Current Understanding of the Genetics and Molecular Mechanisms Regulating Wood Formation in Plants
Abstract
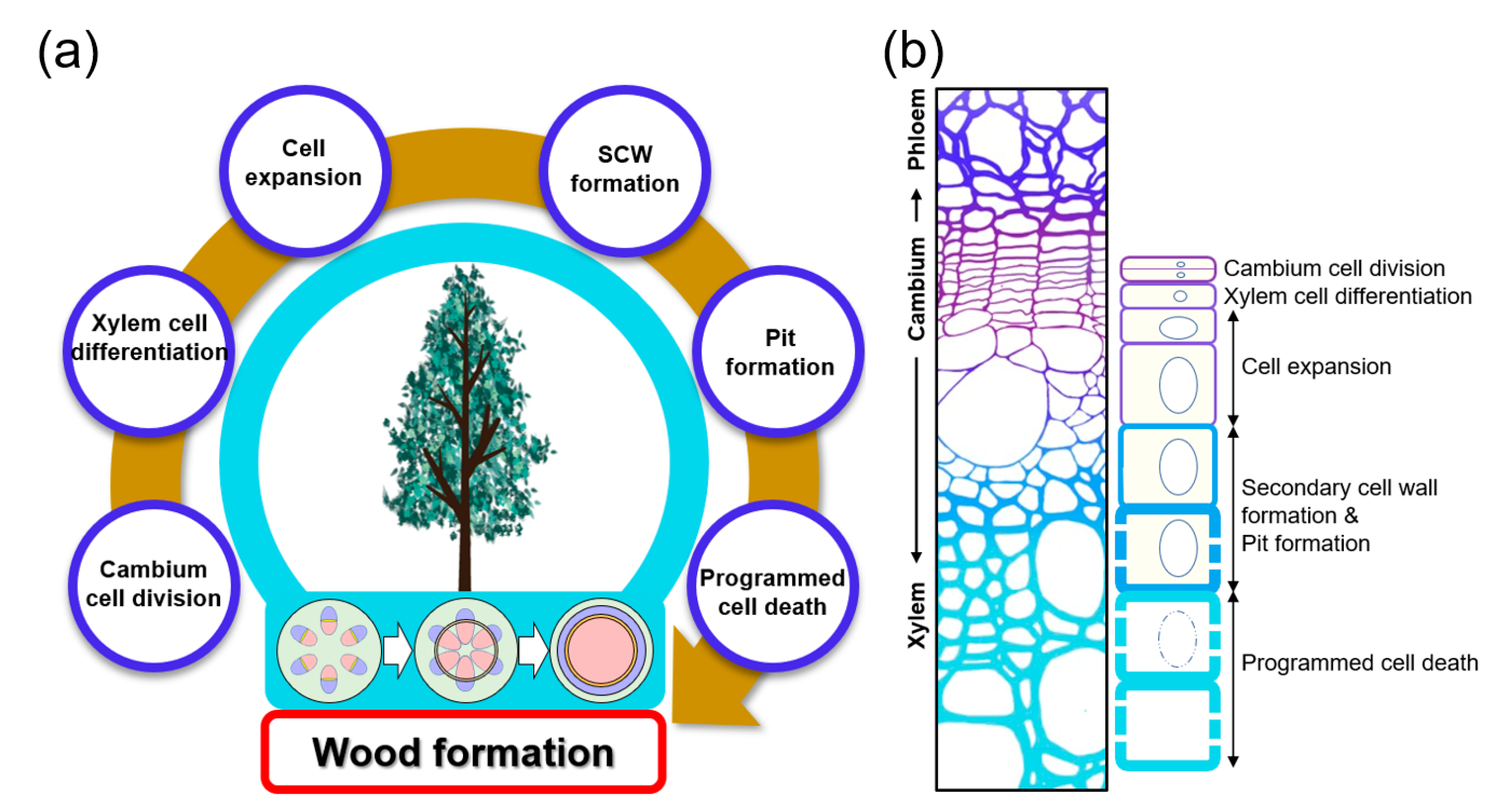
:1. Introduction
2. Vascular Cambium Cell Development and Xylem Cell Differentiation: Initiating Wood Formation
2.1. Regulation by Plant Hormones
2.2. Regulation by Transcription Factors and Signal Peptides
3. Cell Expansion: Determining the Final Shape and Size of the Xylem Cell
4. Pit Formation: Decorating SCW
5. Programmed Cell Death: Finalizing Xylem Differentiation
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Dara Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, M.; Campbell, L.; Turner, S. Secondary cell walls: Biosynthesis and manipulation. J. Exp. Bot. 2016, 67, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Morris, H.; Plavcova, L.; Cvecko, P.; Fichtler, E.; Gillingham, M.; Martínez-Cabrera, H.; McGlinn, D.; Wheeler, E.; Zheng, J.; Ziemińska, K.; et al. A global analysis of parenchyma tissue fractions in secondary xylem of seed plants. New Phytol. 2015, 209, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Rajput, K.S.; Gondaliya, A.D.; Lekhak, M.M.; Yadav, S.R. Structure and Ontogeny of Intraxylary Secondary Xylem and Phloem Development by the Internal Vascular Cambium in Campsis radicans (L.) Seem. (Bignoniaceae). J. Plant Growth Regul. 2018, 37, 755–767. [Google Scholar] [CrossRef]
- Nieminen, K.; Blomster, T.; Helariutta, Y.; Mähönen, A.P. Vascular Cambium Development. Arab. Book 2015, 13, e0177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashima, S.; Sebastian, J.; Lee, J.Y.; Helariutta, Y. Stem cell function during plant vascular development. EMBO J. 2013, 32, 178–193. [Google Scholar] [CrossRef] [Green Version]
- Mizrachi, E.; Myburg, A.A. Systems genetics of wood formation. Curr. Opin. Plant Biol. 2016, 30, 94–100. [Google Scholar] [CrossRef]
- Chiang, M.H.; Greb, T. How to organize bidirectional tissue production? Curr. Opin. Plant Biol. 2019, 51, 15–21. [Google Scholar] [CrossRef]
- Wang, D.; Chen, Y.; Li, W.; Li, Q.; Lu, M.; Zhou, G.; Chai, G. Vascular Cambium: The Source of Wood Formation. Front. Plant Sci. 2021, 12, 700928. [Google Scholar] [CrossRef]
- Lymperopoulos, P.; Msanne, J.; Rabara, R. Phytochrome and Phytohormones: Working in Tandem for Plant Growth and Development. Front. Plant Sci. 2018, 9, 1037. [Google Scholar] [CrossRef] [Green Version]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, J.; Karlberg, A.; Antti, H.; Lopez-Vernaza, M.; Mellerowicz, E.; Perrot-Rechenmann, C.; Sandberg, G.; Bhalerao, R.P. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell 2008, 20, 843–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibañes, M.; Fàbregas, N.; Chory, J.; Caño-Delgado, A.I. Brassinosteroid signaling and auxin transport are required to establish the periodic pattern of Arabidopsis shoot vascular bundles. Proc. Natl. Acad. Sci. USA 2009, 106, 13630–13635. [Google Scholar] [CrossRef] [Green Version]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Alonso Serra, J.; Koskinen, P.; Zhang, J.; Elo, A.; Mähönen, A.P.; Street, N.; et al. Cytokinin and Auxin Display Distinct but Interconnected Distribution and Signaling Profiles to Stimulate Cambial Activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Sánchez Rodríguez, F.; Wu, M.F.; Solé-Gil, A.; Leal Gavarrón, M.; Siligato, R.; Miyashima, S.; et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Fischer, U.; Kucukoglu, M.; Helariutta, Y.; Bhalerao, R.P. The Dynamics of Cambial Stem Cell Activity. Annu. Rev. Plant Biol. 2019, 29, 293–319. