Integrated Transcriptome and Metabolomic Analysis Reveal Anti-Angiogenic Properties of Disarib, a Novel Bcl2-Specific Inhibitor
Abstract
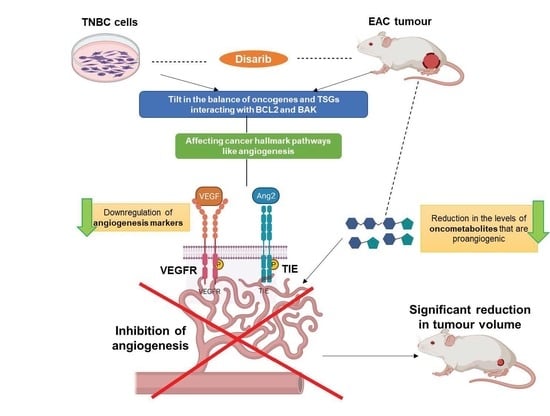
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Animals
2.3. Ehrlich Ascites Breast Adenocarcinoma (EAC) Tumour Model
2.4. RNA Extraction and Library Preparation
2.5. Processing and Alignment of Fastq Files
2.6. Normalisation and Differential Gene Expression Analysis
2.7. Significant Gene List Analysis
2.8. First-Strand cDNA Synthesis
2.9. Real-Time PCR
2.10. Comparative Ct Analysis for Relative Gene Expression Analysis
2.11. Metabolite Analysis through Mass Spectrometry
2.11.1. Sample Preparation
2.11.2. Solvent Preparation
2.11.3. Instrument
2.11.4. Metabolome Analysis
2.11.5. Chorioallantoic Membrane (CAM) Assay for Checking Angiogenesis
2.11.6. Statistical Analysis
3. Results
3.1. Principal Component Analysis Segregated Control, and Disarib Treated EAC Samples
3.2. EAC Samples Displayed Equal Upregulation and Downregulation of Differentially Expressed Genes
3.3. Disarib Modulates the Expression of Oncogenes and Tumour Suppressors in EAC Tumour
3.4. Disarib Induced Shrinkage of EAC Tumours Correlated with Downregulation of Pathways in Cancer Hallmarks
3.5. EAC Tumour Mice Displayed High Levels of Oncometabolite Compared to Normal Animals
3.6. Disarib Treatment Reduced Oncometabolite Levels in EAC Tumours
3.7. Integration of Transcriptome and Metabolome of Disarib Treated EAC Samples Revealed Altered Amino Acid and Purine Metabolism
3.8. Disarib Significantly Reduces Angiogenesis in EAC Tumours
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sawyers, C. Targeted cancer therapy. Nature 2004, 432, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Bouchalova, K.; Svoboda, M.; Kharaishvili, G.; Radova, L.; Bouchal, J.; Trojanec, R.; Koudelakova, V.; Hajduch, M.; Cwiertka, K.; Kolar, Z. BCL2 Protein in Prediction of Relapse in Triple-Negative Breast Cancer (TNBC) Treated with Adjuvant Anthracycline-Based Chemotherapy; American Society of Clinical Oncology: Alexandria, VA, USA, 2012. [Google Scholar]
- Bhargava, V.; Kell, D.L.; van de Rijn, M.; Warnke, R.A. Bcl-2 immunoreactivity in breast carcinoma correlates with hormone receptor positivity. Am. J. Pathol. 1994, 145, 535. [Google Scholar] [PubMed]
- Delbridge, A.R.; Grabow, S.; Strasser, A.; Vaux, D.L. Thirty years of BCL-2: Translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer 2016, 16, 99. [Google Scholar] [CrossRef] [PubMed]
- Radha, G.; Raghavan, S.C. BCL2: A promising cancer therapeutic target. Biochim. Biophys. Acta (BBA) Rev. Cancer 2017, 1868, 309–314. [Google Scholar] [CrossRef]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Tahir, S.K.; Smith, M.L.; Hessler, P.; Rapp, L.R.; Idler, K.B.; Park, C.H.; Leverson, J.D.; Lam, L.T. Potential mechanisms of resistance to venetoclax and strategies to circumvent it. BMC Cancer 2017, 17, 399. [Google Scholar] [CrossRef]
- Choudhary, G.S.; Al-Harbi, S.; Mazumder, S.; Hill, B.T.; Smith, M.R.; Bodo, J.; Hsi, E.D.; Almasan, A. MCL-1 and BCL-xL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell Death Dis. 2015, 6, e1593. [Google Scholar] [CrossRef]
- Bose, P.; Gandhi, V.; Konopleva, M. Pathways and mechanisms of venetoclax resistance. Leuk. Lymphoma 2017, 58, 2026–2039. [Google Scholar] [CrossRef]
- Vartak, S.V.; Iyer, D.; Santhoshkumar, T.R.; Sharma, S.; Mishra, A.; Goldsmith, G.; Srivastava, M.; Srivastava, S.; Karki, S.S.; Surolia, A. Novel BCL2 inhibitor, Disarib induces apoptosis by disruption of BCL2-BAK interaction. Biochem. Pharmacol. 2017, 131, 16–28. [Google Scholar] [CrossRef]
