Microarray-Based Prediction of Polycythemia after Exposure to High Altitudes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Preliminary Processing
2.2. Identification of Differential Expression Genes
2.3. Microarray Time-Course Analysis
2.4. Logistic Prediction Model Using Procedure Logistic
2.5. Analysis of Weighted Gene Co-Expression Network Analysis (WGCNA)
2.6. Evaluation of Immune Cell Types
2.7. Functional and Pathway Enrichment Analysis
3. Results
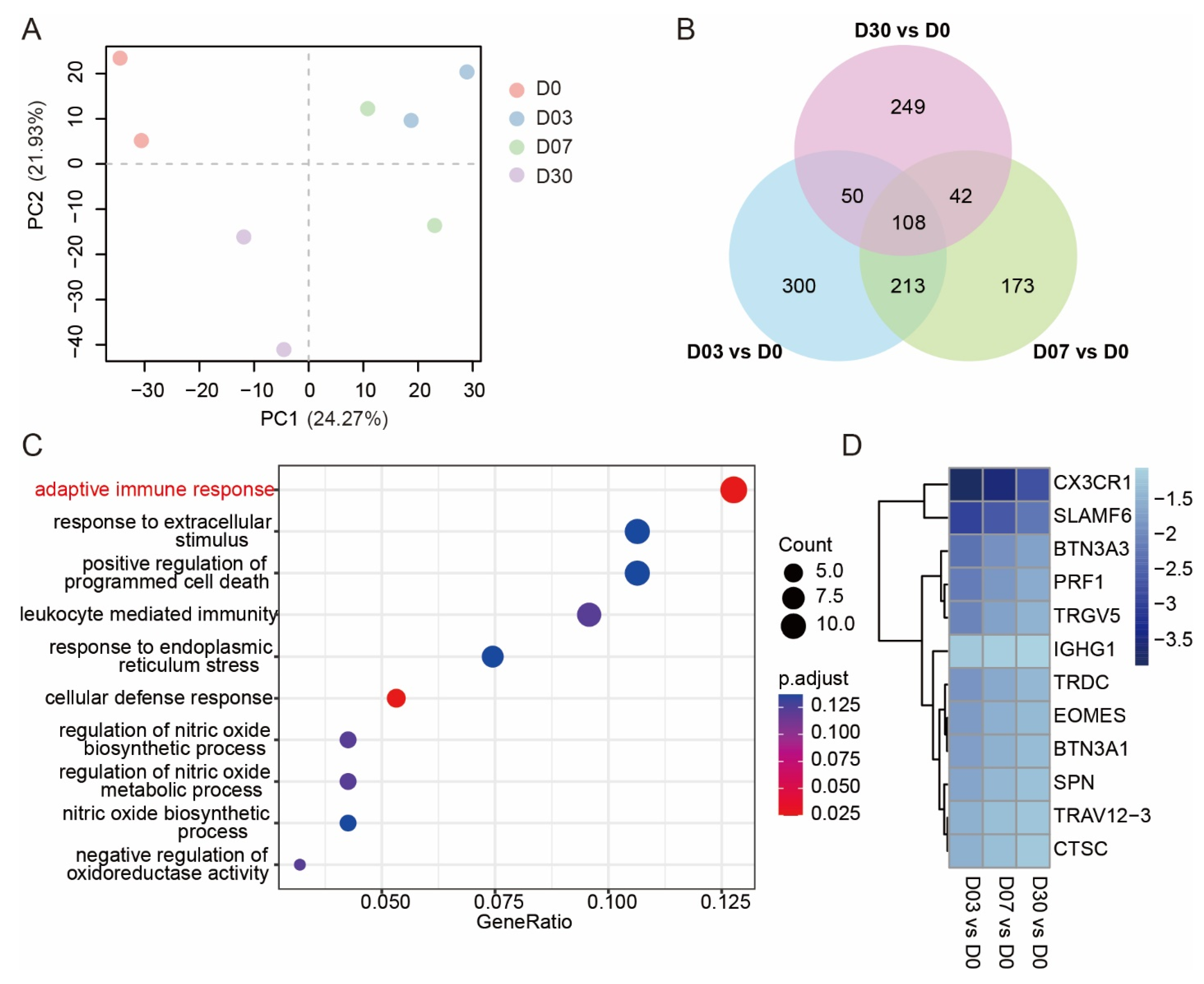
3.1. Exposure to the QTP Weakened the Adaptive Immune Response in Leukocytes
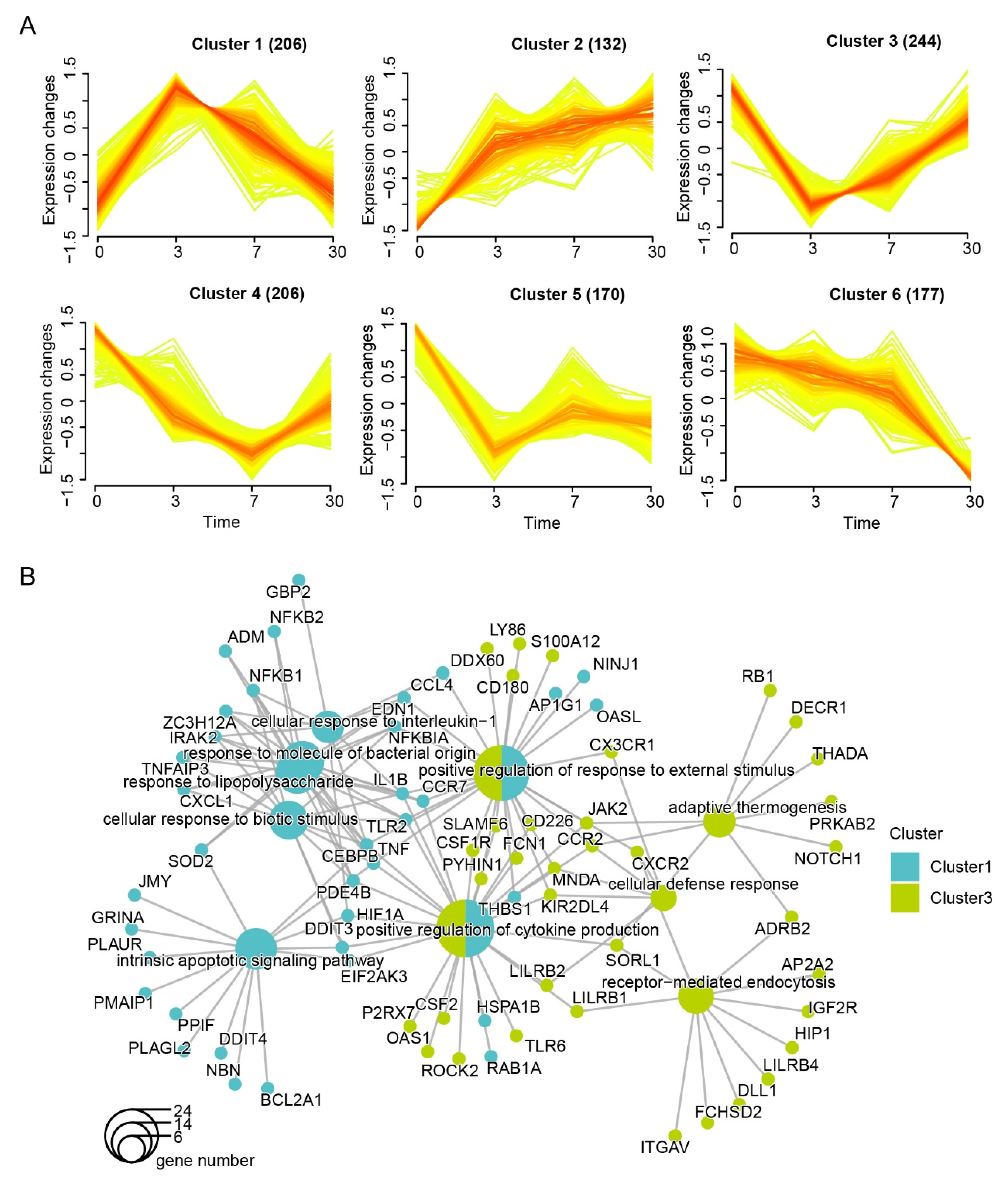
3.2. Dynamic Expression and Functional Characteristics of Leukocytes upon Arrival in the QTP
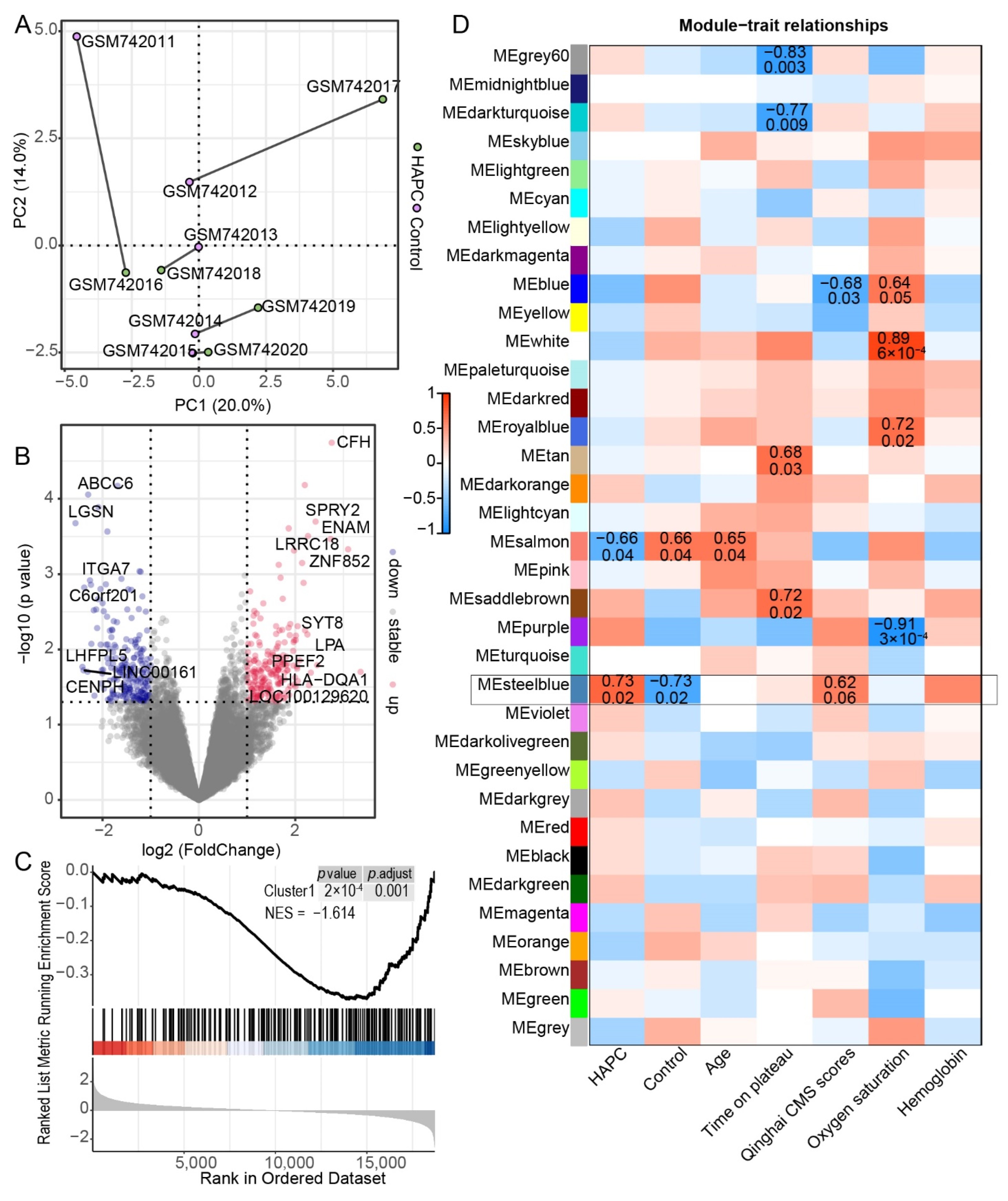
3.3. Gene Expression in Leukocytes of HAPC Patients Exhibited Substantial Individual Variation
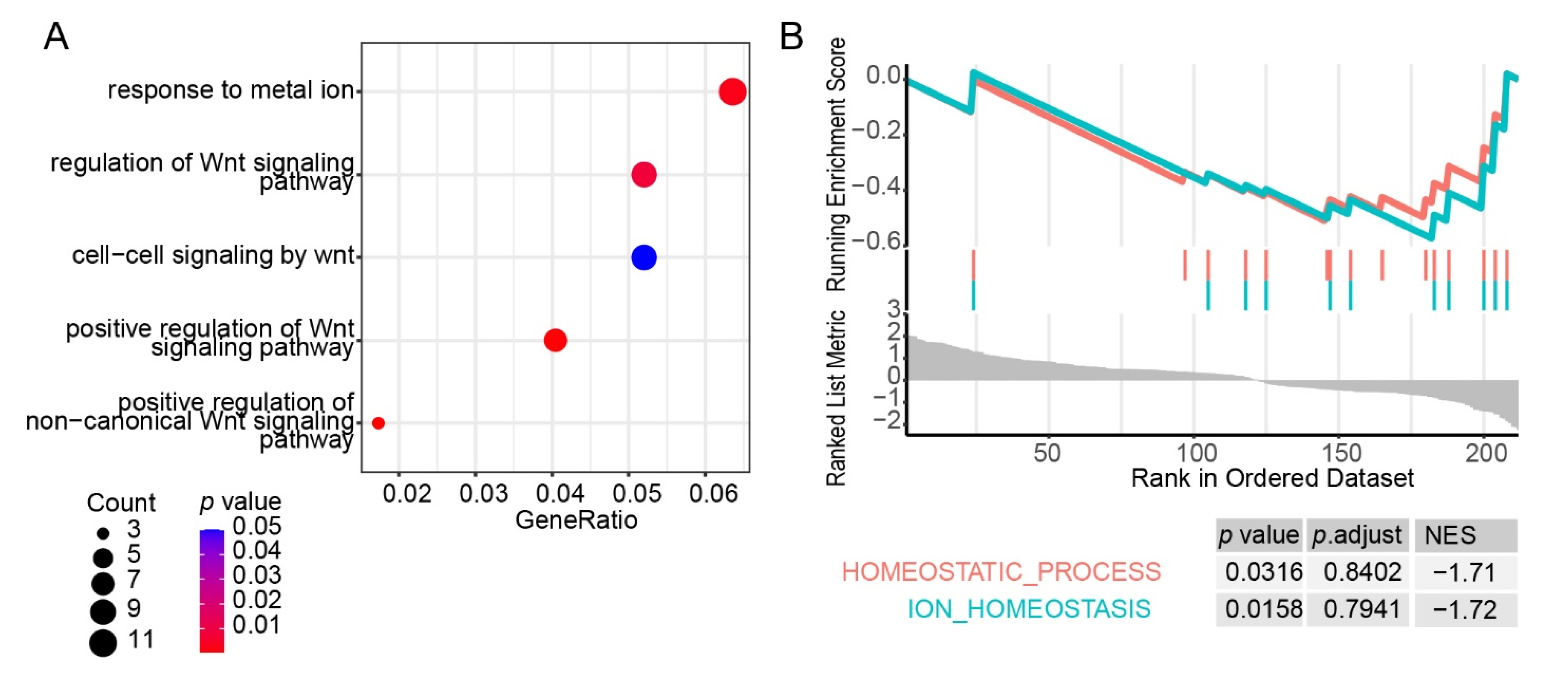
3.4. Construction of a Prediction Model for HAPC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jia, Z.; Zhao, X.; Liu, X.; Zhao, L.; Jia, Q.; Shi, J.; Xu, X.; Hao, L.; Xu, Z.; Zhong, Q.; et al. Impacts of the Plateau Environment on the Gut Microbiota and Blood Clinical Indexes in Han and Tibetan Individuals. mSystems 2020, 5, e00660-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, J.T.; Leon-Velarde, F. Chronic Mountain Sickness: Recent Studies of the Relationship between Hemoglobin Concentration and Oxygen Transport. High Alt. Med. Biol. 2004, 5, 147. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.; Windsor, J. From Mountain to Bedside: Understanding the Clinical Relevance of Human Acclimatisation to High-Altitude Hypoxia. Postgrad. Med. J. 2008, 84, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferson, J.A.; Escudero, E.; Hurtado, M.