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Shen, Y.; He, F.; Fu, X.; Yu, H.; Lu, W.; Li, Y.; Li, C.; Fan, D.; Wang, H.C.; et al. Auxin-mediated Aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 2019, 222, 752–767. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, D.; Xu, P.; Sun, J.; Li, L. A HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 2018, 16, 808–817. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Wang, D.; Liu, Y.; Lu, M.; Zhuang, Y.; Xie, Z.; Wang, C.; Wang, S.; Kong, Y.; Chai, G.; et al. Dual regulation of xylem formation by an auxin-mediated PaC3H17-PaMYB199 module in Populus. New Phytol. 2020, 225, 1545–1561. [Google Scholar] [CrossRef]
- Zheng, S.; He, J.; Lin, Z.; Zhu, Y.; Sun, J.; Li, L. Two MADS-box genes regulate vascular cambium activity and secondary growth by modulating auxin homeostasis in Populus. Plant Commun. 2021, 2, 100134. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tähtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K.; et al. Cytokinin signaling regulates cambial development in poplar. Proc. Natl. Acad. Sci. USA 2008, 105, 20032–20037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi-Ito, K.; Saegusa, M.; Iwamoto, K.; Oda, Y.; Katayama, H.; Kojima, M.; Sakakibara, H.; Fukuda, H. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 2014, 8, 2053–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smet, W.; Sevilem, I.; de Luis Balaguer, M.A.; Wybouw, B.; Mor, E.; Miyashima, S.; Blob, B.; Roszak, P.; Jacobs, T.B.; Boekschoten, M.; et al. DOF2.1 Controls Cytokinin-Dependent Vascular Cell Proliferation Downstream of TMO5/LHW. Curr. Biol. 2019, 4, 520–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [Green Version]
- Kondo, Y.; Ito, T.; Nakagami, H.; Hirakawa, Y.; Saito, M.; Tamaki, T.; Shirasu, K.; Fukuda, H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 2014, 24, 3504. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Gerttula, S.; Li, Z.; Zhao, S.T.; Liu, Y.L.; Liu, Y.; Lu, M.Z.; Groover, A.T. Brassinosteroid regulation of wood formation in poplar. New Phytol. 2020, 225, 1516–1530. [Google Scholar] [CrossRef]
- Lee, J.; Han, S.; Lee, H.Y.; Jeong, B.; Heo, T.Y.; Hyun, T.K.; Kim, K.; Je, B.I.; Lee, H.; Shim, D.; et al. Brassinosteroids facilitate xylem differentiation and wood formation in tomato. Planta 2019, 249, 1391–1403. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.; Park, S.G.; Hwang, H.; Yoo, S.I.; Bae, W.; Kim, E.; Kim, J.; Lee, H.Y.; Heo, T.Y.; et al. Brassinosteroid-BZR1/2-WAT1 module determines the high level of auxin signalling in vascular cambium during wood formation. New Phytol. 2021, 230, 1503–1516. [Google Scholar] [CrossRef]
- Love, J.; Björklund, S.; Vahala, J.; Hertzberg, M.; Kangasjärvi, J.; Sundberg, B. Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. USA 2009, 106, 5984–5989. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Wang, S.; Li, S.; Du, Q.; Qi, L.; Wang, W.; Chen, J.; Wang, H. Activation of ACS7 in Arabidopsis affects vascular development and demonstrates a link between ethylene synthesis and cambial activity. J. Exp. Bot. 2020, 71, 7160–7170. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Kim, H.T.; Choi, Y.I.; Lee, C.; Nguyen, V.P.; Jeon, H.W.; Cho, J.S.; Funada, R.; Pharis, R.P.; Kurepin, L.V.; et al. Overexpression of gibberellin 20-oxidase1 from Pinus densiflora results in enhanced wood formation with gelatinous fiber development in a transgenic hybrid poplar. Tree Physiol. 2015, 35, 1264–1277. [Google Scholar] [PubMed] [Green Version]