- Sharma, S.; Varsha, K.K.; Kumari, S.; Gopalakrishnan, V.; Jose, A.E.; Choudhary, B.; Mantelingu, K.; Raghavan, S.C. Acute toxicity analysis of Disarib, an inhibitor of BCL2. Sci. Rep. 2020, 10, 15188. [Google Scholar] [CrossRef]
- Haas, B.J.; Zody, M.C. Advancing RNA-seq analysis. Nat. Biotechnol. 2010, 28, 421–423. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017, 171, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D. A review of applications of metabolomics in cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeersch, K.A.; Styczynski, M.P. Applications of metabolomics in cancer research. J. Carcinog. 2013, 12, 9. [Google Scholar] [PubMed]
- Hassan, M.A.; Al-Sakkaf, K.; Shait Mohammed, M.R.; Dallol, A.; Al-Maghrabi, J.; Aldahlawi, A.; Ashoor, S.; Maamra, M.; Ragoussis, J.; Wu, W. Integration of transcriptome and metabolome provides unique insights to pathways associated with obese breast cancer patients. Front. Oncol. 2020, 10, 804. [Google Scholar] [CrossRef]
- Iervolino, A.; Trisciuoglio, D.; Ribatti, D.; Candiloro, A.; Biroccio, A.; Zupi, G.; Del Bufalo, D. Bcl-2 overexpression in human melanoma cells increases angiogenesis through VEGF mRNA stabilization and HIF-1mediated transcriptional activity. FASEB J. 2002, 16, 1453–1455. [Google Scholar] [CrossRef]
- Karl, E.; Zhang, Z.; Dong, Z.; Neiva, K.G.; Soengas, M.S.; Koch, A.E.; Polverini, P.J.; Nunez, G.; Nör, J.E. Unidirectional crosstalk between Bcl-xL and Bcl-2 enhances the angiogenic phenotype of endothelial cells. Cell Death Differ. 2007, 14, 1657–1666. [Google Scholar] [CrossRef] [Green Version]
- Tonini, T.; Rossi, F.; Claudio, P.P. Molecular basis of angiogenesis and cancer. Oncogene 2003, 22, 6549–6556. [Google Scholar] [CrossRef] [Green Version]
- Carmeliet, P.; Jain, R.K. Angiogenesis in cancer and other diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef]
- Fantozzi, A.; Gruber, D.C.; Pisarsky, L.; Heck, C.; Kunita, A.; Yilmaz, M.; Meyer-Schaller, N.; Cornille, K.; Hopfer, U.; Bentires-Alj, M. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014, 74, 1566–1575. [Google Scholar] [CrossRef] [Green Version]
- Hicklin, D.J.; Ellis, L.M. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J. Clin. Oncol. 2005, 23, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- Deshayes, F.; Nahmias, C. Angiotensin receptors: A new role in cancer? Trends Endocrinol. Metab. 2005, 16, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, B.D.; Joo, E.; Dong, Z.; Warner, K.; Wang, G.; Nikolovska-Coleska, Z.; Wang, S.; Nör, J.E. Antiangiogenic effect of TW37, a small-molecule inhibitor of Bcl-2. Cancer Res. 2006, 66, 8698–8706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, D.; Vartak, S.V.; Mishra, A.; Goldsmith, G.; Kumar, S.; Srivastava, M.; Hegde, M.; Gopalakrishnan, V.; Glenn, M.; Velusamy, M. Identification of a novel BCL2-specific inhibitor that binds predominantly to the BH1 domain. FEBS J. 2016, 283, 3408–3437. [Google Scholar] [CrossRef] [PubMed]
- Geran, R.I.; Greenberg, N.H.; Macdonald, M.M.; Schumacher, A.M.; Abbott, B.J. Protocols for screening chemical agents and natural products against animal tumors and other biological systems. Cancer Chemother. Rep. 1972, 3, 17–19. [Google Scholar]
