Cytogenetic Analysis of the Members of the Snake Genera Cylindrophis, Eryx, Python, and Tropidophis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples and Species Verification
2.2. Chromosome Preparation and Staining
2.3. Fluorescence In Situ Hybridization (FISH) with Probes for Repetitive Elements
2.4. Comparative Genome Hybridization (CGH)
2.5. Microscopy Analysis
3. Results
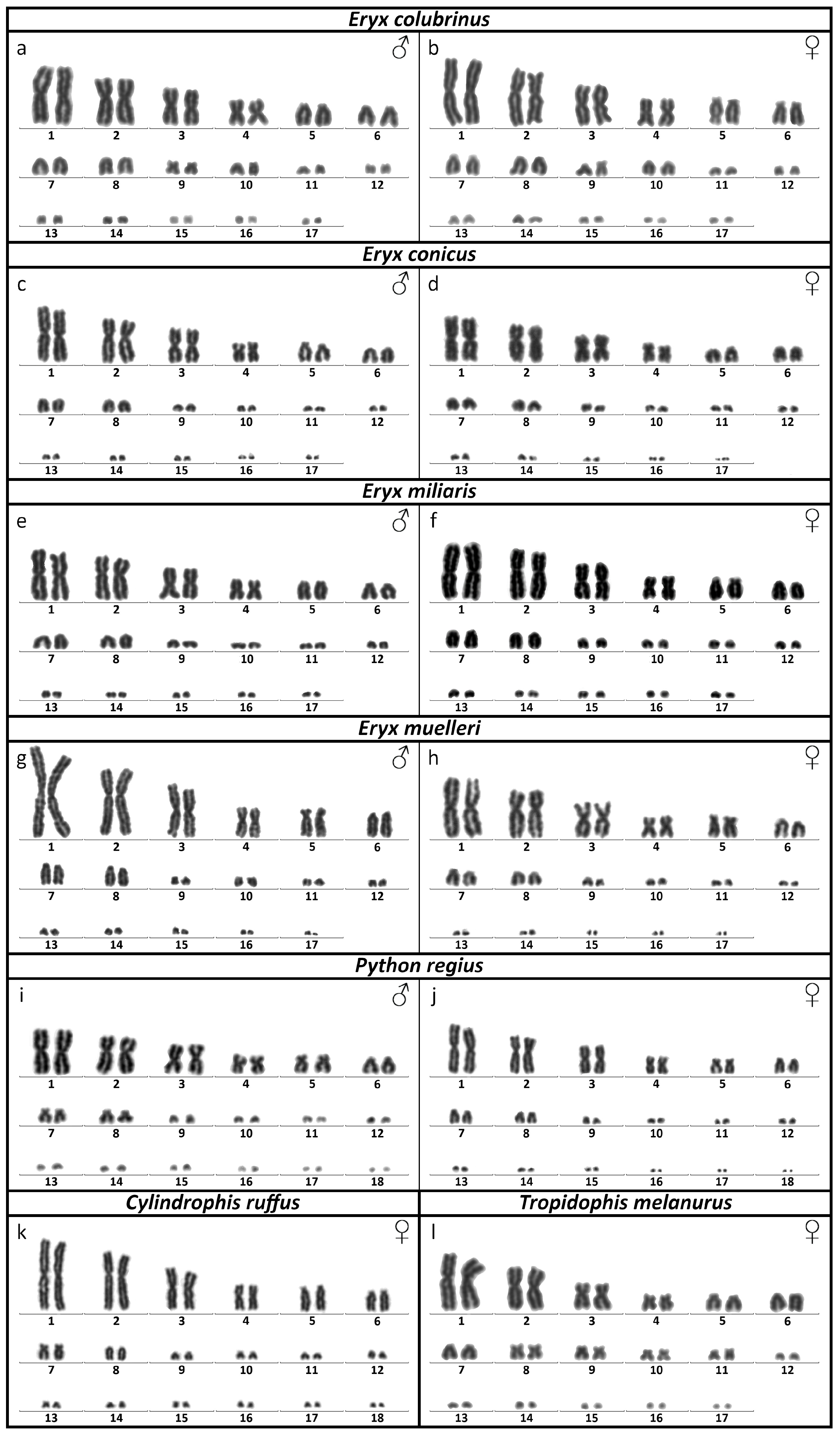
3.1. Karyotype Reconstruction
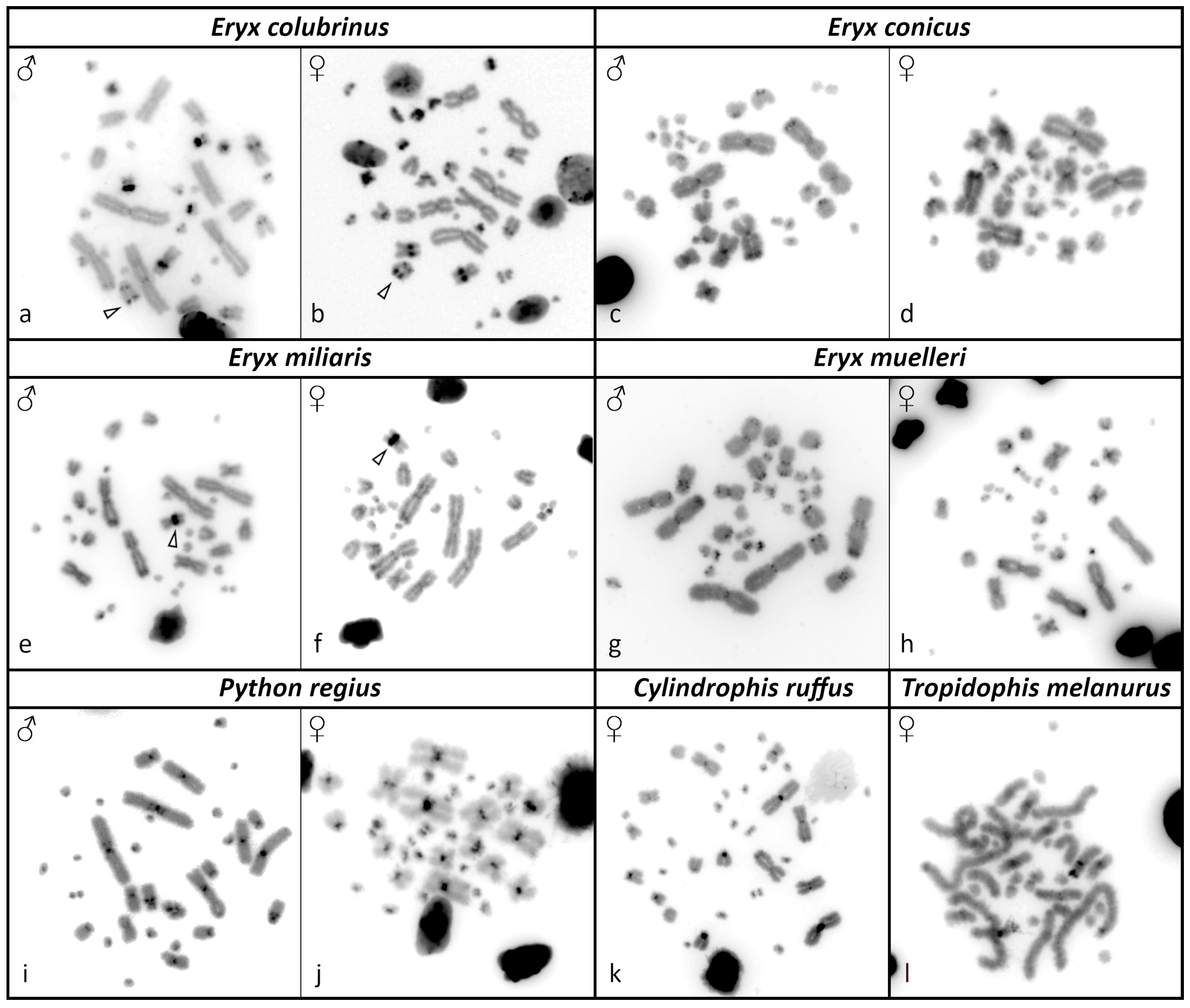
3.2. C-Banding
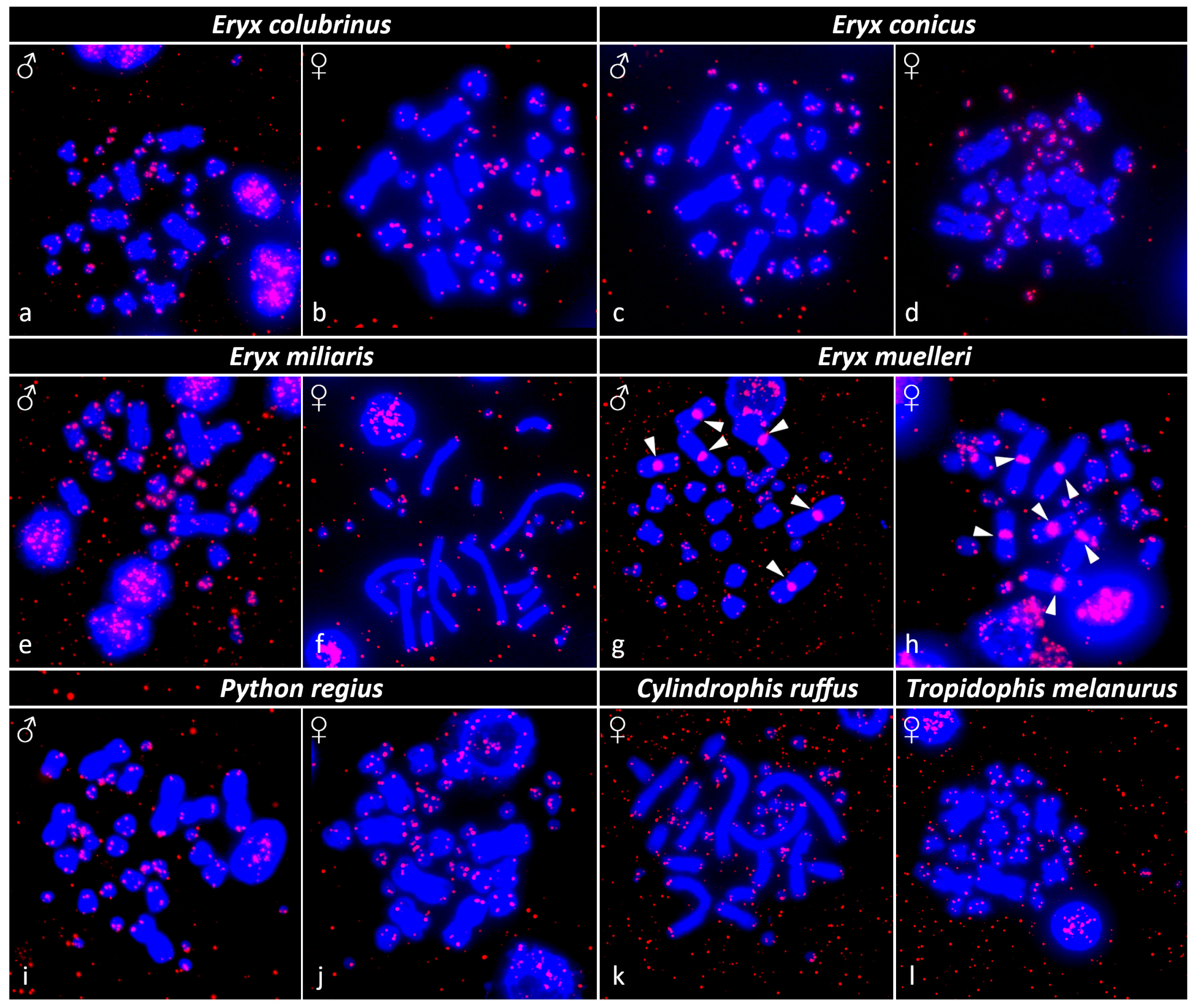
3.3. Fluorescence In Situ Hybridization
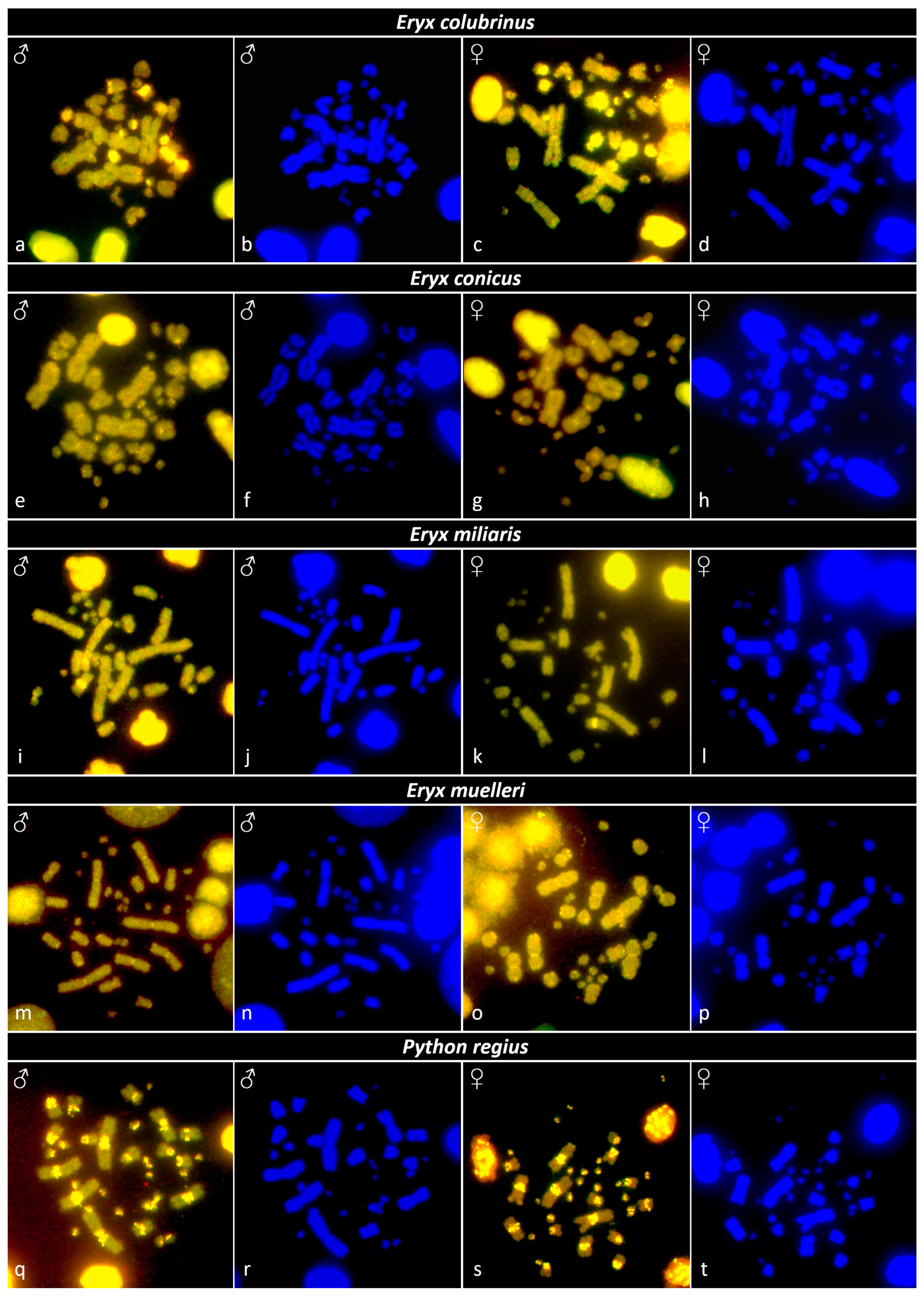
3.4. Comparative Genome Hybridization
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uetz, P.; Freed, P.; Aguilar, R.; Hošek, J. (Eds.) The Reptile Database. 2022. Available online: http://www.reptile-database.org (accessed on 11 April 2022).
- Pyron, R.A.; Burbrink, F.T.; Wiens, J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013, 13, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pyron, R.A.; Wallach, V. Systematics of the blindsnakes (Serpentes: Scolecophidia: Typhlopoidea) based on molecular and morphological evidence. Zootaxa 2014, 3829, 1–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, R.G.; Niemiller, M.L.; Revell, L.J. Toward a Tree-of-Life for the boas and pythons: Multilocus species-level phylogeny with unprecedented taxon sampling. Mol. Phylogenet. Evol. 2014, 71, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wiens, J.J. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 2016, 94, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Beçak, W.; Beçak, M.L. Cytotaxonomy and chromosomal evolution in Serpentes. Cytogenet. Genome Res. 1969, 8, 247–262. [Google Scholar] [CrossRef]
- Olmo, E.; Signorino, G.G. Chromorep: A Reptile Chromosomes Database. 2005. Available online: http://chromorep.univpm.it (accessed on 20 June 2020).
- Oguiura, N.; Ferrarezzi, H.; Batistic, R.F. Cytogenetics and molecular data in snakes: A phylogenetic approach. Cytogenet. Genome Res. 2009, 127, 128–142. [Google Scholar] [CrossRef]
- Valenzuela, N.; Lance, V.A. Temperature-Dependent Sex Determination in Vertebrates; Smithsonian Books: Washington, DC, USA, 2004; pp. 1–194. [Google Scholar]