- Jeon, H.W.; Cho, J.S.; Park, E.J.; Han, K.H.; Choi, Y.I.; Ko, J.H. Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 2016, 14, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Mauriat, M.; Moritz, T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 2009, 58, 989–1003. [Google Scholar] [CrossRef]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; Conde e Silva, N.; Kreis, M.; Deveaux, Y. WOX 14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Nakanomyo, I.; Motose, H.; Iwamoto, K.; Sawa, S.; Dohmae, N.; Fukuda, H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 2006, 313, 842–845. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 2008, 105, 15208–15213. [Google Scholar] [CrossRef] [Green Version]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef] [Green Version]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef] [Green Version]
- Etchells, J.P.; Mishra, L.S.; Kumar, M.; Campbell, L.; Turner, S.R. Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr Biol. 2015, 25, 1050–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucukoglu, M.; Nilsson, J.; Zheng, B.; Chaabouni, S.; Nilsson, O. WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 2017, 215, 642–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Song, D.; Zhang, R.; Luo, L.; Cao, S.; Huang, C.; Sun, J.; Gui, J.; Li, L. A xylem-produced peptide PtrCLE20 inhibits vascular cambium activity in Populus. Plant Biotechnol J. 2020, 18, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, M.; Kondo, Y.; Fukuda, H. BES1 and BZR1 Redundantly Promote Phloem and Xylem Differentiation. Plant Cell Physiol. 2018, 59, 590–600. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Cho, H.; Noh, J.; Qi, J.; Jung, H.J.; Nam, H.; Lee, S.; Hwang, D.; Greb, T.; Hwang, I. BIL1-mediated MP phosphorylation integrates PXY and cytokinin signalling in secondary growth. Nat. Plants 2018, 4, 605–614. [Google Scholar] [CrossRef]
- Robischon, M.; Du, J.; Miura, E.; Groover, A. The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 2011, 155, 1214–1225. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Miura, E.; Robischon, M.; Martinez, C.; Groover, A. The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE 2011, 6, e17458. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Song, D.; Sun, J.; Wang, X.; Li, L. PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 2013, 6, 1331–1343. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhong, R.; Ye, Z.H. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 2014, 9, e105726. [Google Scholar]