- Noaman, E.; El-Din, N.K.B.; Bibars, M.A.; Abou Mossallam, A.A.; Ghoneum, M. Antioxidant potential by arabinoxylan rice bran, MGN-3/biobran, represents a mechanism for its oncostatic effect against murine solid Ehrlich carcinoma. Cancer Lett. 2008, 268, 348–359. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Morvan, M.L.; Vert, J.-P. Supervised quantile normalisation. arXiv 2017, arXiv:1706.00244. [Google Scholar]
- Peng, R.D. R Programming for Data Science; Leanpub: Victoria, BC, Canada, 2016. [Google Scholar]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; O’kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2010, 39, D691–D697. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Manual, I. ProtoScript® First Strand cDNA Synthesis Kit. 2017. Available online: https://www.neb.com/-/media/nebus/files/manuals/manuale6550.pdf?rev=9c51e082b8614a27b3854e503273db87&hash=8BF8FA6D0CE9F0821F865C5250D22CB5 (accessed on 15 September 2021).
- Mohr, S.; Ghanem, E.; Smith, W.; Sheeter, D.; Qin, Y.; King, O.; Polioudakis, D.; Iyer, V.R.; Hunicke-Smith, S.; Swamy, S. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. Rna 2013, 19, 958–970. [Google Scholar] [CrossRef] [Green Version]
- Tzanetakis, I.E.; Keller, K.E.; Martin, R.R. The use of reverse transcriptase for efficient first-and second-strand cDNA synthesis from single-and double-stranded RNA templates. J. Virol. Methods 2005, 124, 73–77. [Google Scholar] [CrossRef]
- Ponchel, F.; Toomes, C.; Bransfield, K.; Leong, F.T.; Douglas, S.H.; Field, S.L.; Bell, S.M.; Combaret, V.; Puisieux, A.; Mighell, A.J. Real-time PCR based on SYBR-Green I fluorescence: An alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol. 2003, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Deepak, S.A.; Kottapalli, K.R.; Rakwal, R.; Oros, G.; Rangappa, K.S.; Iwahashi, H.; Masuo, Y.; Agrawal, G.K. Real-time PCR: Revolutionizing detection and expression analysis of genes. Curr. Genom. 2007, 8, 234–251. [Google Scholar] [CrossRef]
- Schmittgen, T.D. Real-time quantitative PCR. Methods 2001, 4, 383–385. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Lindahl, A.; Sääf, S.; Lehtiö, J.; Nordström, A. Tuning metabolome coverage in reversed phase LC–MS metabolomics of MeOH extracted samples using the reconstitution solvent composition. Anal. Chem. 2017, 89, 7356–7364. [Google Scholar] [CrossRef] [Green Version]
- Want, E.J.; O’Maille, G.; Smith, C.A.; Brandon, T.R.; Uritboonthai, W.; Qin, C.; Trauger, S.A.; Siuzdak, G. Solvent-dependent metabolite distribution, clustering, and protein extraction for serum profiling with mass spectrometry. Anal. Chem. 2006, 78, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Pulukool, S.K.; Bhagavatham, S.K.S.; Kannan, V.; Sukumar, P.; Dandamudi, R.B.; Ghaisas, S.; Kunchala, H.; Saieesh, D.; Naik, A.A.; Pargaonkar, A. Elevated dimethylarginine, ATP, cytokines, metabolic remodeling involving tryptophan metabolism and potential microglial inflammation characterize primary open angle glaucoma. Sci. Rep. 2021, 11, 9766. [Google Scholar] [CrossRef] [PubMed]
- Bhagavatham, S.K.S.; Khanchandani, P.; Kannan, V.; Potikuri, D.; Sridharan, D.; Pulukool, S.K.; Naik, A.A.; Dandamudi, R.B.; Divi, S.M.; Pargaonkar, A. Adenosine deaminase modulates metabolic remodeling and orchestrates joint destruction in rheumatoid arthritis. Sci. Rep. 2021, 11, 15129. [Google Scholar] [CrossRef]
- Borg, D.; Tverdovsky, A.; Stripp, R. A fast and comprehensive analysis of 32 synthetic cannabinoids using agilent triple quadrupole LC–MS-MS. J. Anal. Toxicol. 2017, 41, 6–16. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef]