-E.; Kelly, J.P.; Swenson, E.R.; Wener, M.H.; Burnier, M.; Maillard, M.; Schreiner, G.F.; Schoene, R.B.; et al. Hyperuricemia, Hypertension, and Proteinuria Associated with High-Altitude Polycythemia. Am. J. Kidney Dis. 2002, 39, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Viault, F. On the Large Increase in the Number of Red Cells in the Blood of the Inhabitants of the High Plateaus of South America. In High Altitude Physiology; West, J.B., Ed.; Hutchinson Ross: Stroudsberg, PA, USA, 1981; pp. 333–334. [Google Scholar]
- Jiang, C.; Liu, F.; Luo, Y.; Li, P.; Chen, J.; Xu, G.; Wang, Y.; Li, X.; Huang, J.; Gao, Y. Gene Expression Profiling of High Altitude Polycythemia in Han Chinese Migrating to the Qinghai-Tibetan Plateau. Mol. Med. Rep. 2012, 5, 287–293. [Google Scholar] [CrossRef]
- Chen, K.; Li, N.; Fan, F.; Geng, Z.; Zhao, K.; Wang, J.; Zhang, Y.; Tang, C.; Wang, X.; Meng, X. Tibetan Medicine Duoxuekang Capsule Ameliorates High-Altitude Polycythemia Accompanied by Brain Injury. Front. Pharmacol. 2021, 12, 680636. [Google Scholar] [CrossRef]
- Windsor, J.S.; Rodway, G.W. Heights and Haematology: The Story of Haemoglobin at Altitude. Postgrad. Med. J. 2007, 83, 148–151. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Ma, L.; Zhang, Z.; Li, Y.; Hao, M.; Zhao, Z.; Zhao, Y.; Liu, F.; Liu, L.; Luo, X.; et al. Associations of High-Altitude Polycythemia with Polymorphisms in PIK3CD and COL4A3 in Tibetan Populations. Hum. Genomics 2018, 12, 37. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-S.; Huang, H.; Zhou, S.-M.; Tian, H.; Li, P. Excessive Iron Availability Caused by Disorders of Interleukin-10 and Interleukin-22 Contributes to High Altitude Polycythemia. Front. Physiol. 2018, 9, 548. [Google Scholar] [CrossRef]