- Mitsuda, N.; Iwase, A.; Yamamoto, H.; Yoshida, M.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. NAC Transcription Factors, NST1 and NST3, Are Key Regulators of the Formation of Secondary Walls in Woody Tissues of Arabidopsis. Plant Cell 2007, 19, 270–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.T.; Endo, H.; Sano, R.; Kurata, T.; Yamaguchi, M.; Ohtani, M.; Demura, T. Transcription Factors VND1-VND3 Contribute to Cotyledon Xylem Vessel Formation. Plant Physiol. 2018, 176, 773–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtani, M.; Nishikubo, N.; Xu, B.; Yamaguchi, M.; Mitsuda, N.; Goué, N.; Shi, F.; Ohme-Takagi, M.; Demura, T. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 2011, 67, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yoo, C.G.; Rottmann, W.; Winkeler, K.A.; Collins, C.M.; Gunter, L.E.; Jawdy, S.S.; Yang, X.; Pu, Y.; Ragauskas, A.J.; et al. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biol. 2019, 11, 486. [Google Scholar] [CrossRef] [Green Version]
- Takata, N.; Awano, T.; Nakata, M.T.; Sano, Y.; Sakamoto, S.; Mitsuda, N.; Taniguchi, T. Populus NST/SND orthologs are key regulators of secondary cell wall formation in wood fibers, phloem fibers and xylem ray parenchyma cells. Tree Physiol. 2019, 39, 514–525. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Kato, K.; Demura, T. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011, 66, 579–590. [Google Scholar] [CrossRef]
- Endo, H.; Yamaguchi, M.; Tamura, T.; Nakano, Y.; Nishikubo, N.; Yoneda, A.; Kato, K.; Kubo, M.; Kajita, S.; Katayama, Y.; et al. Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 2015, 56, 242–254. [Google Scholar] [CrossRef] [Green Version]
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef] [Green Version]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2019, 56, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.C.; Ko, J.H.; Han, K.H. Identification of a cis-acting regulatory motif recognized by MYB46, a master regulator of secondary wall biosynthesis. Plant Mol. Biol. 2012, 78, 489–501. [Google Scholar] [CrossRef]
- Ko, J.H.; Jeon, H.W.; Kim, W.C.; Han, K.H. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.H.; Lee, K.H.; Du, Q.; Yang, S.; Yuan, B.; Qi, L.; Wang, H. A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytol. 2020, 226, 59–74. [Google Scholar] [CrossRef]
- Tang, N.; Shahzad, Z.; Lonjon, F.; Loudet, O.; Vailleau, F.; Maurel, C. Natural variation at XND1 impacts root hydraulics and trade-off for stress responses in Arabidopsis. Nat. Commun. 2018, 9, 3884. [Google Scholar] [CrossRef] [Green Version]
- Grant, E.H.; Fujino, T.; Beers, E.P.; Brunner, A.M. Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 2010, 232, 337–352. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Ohtani, M.; Mitsuda, N.; Kubo, M.; Ohme-Takagi, M.; Fukuda, H.; Demura, T. VND-INTERACTING2, a NAC Domain Transcription Factor, Negatively Regulates Xylem Vessel Formation in Arabidopsis. Plant Cell 2010, 22, 1249–1263. [Google Scholar] [CrossRef] [Green Version]
- Ailizati, A.; Nagahage, I.S.P.; Miyagi, A.; Ishikawa, T.; Kawai-Yamada, M.; Demura, T.; Yamaguchi, M. An Arabidopsis NAC domain transcriptional activator VND7 negatively regulates VNI2 expression. Plant Biotech. 2021, 38, 415–420. [Google Scholar] [CrossRef]
- Yuan, S.; Wu, Y.; Cosgrove, D.J. A Fungal Endoglucanase with Plant Cell Wall Extension Activity. Plant Physiol. 2001, 127, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Gray-Mitsumune, M.; Blomquist, K.; McQueen-Mason, S.; Teeri, T.T.; Sundberg, B.; Mellerowicz, E.J. Ectopic expression of a wood-abundant expansin PttEXPA1 promotes cell expansion in primary and secondary tissues in aspen. Plant Biotechnol. J. 2007, 6, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Yennawar, N.H.; Li, L.C.; Dudzinski, D.M.; Tabuchi, A.; Cosgrove, D.J. Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc. Natl. Acad. Sci. USA 2006, 103, 14664–14671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishitani, K.; Tominaga, R. (1992) Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 1992, 267, 21058–21064. [Google Scholar] [CrossRef]