- Singh, L.; Sharma, T.; Ray-Chaudhuri, S.P. Multiple sex-chromosomes in the common Indian Krait, Bungarus caeruleus Schneider. Chromosoma 1970, 31, 386–391. [Google Scholar] [CrossRef]
- Singh, L. Evolution of karyotypes in snakes. Chromosoma 1972, 38, 185–236. [Google Scholar] [CrossRef]
- Singh, L. Multiple W chromosome in a sea snake, Enhydrina schistosa Daudin. Experientia 1972, 28, 95–97. [Google Scholar] [CrossRef]
- Mengden, G.A.; Stock, A.D. Chromosomal evolution in Serpentes; a comparison of G and C chromosome banding patterns of some colubrid and boid genera. Chromosoma 1980, 79, 53–64. [Google Scholar] [CrossRef]
- Rovatsos, M.; Vukić, J.; Lymberakis, P.; Kratochvíl, L. Evolutionary stability of sex chromosomes in snakes. Proc. R Soc. B Biol. Sci. 2015, 282, 20151992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augstenová, B.; Mazzoleni, S.; Kratochvíl, L.; Rovatsos, M. Evolutionary dynamics of the W chromosome in caenophidian snakes. Genes 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augstenová, B.; Johnson Pokorná, M.; Altmanová, M.; Frynta, D.; Rovatsos, M.; Kratochvíl, L. ZW, XY, and yet ZW: Sex chromosome evolution in snakes even more complicated. Evolution 2018, 72, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Kumazawa, Y.; Ota, H.; Nishida, C.; Matsuda, Y. Karyotype analysis of four blind snake species (Reptilia: Squamata: Scolecophidia) and karyotypic changes in Serpentes. Cytogenet. Genome Res. 2019, 157, 98–106. [Google Scholar] [CrossRef]
- Beçak, W.; Beçak, M.L.; Nazareth, H.R.S.; Ohno, S. Close karyological kinship between the reptilian suborder Serpentes and the class Aves. Chromosoma 1964, 15, 606–617. [Google Scholar] [CrossRef]
- Mengden, G.A. Chromosomal Evolution and the Phylogeny of Elapid Snakes. Ph.D. Thesis, Australian National University, Canberra, Australia, 1982. [Google Scholar]
- Matsubara, K.; Tarui, H.; Toriba, M.; Yamada, K.; Nishida-Umehara, C.; Agata, K.; Matsuda, Y. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 18190–18195. [Google Scholar] [CrossRef] [Green Version]
- Mezzasalma, M.; Andreone, F.; Glaw, F.; Petraccioli, A.; Odierna, G.; Guarino, F.M. A karyological study of three typhlopid species with some inferences on chromosome evolution in blindsnakes (Scolecophidia). Zool. Anz. 2016, 264, 34–40. [Google Scholar] [CrossRef]