- Xu, J.; Yang, Y.-Z.; Tang, F.; Ga, Q.; Tana, W.; Ge, R.-L. EPAS1 Gene Polymorphisms Are Associated With High Altitude Polycythemia in Tibetans at the Qinghai-Tibetan Plateau. Wilderness Environ. Med. 2015, 26, 288–294. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Zhang, Y.; Zhang, Z.; Zhao, Y.; Fan, X.; Ma, L.; Zhang, Y.; He, H.; Kang, L. Associations of High Altitude Polycythemia with Polymorphisms in EPHA2 and AGT in Chinese Han and Tibetan Populations. Oncotarget 2017, 8, 53234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Li, X.; Fan, X.; Fan, F.; Lei, H.; Li, C. Gene Expression Profile Reveals Hematopoietic-Related Molecule Changes in Response to Hypoxic Exposure. DNA Cell Biol. 2020, 39, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; E Futschik, M. Mfuzz: A Software Package for Soft Clustering of Microarray Data. Bioinformation 2007, 2, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.G.; Chin, K.-V. Logistic Regression for Disease Classification Using Microarray Data: Model Selection in a Large p and Small n Case. Bioinformatics 2007, 23, 1945–1951. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochgerner, M.; Sturm, E.M.; Schnoegl, D.; Kwapiszewska, G.; Olschewski, H.; Marsh, L.M. Low Oxygen Levels Decrease Adaptive Immune Responses and Ameliorate Experimental Asthma in Mice. Allergy 2022, 77, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Hoki, T.; Oba, T.; Jain, V.; Chen, H.; Attwood, K.; Battaglia, S.; George, S.; Chatta, G.; Puzanov, I.; et al. T-Cell CX3CR1 Expression as a Dynamic Blood-Based Biomarker of Response to Immune Checkpoint Inhibitors. Nat. Commun. 2021, 12, 1402. [Google Scholar] [CrossRef]