- Nishikubo, N.; Takahashi, J.; Roos, A.A.; Derba-Maceluch, M.; Piens, K.; Brumer, H.; Teeri, T.T.; Stålbrand, H.; Mellerowicz, E.J. Xyloglucan endo-Transglycosylase-Mediated Xyloglucan Rearrangements in Developing Wood of Hybrid Aspen. Plant Physiol. 2011, 155, 399–413. [Google Scholar] [CrossRef] [Green Version]
- Pelloux, J.; Rustérucci, C.; Mellerowicz, E.J. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12, 267–277. [Google Scholar] [CrossRef]
- Siedlecka, A.; Wiklund, S.; Péronne, M.A.; Micheli, F.; Lesniewska, J.; Sethson, I.; Edlund, U.; Richard, L.; Sundberg, B.; Mellerowicz, E.J. Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol. 2008, 146, 554–565. [Google Scholar] [CrossRef] [Green Version]
- Gou, J.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef] [Green Version]
- Seyfferth, C.; Wessels, B.A.; Vahala, J.; Kangasjärvi, J.; Delhomme, N.; Hvidsten, T.R.; Tuominen, H.; Lundberg-Felten, J. Populus PtERF85 Balances Xylem Cell Expansion and Secondary Cell Wall Formation in Hybrid Aspen. Cells 2021, 10, 1971. [Google Scholar] [CrossRef]
- Nookaraju, A.; Pandey, S.K.; Ahlawat, Y.K.; Joshi, C.P. Understanding the Modus Operandi of Class II KNOX Transcription Factors in Secondary Cell Wall Biosynthesis. Plants 2022, 11, 493. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, L. Multi-layered Regulation of Plant Cell Wall Thickening. Plant Cell Physiol. 2021, 62, 1867–1873. [Google Scholar] [CrossRef]
- Xiao, R.; Zhang, C.; Guo, X.; Li, H.; Lu, H. MYB Transcription Factors and Its Regulation in Secondary Cell Wall Formation and Lignin Biosynthesis during Xylem Development. Int J. Mol. Sci. 2021, 22, 3560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, M.; Tuskan, G.A.; Muchero, W.; Chen, J.-G. Recent Advances in the Transcriptional Regulation of Secondary Cell Wall Biosynthesis in the Woody Plants. Front. Plant Sci. 2018, 9, 1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowell, E.F.; Bischoff, V.; Desprez, T.; Rolland, A.; Stierhof, Y.D.; Schumacher, K.; Gonneau, M.; Höfte, H.; Vernhettes, S. Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 2009, 21, 1141–1154. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, R.; Lindeboom, J.J.; Paredez, A.R.; Emons, A.M.; Ehrhardt, D.W. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009, 11, 797–806. [Google Scholar] [CrossRef]
- Shoji, T.; Narita, N.N.; Hayashi, K.; Asada, J.; Hamada, T.; Sonobe, S.; Nakajima, K.; Hashimoto, T. Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis. Plant Physiol. 2004, 136, 3933–3944. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Xu, T.; Zhu, L.; Wen, M.; Yang, Z. A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol. 2009, 19, 1827–1832. [Google Scholar] [CrossRef] [Green Version]
- Wightman, R.; Chomicki, G.; Kumar, M.; Carr, P.; Turner, S.R. SPIRAL2 determines plant microtubule organization by modulating microtubule severing. Curr. Biol. 2013, 23, 1902–1907. [Google Scholar] [CrossRef] [Green Version]