- Viana, P.F.; Ribeiro, L.B.; Souza, G.M.; Chalkidis, H.M.; Gross, M.C.; Feldberg, E. Is the karyotype of neotropical boid snakes really conserved? Cytotaxonomy, chromosomal rearrangements and karyotype organization in the Boidae family. PLoS ONE 2016, 11, e0160274. [Google Scholar] [CrossRef]
- Augstenová, B.; Mazzoleni, S.; Kostmann, A.; Altmanová, M.; Frynta, D.; Kratochvíl, L.; Rovatsos, M. Cytogenetic analysis did not reveal differentiated sex chromosomes in ten species of boas and pythons (Reptilia: Serpentes). Genes 2019, 10, 934. [Google Scholar] [CrossRef] [Green Version]
- Vicoso, B.; Emerson, J.J.; Zektser, Y.; Mahajan, S.; Bachtrog, D. Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013, 11, e1001643. [Google Scholar] [CrossRef] [Green Version]
- Ohno, S. Sex. Chromosomes and Sex-Linked Genes; Springer: Berlin, Germany, 1967; pp. 1–167. [Google Scholar]
- Booth, W.; Johnson, D.H.; Moore, S.; Schal, C.; Vargo, E.L. Evidence for viable, non-clonal but fatherless Boa constrictors. Biol. Lett. 2011, 7, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.; Castoe, T.A.; Nielsen, S.V.; Banks, J.L.; Card, D.C.; Schield, D.R.; Schuett, G.W.; Booth, W. The discovery of XY sex chromosomes in a boa and python. Curr. Biol. 2017, 27, 2148–2153.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovatsos, M.; Kratochvíl, L.; Altmanová, M.; Johnson Pokorná, M. Interstitial telomeric motifs in squamate reptiles: When the exceptions outnumber the rule. PLoS ONE 2015, 10, e0134985. [Google Scholar] [CrossRef] [Green Version]
- O’Meally, D.; Patel, H.R.; Stiglec, R.; Sarre, S.D.; Georges, A.; Graves, J.A.M.; Ezaz, T. Non-homologous sex chromosomes of birds and snakes share repetitive sequences. Chromosome Res. 2010, 18, 787–800. [Google Scholar] [CrossRef]
- Literman, R.; Badenhorst, D.; Valenzuela, N. qPCR-based molecular sexing by copy number variation in rRNA genes and its utility for sex identification in soft-shell turtles. Methods Ecol. Evol. 2014, 5, 872–880. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.; Montiel, E.E.; Valenzuela, N. Discovery of putative XX/XY male heterogamety in Emydura subglobosa turtles exposes a novel trajectory of sex chromosome evolution in Emydura. Cytogenet. Genome Res. 2019, 158, 160–169. [Google Scholar] [CrossRef]