- Chen, Y.; Gaber, T. Hypoxia/HIF Modulates Immune Responses. Biomedicines 2021, 9, 260. [Google Scholar] [CrossRef]
- Coimbra-Costa, D.; Alva, N.; Duran, M.; Carbonell, T.; Rama, R. Oxidative Stress and Apoptosis after Acute Respiratory Hypoxia and Reoxygenation in Rat Brain. Redox Biol. 2017, 12, 216–225. [Google Scholar] [CrossRef]
- Kosanovic, D.; Platzek, S.M.; Petrovic, A.; Sydykov, A.; Maripov, A.; Mamazhakypov, A.; Sartmyrzaeva, M.; Muratali Uulu, K.; Cholponbaeva, M.; Toktosunova, A.; et al. Circulating Apoptotic Signals during Acute and Chronic Exposure to High Altitude in Kyrgyz Population. Front. Physiol. 2019, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Yu, Q.; Zeng, D.; Shen, Z.; Li, J.; Zhu, L.; Zhang, X.; Xu, Q.; Song, H.; Kong, P. Serum Inflammatory Factor Profiles in the Pathogenesis of High-Altitude Polycythemia and Mechanisms of Acclimation to High Altitudes. Mediat. Inflamm. 2021, 2021, 8844438. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Wu, G.; Xu, G.; Sun, B.-D.; Gao, Y.-Q. Hematological Risk Factors for High-Altitude Headache in Chinese Men Following Acute Exposure at 3700 m. Front. Physiol. 2017, 8, 801. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, B.K.; Steensberg, A. Exercise and Hypoxia: Effects on Leukocytes and Interleukin-6-Shared Mechanisms? Med. Sci. Sports Exerc. 2002, 34, 2004–2013. [Google Scholar] [CrossRef]