- Kalivodová, A.; Hron, K.; Filzmoser, P.; Najdekr, L.; Janečková, H.; Adam, T. PLS-DA for compositional data with application to metabolomics. J. Chemom. 2015, 29, 21–28. [Google Scholar] [CrossRef]
- Ribatti, D. The chick embryo chorioallantoic membrane (CAM) assay. Reprod. Toxicol. 2017, 70, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Tufan, A.C.; Satiroglu-Tufan, N.L. The chick embryo chorioallantoic membrane as a model system for the study of tumor angiogenesis, invasion and development of anti-angiogenic agents. Curr. Cancer Drug Targets 2005, 5, 249–266. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, R.A.; Gannon, N.P.; Garcia-Smith, R.; Licon-Munoz, Y.; Barberena, M.A.; Bisoffi, M.; Trujillo, K.A. β-alanine suppresses malignant breast epithelial cell aggressiveness through alterations in metabolism and cellular acidity in vitro. Mol. Cancer 2014, 13, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stork, P.J.; Schmitt, J.M. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002, 12, 258–266. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: Mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef] [Green Version]
- Vandenberg, C.J.; Cory, S. ABT-199, a new Bcl-2–specific BH3 mimetic, has in vivo efficacy against aggressive Myc-driven mouse lymphomas without provoking thrombocytopenia. Blood J. Am. Soc. Hematol. 2013, 121, 2285–2288. [Google Scholar]
- Cang, S.; Iragavarapu, C.; Savooji, J.; Song, Y.; Liu, D. ABT-199 (venetoclax) and BCL-2 inhibitors in clinical development. J. Hematol. Oncol. 2015, 8, 129. [Google Scholar] [CrossRef]
- Sledge, G.W., Jr.; Miller, K.D. Exploiting the hallmarks of cancer: The future conquest of breast cancer. Eur. J. Cancer 2003, 39, 1668–1675. [Google Scholar] [CrossRef]
- Majidinia, M.; Yousefi, B. DNA repair and damage pathways in breast cancer development and therapy. DNA Repair 2017, 54, 22–29. [Google Scholar] [CrossRef]
- Caldon, C.E.; Daly, R.J.; Sutherland, R.L.; Musgrove, E.A. Cell cycle control in breast cancer cells. J. Cell. Biochem. 2006, 97, 261–274. [Google Scholar] [CrossRef]
- Wang, L.; Li, J.; Zheng, Z.; Liu, H.; Du, G.; Li, S. Antitumor activities of a novel indolin-2-ketone compound, Z24: More potent inhibition on bFGF-induced angiogenesis and bcl-2 over-expressing cancer cells. Eur. J. Pharmacol. 2004, 502, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, B.D.; Nör, J.E. Small-molecule inhibitors reveal a new function for Bcl-2 as a proangiogenic signaling molecule. Small Mol. Inhib. Protein Protein Interact. 2010, 348, 115–137. [Google Scholar]
- Harmey, J.H.; Bouchier-Hayes, D. Vascular endothelial growth factor (VEGF), a survival factor for tumour cells: Implications for anti-angiogenic therapy. Bioessays 2002, 24, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Khosravi Shahi, P.; Soria Lovelle, A.; Perez Manga, G. Tumoral angiogenesis and breast cancer. Clin. Transl. Oncol. 2009, 11, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Longatto Filho, A.; Lopes, J.M.; Schmitt, F.C. Angiogenesis and breast cancer. J. Oncol. 2010, 2010, 576384. [Google Scholar] [CrossRef]
- Zhu, X.; Zhou, W. The emerging regulation of VEGFR-2 in triple-negative breast cancer. Front. Endocrinol. 2015, 6, 159. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.-D.; Liu, Y.; Zhang, Z.-Y.; Liu, G.-Y.; Xu, J.-H.; Liu, L.-Y.; Hu, Y.-M. Expression and prognostic significance of VEGFR-2 in breast cancer. Pathol. Res. Pract. 2015, 211, 539–543. [Google Scholar] [CrossRef]