- Singchat, W.; Kraichak, E.; Tawichasri, P.; Tawan, T.; Suntronpong, A.; Sillapaprayoon, S.; Phatcharakullawarawat, R.; Muangmai, N.; Suntrarachun, S.; Baicharoen, S.; et al. Dynamics of telomere length in captive Siamese cobra (Naja kaouthia) related to age and sex. Ecol. Evol. 2019, 9, 6366–6377. [Google Scholar] [CrossRef] [Green Version]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 2020, 10, 4276. [Google Scholar] [CrossRef] [Green Version]
- Rovatsos, M.; Johnson Pokorná, M.; Altmanová, M.; Kratochvíl, L. Female heterogamety in Madagascar chameleons (Squamata: Chamaeleonidae: Furcifer): Differentiation of sex and neo-sex chromosomes. Sci. Rep. 2015, 5, 13196. [Google Scholar] [CrossRef] [Green Version]
- Nagy, Z.T.; Sonet, G.; Glaw, F.; Vences, M. First large-scale DNA barcoding assessment of reptiles in the biodiversity hotspot of Madagascar, based on newly designed COI primers. PLoS ONE 2012, 7, e34506. [Google Scholar] [CrossRef]
- Burbrink, F.T.; Lawson, R.; Slowinski, J.B. Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): A critique of the subspecies concept. Evolution 2000, 54, 2107–2118. [Google Scholar] [CrossRef] [PubMed]
- de Queiroz, A.; Lawson, R.; Lemos-Espinal, J.A. Phylogenetic relationships of North American garter snakes (Thamnophis) based on four mitochondrial genes: How much DNA sequence is enough? Mol. Phyl. Evol. 2002, 22, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Koubová, M.; Johnson Pokorná, M.; Rovatsos, M.; Farkačová, K.; Altmanová, M.; Kratochvíl, L. Sex determination in Madagascar geckos of the genus Paroedura (Squamata: Gekkonidae): Are differentiated sex chromosomes indeed so evolutionary stable? Chromosome Res. 2014, 22, 441–452. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Augstenová, B.; Clemente, L.; Auer, M.; Fritz, U.; Praschag, P.; Protiva, T.; Velenský, P.; Kratochvíl, L.; Rovatsos, M. Turtles of the genera Geoemyda and Pangshura (Testudines: Geoemydidae) lack differentiated sex chromosomes: The end of a 40-year error cascade for Pangshura. PeerJ 2019, 7, e6241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Ijdo, J.W.; Baldini, A.; Ward, D.C.; Reeders, S.T.; Wells, R.A. Origin of human chromosome 2: An ancestral telomere-telomere fusion. Proc. Natl. Acad. Sci. USA 1991, 88, 9051–9055. [Google Scholar] [CrossRef] [Green Version]