- Wang, X.; Huang, L.; Gao, H. Effects of Hypoxic Preconditioning Combined with Altitude Training on CD55, CD59 and the Immune Function of Swimmers. Ann. Palliat. Med. 2021, 10, 509–517. [Google Scholar] [CrossRef]
- Scharping, N.E.; Rivadeneira, D.B.; Menk, A.V.; Vignali, P.D.A.; Ford, B.R.; Rittenhouse, N.L.; Peralta, R.; Wang, Y.; Wang, Y.; DePeaux, K.; et al. Mitochondrial Stress Induced by Continuous Stimulation under Hypoxia Rapidly Drives T Cell Exhaustion. Nat. Immunol. 2021, 22, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Shibahara, S.; Han, F.; Li, B.; Takeda, K. Hypoxia and Heme Oxygenases: Oxygen Sensing and Regulation of Expression. Antioxid Redox Signal 2007, 9, 2209–2225. [Google Scholar] [CrossRef]
- Tang, F.; Feng, L.; Li, R.; Wang, W.; Liu, H.; Yang, Q.; Ge, R.-L. Inhibition of Suicidal Erythrocyte Death by Chronic Hypoxia. High Alt. Med. Biol. 2019, 20, 112–119. [Google Scholar] [CrossRef]
- Scheinfeldt, L.B.; Soi, S.; Thompson, S.; Ranciaro, A.; Woldemeskel, D.; Beggs, W.; Lambert, C.; Jarvis, J.P.; Abate, D.; Belay, G.; et al. Genetic Adaptation to High Altitude in the Ethiopian Highlands. Genome Biol. 2012, 13, R1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gersten, M.; Zhou, D.; Azad, P.; Haddad, G.G.; Subramaniam, S. Wnt Pathway Activation Increases Hypoxia Tolerance during Development. PLoS ONE 2014, 9, e103292. [Google Scholar] [CrossRef]
- Shipe, M.E.; Deppen, S.A.; Farjah, F.; Grogan, E.L. Developing Prediction Models for Clinical Use Using Logistic Regression: An Overview. J. Thorac. Dis. 2019, 11, S574–S584. