalia bipunctata L.). J. Insect Physiol. 2000, 46, 379–391. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, P.; Jia, H.; Xue, H.; Zeng, Y.; Tian, L.; Hu, X.; Chang, S.; Jiang, Y.; Yu, J. Expression of Modified Snowdrop Lectin (Galanthus nivalis Agglutinin) Protein Confers Aphids and Plutella xylostella Resistance in Arabidopsis and Cotton. Genes 2022, 13, 1169. https://doi.org/10.3390/genes13071169
He P, Jia H, Xue H, Zeng Y, Tian L, Hu X, Chang S, Jiang Y, Yu J. Expression of Modified Snowdrop Lectin (Galanthus nivalis Agglutinin) Protein Confers Aphids and Plutella xylostella Resistance in Arabidopsis and Cotton. Genes. 2022; 13(7):1169. https://doi.org/10.3390/genes13071169
Chicago/Turabian StyleHe, Peng, Huanhuan Jia, Hui Xue, Yuechen Zeng, Lili Tian, Xiaoli Hu, Shufen Chang, Yanli Jiang, and Jianing Yu. 2022. "Expression of Modified Snowdrop Lectin (Galanthus nivalis Agglutinin) Protein Confers Aphids and Plutella xylostella Resistance in Arabidopsis and Cotton" Genes 13, no. 7: 1169. https://doi.org/10.3390/genes13071169
APA StyleHe, P., Jia, H., Xue, H., Zeng, Y., Tian, L., Hu, X., Chang, S., Jiang, Y., & Yu, J. (2022). Expression of Modified Snowdrop Lectin (Galanthus nivalis Agglutinin) Protein Confers Aphids and Plutella xylostella Resistance in Arabidopsis and Cotton. Genes, 13(7), 1169. https://doi.org/10.3390/genes13071169