- Endow, S.A. Polytenization of the ribosomal genes on the X and Y chromosomes of Drosophila melanogaster. Genetics 1982, 100, 375–385. [Google Scholar] [CrossRef]
- Rovatsos, M.; Altmanová, M.; Augstenová, B.; Mazzoleni, S.; Velenský, P.; Kratochvíl, L. ZZ/ZW sex determination with multiple neo-sex chromosomes is common in Madagascan chameleons of the genus Furcifer (Reptilia: Chamaeleonidae). Genes 2019, 10, 1020. [Google Scholar] [CrossRef] [Green Version]
- Gorman, G.C.; Gress, F. Chromosome cytology of four boid snakes and a varanid lizard, with comments on the cytosystematics of primitive snakes. Herpetologica 1970, 26, 308–317. [Google Scholar]
- Sharma, O.P.; Kour, G. On the Chromosomes of four species of Indian snakes. Cytologia 2005, 70, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Singh, L.; Sharma, T.; Ray-Chaudhuri, S. Chromosomes and the classification of the snakes of the family Boidae. Cytogenet. Genome Res. 1968, 7, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Beçak, W.; Beçak, M.L. W-sex chromatin fluorescence in snakes. Experientia 1972, 28, 228–229. [Google Scholar] [CrossRef] [PubMed]
- Heneen, W.K.; Habib, Z.A.; Röhme, D. Heteromorphism of constitutive heterochromatin in carcinoma and dysplasia of the uterine cervix. Eur. J. Obstet. Gynecol. 1980, 10, 173–182. [Google Scholar] [CrossRef]
- Freitas, L.; Seuánez, H. Chromosome heteromorphisms in Cebus apella. J. Hum. Evol. 1982, 11, 173–180. [Google Scholar] [CrossRef]
- Haaf, T.; Schmid, M. Chromosome heteromorphisms in the gorilla karyotype: Analyses with distamycin A/DAPI, quinacrine and 5-azacytidine. Heredity 1987, 78, 287–292. [Google Scholar] [CrossRef]
- Bressa, M.J.; Franco, M.J.; Toscani, M.A.; Papeschi, A.G. Heterochromatin heteromorphism in Holhymenia rubiginosa (Heteroptera: Coreidae). Eur. J. Entomol. 2008, 105, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, G.; Barbosa, L.M.; Prizon-Nakajima, A.C.; de Paiva, S.; Vieira, M.; Gallo, R.B.; Borin-Carvalho, L.A.; da Rosa, R.; Wadzki, C.; Dos Santos, I.; et al. Constitutive heterochromatin heteromorphism in the Neotropical armored catfish Hypostomus regain (Ihering, 1905) (Loricariidae, Hypostominae) from the Paraguay River basin (Mato Grosso do Sul, Brazil). Comp. Cytogenet. 