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Meydani, S.N. Age-Associated Changes in Immune and Inflammatory Responses: Impact of Vitamin E Intervention. J. Leukoc. Biol. 2008, 84, 900–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Liu, F.; Ye, S.; Jiang, P.; Yu, X.; Xu, J.; Du, X.; Ma, L.; Cao, H.; Yuan, C.; et al. Plasma Proteome Profiling of High-Altitude Polycythemia Using TMT-Based Quantitative Proteomics Approach. J. Proteom. 2019, 194, 60–69. [Google Scholar] [CrossRef]
- Kapolka, N.J.; Isom, D.G. HCAR3: An Underexplored Metabolite Sensor. Nat. Rev. Drug Discov. 2020, 19, 745. [Google Scholar] [CrossRef]
- McGuire Sams, C.; Shepp, K.; Pugh, J.; Bishop, M.R.; Merner, N.D. Rare and Potentially Pathogenic Variants in Hydroxycarboxylic Acid Receptor Genes Identified in Breast Cancer Cases. BMC Med. Genomics 2021, 14, 284. [Google Scholar] [CrossRef]
- Wnorowski, A.; Wnorowska, S.; Kurzepa, J.; Parada-Turska, J. Alterations in Kynurenine and NAD+ Salvage Pathways during the Successful Treatment of Inflammatory Bowel Disease Suggest HCAR3 and NNMT as Potential Drug Targets. Int. J. Mol. Sci. 2021, 22, 13497. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, S.J.; Munster, S.K.; Uyhelji, H.A.; Hutchings, D.C.; White, V.L.; Booth, J.L.; Burian, D.M.; Metcalf, J.P. Transcriptional Responses to Altitude-Induced Hypoxia in Bronchial Epithelium, Broncho-Alveolar Cells, and Blood Differ between Smokers and Nonsmokers. United States, Department of Transportation, Federal Aviation Administration, Office of Aviation, Civil Aerospace Medical Institute: Washington, DC, USA; Venesco LLC: Chantilly, VA, USA, 2021. Available online: https://www.faa.gov/data_research/research/med_humanfacs/oamtechreports/2020s/media/202125.pdf (accessed on 17 March 2022).