- Oda, Y.; Mimura, T.; Hasezawa, S. Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 2005, 137, 1027–1036. [Google Scholar] [CrossRef] [Green Version]
- Pesquet, E.; Korolev, A.V.; Calder, G.; Lloyd, C.W. The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr. Biol. 2010, 20, 744–749. [Google Scholar] [CrossRef] [Green Version]
- Oda, Y.; Iida, Y.; Kondo, Y.; Fukuda, H. Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr. Biol. 2010, 20, 1197–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derbyshire, P.; Ménard, D.; Green, P.; Saalbach, G.; Buschmann, H.; Lloyd, C.W.; Pesquet, E. Proteomic Analysis of Microtubule Interacting Proteins over the Course of Xylem Tracheary Element Formation in Arabidopsis. Plant Cell 2015, 27, 2709–2726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, Y.; Fukuda, H. Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 2012, 337, 1333–1336. [Google Scholar] [CrossRef]
- Oda, Y.; Fukuda, H. Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell 2013, 25, 4439–4450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mucha, E.; Hoefle, C.; Huckelhoven, R.; Berken, A. RIP3 and AtKinesin-13A—A novel interaction linking Rho proteins of plants to microtubules. Eur. J. Cell Biol. 2010, 89, 906–916. [Google Scholar] [CrossRef]
- Sugiyama, Y.; Wakazaki, M.; Toyooka, K.; Fukuda, H.; Oda, Y. A Novel Plasma Membrane-Anchored Protein Regulates Xylem Cell-Wall Deposition through Microtubule-Dependent Lateral Inhibition of Rho GTPase Domains. Curr. Biol. 2017, 27, 2522–2528. [Google Scholar] [CrossRef]
- Sasaki, T.; Fukuda, H.; Oda, Y. CORTICAL MICROTUBULE DISORDERING1 Is Required for Secondary Cell Wall Patterning in Xylem Vessels. Plant Cell 2017, 29, 3123–3139. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, Y.; Nagashima, Y.; Wakazaki, M.; Sato, M.; Toyooka, K.; Fukuda, H.; Oda, Y. A Rho-actin signaling pathway shapes cell wall boundaries in Arabidopsis xylem vessels. Nat. Commun. 2019, 10, 468. [Google Scholar] [CrossRef]
- Obara, K.; Kuriyama, H.; Fukuda, H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in Zinnia. Plant Physiol. 2001, 125, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Ito, J.; Fukuda, H. ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 2002, 14, 3201–3211. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Craig, J.C.; Petzold, H.E.; Dickerman, A.W.; Beers, E.P. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 2005, 138, 803–818. [Google Scholar] [CrossRef] [Green Version]
- Avci, U.; Earl Petzold, H.; Ismail, I.O.; Beers, E.P.; Haigler, C.H. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 2008, 56, 303–315. [Google Scholar] [CrossRef]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol Chem. 2008, 283, 774–783. [Google Scholar] [CrossRef] [Green Version]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Turner, S.; Gallois, P.; Brown, D. Tracheary element differentiation. Annu. Rev. Plant Biol. 2007, 58, 407–433. [Google Scholar] [CrossRef] [Green Version]