2019, 13, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Sims, S.H.; Macgregor, H.C.; Pellatt, P.S.; Horner, H.A. Chromosome 1 in crested and marbled newts (Triturus). Chromosoma 1984, 89, 169–185. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Aprea, G.; Glaw, F.; Odierna, G.; Guarino, F.M. When can chromosomes drive speciation? The peculiar case of the Malagasy tomato frogs (genus Dyscophus). Zool. Anz. 2017, 268, 41–46. [Google Scholar] [CrossRef]
- Batistic, R.F.; Ferrarezzi, H.; Soma, M. O cariótipo de Tropidophis paucisquamis e suas afinidades com outras famílias. Resumos do III Simpósio do Programa Biota/FAPESP Universidade Federal de São Carlos. 2002. Available online: https://www.biota.org.br/publi/banco/index?show+91144174 (accessed on 15 May 2022).
- Mezzasalma, M.; Andreone, F.; Branch, W.R.; Glaw, F.; Guarino, F.M.; Nagy, Z.T.; Odierna, G.; Aprea, G. Chromosome evolution in pseudoxyrhophiine snakes from Madagascar: A wide range of karyotypic variability. Biol. J. Linn. Soc. 2014, 112, 450–460. [Google Scholar] [CrossRef]
- Porter, C.A.; Hamilton, M.J.; Sites, J.W., Jr.; Baker, R.J. Location of ribosomal DNA in chromosomes of squamate reptiles: Systematic and evolutionary implications. Herpetologica 1991, 47, 271–280. [Google Scholar]
- Porter, C.; Haiduk, M.; De Queiroz, K. Evolution and Phylogenetic significance of ribosomal gene location in chromosomes of squamate reptiles. Copeia 1994, 1994, 302–313. [Google Scholar] [CrossRef]
- Viana, P.F.; Ezaz, T.; de Bello Cioffi, M.; Jackson Almeida, B.; Feldberg, E. Evolutionary insights of the ZW sex chromosomes in snakes: A new chapter added by the amazonian puffing snakes of the genus Spilotes. Genes 2019, 10, 288. [Google Scholar] [CrossRef] [Green Version]
- Hernando, A.; García, J.A. Standard karyotype and nucleolus organizer region of Neotropical blindsnake Typhlops brongersmianus, Serpentes: Typhlopidae. Acta Herpetol. 2007, 2, 117–120. [Google Scholar]
- Bruschi, D.; Rivera, M.; Lima, A.; Zúñiga, A.; Recco-Pimentel, S. Interstitial Telomeric Sequences (ITS) and major rDNA mapping reveal insights into the karyotypical evolution of Neotropical leaf frogs species (Phyllomedusa, Hylidae, Anura). Mol. Cytogenet. 2014, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Augstenová, B.; Pensabene, E.; Kratochvíl, L.; Rovatsos, M. Cytogenetic evidence for sex chromosomes and karyotype evolution in anguimorphan lizards. Cells 2021, 10, 1612. [Google Scholar] [CrossRef]