- Yang, Y.; Xu, X. Identification of Key Genes in Coronary Artery Disease: An Integrative Approach Based on Weighted Gene Co-Expression Network Analysis and Their Correlation with Immune Infiltration. Aging 2021, 13, 8306–8319. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ahlford, A.; Järvinen, T.M.; Nordmark, G.; Eloranta, M.-L.; Gunnarsson, I.; Svenungsson, E.; Padyukov, L.; Sturfelt, G.; Jönsen, A.; et al. Genes Identified in Asian SLE GWASs Are Also Associated with SLE in Caucasian Populations. Eur. J. Hum. Genet. 2013, 21, 994–999. [Google Scholar] [CrossRef] [PubMed]

| q | 1 | 10 | 20 | 50 | 100 |
|---|---|---|---|---|---|
| Optimal τ | 4.502 | 2.316 | 0.559 | 0.379 | 0.214 |
| prediction error | 0.059 | 0.058 | 0.056 | 0.051 | 0.051 |
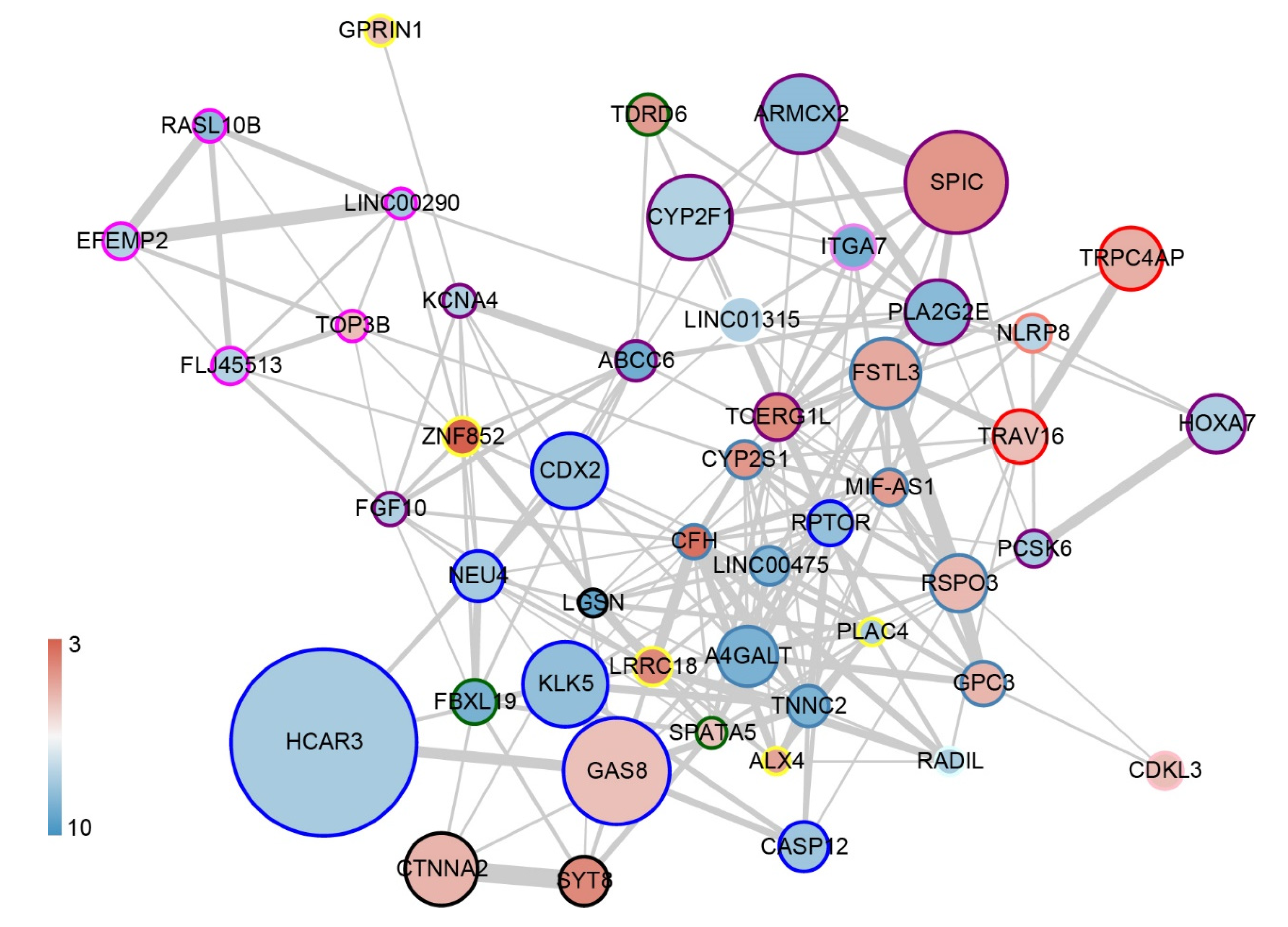
| Gene | Intercept | PLA2G2E | TNNC2 | GAS8 | RPTOR | GPC3 |
|---|---|---|---|---|---|---|
| coefficient | 3.405 | −0.078 | −0.081 | 0.041 | −0.067 | 0.056 |
| Gene | CFH | ZNF852 | NLRP8 | TCERG1L | FLJ45513 | ARMCX2 |
| coefficient | 0.107 | 0.122 | −0.047 | 0.086 | −0.042 | −0.068 |
| Gene | MIF-AS1 | A4GALT | ABCC6 | LGSN | RADIL | NEU4 |
| coefficient | 0.073 | −0.081 | −0.096 | −0.105 | −0.055 | −0.052 |
| Gene | LINC00290 | PCSK6 | FGF10 | SPIC | LRRC18 | EFEMP2 |