- Courtois-Moreau, C.L.; Pesquet, E.; Sjödin, A.; Muñiz, L.; Bollhöner, B.; Kaneda, M.; Samuels, L.; Jansson, S.; Tuominen, H. A unique program for cell death in xylem fibers of Populus stem. Plant J. 2009, 58, 260–274. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Goué, N.; Igarashi, H.; Ohtani, M.; Nakano, Y.; Mortimer, J.C.; Nishikubo, N.; Kubo, M.; Katayama, Y.; Kakegawa, K.; et al. VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 2010, 153, 906–914. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Tong, S.; Jiang, Y.; Ai, F.; Feng, Y.; Zhang, J.; Gong, J.; Qin, J.; Zhang, Y.; Zhu, Y.; et al. Transcriptional landscape of highly lignified poplar stems at single-cell resolution. Genome Biol. 2021, 22, 319. [Google Scholar] [CrossRef]
- Li, H.; Dai, X.; Huang, X.; Xu, M.; Wang, Q.; Yan, X.; Sederoff, R.R.; Li, Q. Single-cell RNA sequencing reveals a high-resolution cell atlas of xylem in Populus. J. Integr. Plant Biol. 2021, 63, 1906–1921. [Google Scholar] [CrossRef]

| Gene Name | Function | Studied Plant Species | Reference |
|---|---|---|---|
| Regulation by plant hormones | |||
| PtoIAA9-PtoARF5 module | Regulate vascular cambium cell division and secondary xylem development | Populus tomentosa | [16,17,18] |
| PtoHB7 and PtrHB8 | Secondary xylem cell differentiation, direct target of PtoIAA9-PtoARF5 module | Populus tomentosa | [18] |
| PtrHB4 | Increase cambium development | Populus trichocarpa | [19] |
| PaC3H17-PaMYB199 module | Regulate cambium cell division | Populus trichocarpa | [20] |
| VCM1 and VCM2 | Knockdown mutant enhanced vascular cambium proliferation and xylem differentiation | Populus deltoides × P. euramericana | [21] |
| AtCKX2 | Reduce cytokinin signaling and cambium cell growth | P. tremula × tremuloides | [22] |
| IPT7 | Key enzymes in the biosynthesis of major bioactive cytokinins, increase cambium cell division | Populus tremula × tremuloides | [14] |
| LHW-TMO5 module | Induce vascular cell proliferation | Arabidopsis thaliana | [23,24] |
| LOG3 and LOG4 | Activate vascular cell division | Arabidopsis thaliana | [23] |
| DOF2.1 | Cytokinin-dependent vascular cell proliferation | Arabidopsis thaliana | [24] |
| BRL1 and BRL3 | Reduce xylem formation and increase phloem development | Arabidopsis thaliana | [25] |
| BES1 | Promotes xylem differentiation from procambial cells | Arabidopsis thaliana | [26] |
| SlGSK3 | Reduce xylem differentiation | Solanum lycopersicum | [28] |
| SlBRI1 | Promote xylem differentiation | Solanum lycopersicum | [28] |
| SlWAT1 | Increase secondary xylem development | Solanum lycopersicum | [29] |
| ACS7 | acs7-d mutant enhanced cambium activity and reduced fiber cell wall development | Arabidopsis thaliana | [31] |
| GA20ox1 | GA over production and cambium proliferation | P. alba × P. tremula var. glandulosa | [33,34] |
| Regulation by transcription factors and signal peptides | |||
| CLE41/44(TDIF)-PXY module | Maintaining the cambium cell population via cell division and inhibiting xylem cell differentiation | Arabidopsis thaliana, Populus tremula × P. tremuloides | [39,40,41] |
| WOX4 | Regulation of cambium activity | Arabidopsis thaliana, Populus tremula L. × P. tremuloides | [16,42] |
| PttCLE41b and PttCLE41-like | Reduce cell division and defect vascular tissue pattern | Populus tremula L. × P. tremuloides | [41,42] |
| PtrCLE20 | Reduce cambium cell activity | Populus trichocarpa | [43] |
| BIN2, BIL1 and BIL2 | Inhibit xylem differentiation by suppression of BES1 and BZR1 | Arabidopsis thaliana | [26,44,45] |
| ARR7 and ARR15 | Supress cambium cell division | Arabidopsis thaliana | [45] |