- Clemente, L.; Mazzoleni, S.; Pensabene Bellavia, E.; Augstenová, B.; Auer, M.; Praschag, P.; Protiva, T.; Velenský, P.; Wagner, P.; Fritz, U.; et al. Interstitial telomeric repeats are rare in turtles. Genes 2020, 11, 657. [Google Scholar] [CrossRef]
- Camper, J.; Hanks, B. Variation in the nucleolus organizer region among New World snakes. J. Herpetol. 1995, 29, 468–471. [Google Scholar] [CrossRef]
- Rovatsos, M.; Pokorná, M.J.; Kratochvíl, L. Differentiation of sex chromosomes and karyotype characterisation in the dragon snake Xenodermus javanicus (Squamata: Xenodermatidae). Cytogenet. Genome Res. 2015, 147, 48–54. [Google Scholar] [CrossRef]
- Backström, N.; Forstmeier, W.; Schielzeth, H.; Mellenius, H.; Nam, K.; Bolund, E.; Webster, M.T.; Öst, T.; Schneider, M.; Kempenaers, B.; et al. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010, 20, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Groenen, M.A.M.; Wahlberg, P.; Foglio, M.; Cheng, H.H.; Megens, H.J.; Crooijmans, R.P.M.A.; Besnier, F.; Lathrop, M.; Muir, W.M.; Wong, G.K.; et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009, 19, 510–519. [Google Scholar] [CrossRef] [Green Version]
- Schield, D.R.; Pasquesi, G.I.M.; Perry, B.W.; Adams, R.H.; Nikolakis, Z.L.; Westfall, A.K.; Orton, R.W.; Meik, J.M.; MacKessy, S.P.; Castoe, T.A.; et al. Snake recombination landscapes are concentrated in functional regions despite PRDM9. Mol. Biol. Evol. 2020, 37, 1272–1294. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Nergadze, S.G.; Giulotto, E. Human intrachromosomal telomeric-like repeats: Sequence organization and mechanisms of origin. Chromosoma 2001, 110, 75–82. [Google Scholar] [CrossRef]
- Mallery, C.S., Jr.; Carrillo, M.M. A case study of sex-linkage in Python regius (Serpentes: Boidae), with new insights into sex determination in Henophidia. Phyllomedusa 2016, 15, 29–42. [Google Scholar] [CrossRef] [Green Version]
- Booth, W.; Schuett, G.W.; Ridgway, A.; Buxton, D.W.; Castoe, T.A.; Bastone, G.; Bennett, C.; McMahan, W. New insights on facultative parthenogenesis in pythons. Biol. J. Linn. Soc. 2014, 112, 461–468. [Google Scholar] [CrossRef]