| coefficient | −0.057 | −0.064 | −0.048 | 0.067 | 0.086 | −0.048 |
| Gene | LINC01315 | CYP2F1 | CDX2 | RASL10B | KLK5 | RSPO3 |
| coefficient | −0.045 | −0.044 | −0.059 | −0.070 | −0.057 | 0.050 |
| Gene | PLAC4 | TDRD6 | ITGA7 | KCNA4 | SPATA5 | HCAR3 |
| coefficient | −0.054 | 0.073 | −0.082 | −0.053 | 0.042 | −0.048 |
| Gene | RBMS3-AS3 | HOXA7 | CTNNA2 | FBXL19 | LINC00475 | SYT8 |
| coefficient | −0.065 | −0.056 | 0.047 | −0.082 | −0.072 | 0.080 |
| Gene | CYP2S1 | CDKL3 | ALX4 | GPRIN1 | TRAV16 | CASP12 |
| coefficient | 0.084 | 0.053 | 0.069 | 0.051 | 0.044 | −0.057 |
| Gene | TRPC4AP | FSTL3 | TOP3B | |||
| coefficient | 0.050 | 0.056 | 0.050 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Liu, D.; Song, P.; Jiang, F.; Zhang, T. Microarray-Based Prediction of Polycythemia after Exposure to High Altitudes. Genes 2022, 13, 1193. https://doi.org/10.3390/genes13071193
Wang H, Liu D, Song P, Jiang F, Zhang T. Microarray-Based Prediction of Polycythemia after Exposure to High Altitudes. Genes. 2022; 13(7):1193. https://doi.org/10.3390/genes13071193
Chicago/Turabian StyleWang, Haijing, Daoxin Liu, Pengfei Song, Feng Jiang, and Tongzuo Zhang. 2022. "Microarray-Based Prediction of Polycythemia after Exposure to High Altitudes" Genes 13, no. 7: 1193. https://doi.org/10.3390/genes13071193