| PopREVOLUTA | Involved in vascular cambium initiation | Populus trichocarpa | [46] |
| PtrHB5 and PtrHB7 | Induce cambium activity and xylem differentiation | Populus tomentosa, Populus alba × P. tremula, Populus × euramericana | [18,47,48] |
| VNDs (VND1–7) | Involved in xylem differentiation | Arabidopsis thaliana, Populus trichocarpa, Dactylis glomerata L | [54,55,56,57,62] |
| MYB46 and MYB83 | Master regulators of SCW biosynthesis | Arabidopsis thaliana | [63,64] |
| XVP | Negative regulator of the TDIF-PXY module | Arabidopsis thaliana | [65] |
| XND1 and PopNAC122 | Mutant showed improved xylem differentiation | Arabidopsis thaliana | [66] |
| VNI2 | Inhibition of xylem vessel development | Arabidopsis thaliana | [69] |
| Cell expansion | |||
| XTH | Involved in cell wall loosening | Populus tremula × tremuloides | [74] |
| PMEs and PAEs | Regulate cell wall loosening through pectin modification | Populus trichocarpa | [77] |
| PtERF85 | Contributes to the transition of fiber cells from elongation to secondary cell wall deposition | Populus tremula L. × P. tremuloides | [78] |
| Pit formation | |||
| RIC1 and SPL2 | Regulating the movement of transverse cortical microtubules | Arabidopsis thaliana | [88] |
| MAP70-5, MAP65, AIR9, CSI1 and MIDD1 | Involved in regulating secondary cell wall patterns | Arabidopsis thaliana | [92] |
| ROP11 | Direct formation of cell wall pits in metaxylem vessel cells through interaction with cortical microtubules | Arabidopsis thaliana | [93] |
| ROPGEF4 | Regulates the formation of ROP-activated domains | Arabidopsis thaliana | [93] |
| ROPGAP3 | Positively regulates pit formation | Arabidopsis thaliana | [93] |
| Kinesin-13A | Microtubule degradation through the active ROP-MIDD1 cascade | Arabidopsis thaliana | [94,95,96] |
| CORD1 and CORD2 | Mutants resulted in an irregular secondary cell wall with small pits in xylem cells | Arabidopsis thaliana | [97] |
| BDR1 | Interacts with F-actin and promotes actin assembly at pit boundaries. | Arabidopsis thaliana | [98] |
| WAL | Interacts with WAL as an ROP effector | Arabidopsis thaliana | [98] |
| Programmed cell death | |||
| BFN1 | Involved in nucleic acid degradation to facilitate nucleotide and phosphate recovery during senescence. | Zinnia elegans | [100] |
| XCP1 and XCP2 | Function in micro-autolysis within the intact central vacuole | Arabidopsis thaliana | [102] |
| AtMC9 | Induce xylem cell death | Arabidopsis thaliana | [104] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.-H.; Bae, E.-K.; Lee, H.; Ko, J.-H. Current Understanding of the Genetics and Molecular Mechanisms Regulating Wood Formation in Plants. Genes 2022, 13, 1181. https://doi.org/10.3390/genes13071181
Kim M-H, Bae E-K, Lee H, Ko J-H. Current Understanding of the Genetics and Molecular Mechanisms Regulating Wood Formation in Plants. Genes. 2022; 13(7):1181. https://doi.org/10.3390/genes13071181
Chicago/Turabian StyleKim, Min-Ha, Eun-Kyung Bae, Hyoshin Lee, and Jae-Heung Ko. 2022. "Current Understanding of the Genetics and Molecular Mechanisms Regulating Wood Formation in Plants" Genes 13, no. 7: 1181. https://doi.org/10.3390/genes13071181
APA StyleKim, M.-H., Bae, E.-K., Lee, H., & Ko, J.-H. (2022). Current Understanding of the Genetics and Molecular Mechanisms Regulating Wood Formation in Plants. Genes, 13(7), 1181. https://doi.org/10.3390/genes13071181