- Kratochvíl, L.; Stöck, M.; Rovatsos, M.; Bullejos, M.; Herpin, A.; Jeffries, D.L.; Peichel, C.L.; Perrin, N.; Valenzuela, N.; Pokorná, M.J. Expanding the classical paradigm: What we have learnt from vertebrates about sex chromosome evolution. Phil. Trans. R. Soc. B 2021, 376, 20200097. [Google Scholar] [CrossRef]
- Gamble, T.; Coryell, J.; Ezaz, T.; Lynch, J.; Scantlebury, D.P.; Zarkower, D. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 2015, 32, 1296–1309. [Google Scholar] [CrossRef] [Green Version]
- Keating, S.E.; Blumer, M.; Grismer, L.L.; Lin, A.; Nielsen, S.V.; Thura, M.K.; Wood, P.L., Jr.; Quah, E.S.H.; Gamble, T. Sex chromosome turnover in bent-toed geckos (Cyrtodactylus). Genes 2021, 12, 116. [Google Scholar] [CrossRef]
- Kostmann, A.; Augstenová, B.; Frynta, D.; Kratochvíl, L.; Rovatsos, M. Cytogenetically elusive sex chromosomes in scincoidean lizards. Int. J. Mol. Sci. 2021, 22, 8670. [Google Scholar] [CrossRef]

| Family | Species | Sex | |
|---|---|---|---|
| ♂ | ♀ | ||
| Cylindrophiidae | Cylindrophis ruffus | 0 | 1 |
| Erycidae | Eryx colubrinus | 2 | 2 |
| Eryx conicus | 1 | 1 | |
| Eryx miliaris | 3 | 3 | |
| Eryx muelleri | 1 | 1 | |
| Pythonidae | Python regius | 3 | 4 |
| Tropidophiidae | Tropidophis melanurus | 0 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Charvát, T.; Augstenová, B.; Frynta, D.; Kratochvíl, L.; Rovatsos, M. Cytogenetic Analysis of the Members of the Snake Genera Cylindrophis, Eryx, Python, and Tropidophis. Genes 2022, 13, 1185. https://doi.org/10.3390/genes13071185
Charvát T, Augstenová B, Frynta D, Kratochvíl L, Rovatsos M. Cytogenetic Analysis of the Members of the Snake Genera Cylindrophis, Eryx, Python, and Tropidophis. Genes. 2022; 13(7):1185. https://doi.org/10.3390/genes13071185
Chicago/Turabian StyleCharvát, Tomáš, Barbora Augstenová, Daniel Frynta, Lukáš Kratochvíl, and Michail Rovatsos. 2022. "Cytogenetic Analysis of the Members of the Snake Genera Cylindrophis, Eryx, Python, and Tropidophis" Genes 13, no. 7: 1185. https://doi.org/10.3390/genes13071185
APA StyleCharvát, T., Augstenová, B., Frynta, D., Kratochvíl, L., & Rovatsos, M. (2022). Cytogenetic Analysis of the Members of the Snake Genera Cylindrophis, Eryx, Python, and Tropidophis. Genes, 13(7), 1185. https://doi.org/10.3390/genes13071185