- Tiainen, L.; Korhonen, E.A.; Leppänen, V.-M.; Luukkaala, T.; Hämäläinen, M.; Tanner, M.; Lahdenperä, O.; Vihinen, P.; Jukkola, A.; Karihtala, P. High baseline Tie1 level predicts poor survival in metastatic breast cancer. BMC Cancer 2019, 19, 732. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.-Y.; Li, C.-F.; Kuo, C.-C.; Tsai, K.K.; Hou, M.-F.; Hung, W.-C. Cancer/stroma interplay via cyclooxygenase-2 and indoleamine 2,3-dioxygenase promotes breast cancer progression. Breast Cancer Res. 2014, 16, 410. [Google Scholar] [CrossRef] [Green Version]
- Morettin, A.; Baldwin, R.M.; Côté, J. Arginine methyltransferases as novel therapeutic targets for breast cancer. Mutagenesis 2015, 30, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carracedo, A.; Cantley, L.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Khatami, F.; Aghamir, S.M.K.; Tavangar, S.M. Oncometabolites: A new insight for oncology. Mol. Genet. Genom. Med. 2019, 7, e873. [Google Scholar] [CrossRef] [PubMed]
- Samson, F.P.; Patrick, A.T.; Fabunmi, T.E.; Yahaya, M.F.; Madu, J.; He, W.; Sripathi, S.R.; Tyndall, J.; Raji, H.; Jee, D. Oleic acid, cholesterol, and linoleic acid as angiogenesis initiators. ACS omega 2020, 5, 20575–20585. [Google Scholar] [CrossRef] [PubMed]
- Albini, A.; Bruno, A.; Baci, D.; Gallazzi, M.; Tramacere, M. Acetyl-L-carnitine (ALCAR) inhibits angiogenesis, migration and macrophage recruitment in prostatic cancer cells. Cancer Res. 2019, 79, 5086. [Google Scholar] [CrossRef]
- Baci, D.; Bruno, A.; Bassani, B.; Tramacere, M.; Mortara, L.; Albini, A.; Noonan, D.M. Acetyl-l-carnitine is an anti-angiogenic agent targeting the VEGFR2 and CXCR4 pathways. Cancer Lett. 2018, 429, 100–116. [Google Scholar] [CrossRef]
- Alkhatabi, H.A.; Zohny, S.F.; Shait Mohammed, M.R.; Choudhry, H.; Rehan, M.; Ahmad, A.; Ahmed, F.; Khan, M.I. Venetoclax-Resistant MV4-11 Leukemic Cells Activate PI3K/AKT Pathway for Metabolic Reprogramming and Redox Adaptation for Survival. Antioxidants 2022, 11, 461. [Google Scholar] [CrossRef]
- Sun, C.; Li, T.; Song, X.; Huang, L.; Zang, Q.; Xu, J.; Bi, N.; Jiao, G.; Hao, Y.; Chen, Y. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. USA 2019, 116, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Feun, L.; You, M.; Wu, C.J.; Kuo, M.T.; Wangpaichitr, M.; Spector, S.; Savaraj, N. Arginine deprivation as a targeted therapy for cancer. Curr. Pharm. Des. 2008, 14, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Rath, M.; Müller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagin, J.A.; Petrini, J.H. Oncogene-induced DNA damage: Cyclic AMP steps into the ring. J. Clin. Investig. 2020, 130, 5668–5670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kong, Q.; Wang, J.; Jiang, Y.; Hua, H. Complex roles of cAMP–PKA–CREB signaling in cancer. Exp. Hematol. Oncol. 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Vincent, E.E.; Poffenberger, M.C.; Jones, R.G. The AMP-activated protein kinase (AMPK) and cancer: Many faces of a metabolic regulator. Cancer Lett. 2015, 356, 165–170. [Google Scholar] [CrossRef]
- Shaw, J.H.; Wolfe, R.R. Fatty acid and glycerol kinetics in septic patients and in patients with gastrointestinal cancer. The response to glucose infusion and parenteral feeding. Ann. Surg. 1987, 205, 368. [Google Scholar] [CrossRef]
- Yu, X.-F.; Ni, Q.-C.; Chen, J.-P.; Xu, J.-F.; Jiang, Y.; Yang, S.-Y.; Ma, J.; Gu, X.-L.; Wang, H.; Wang, Y.-Y. Knocking down the expression of adenylate cyclase-associated protein 1 inhibits the proliferation and migration of breast cancer cells. Exp. Mol. Pathol. 2014, 96, 188–194. [Google Scholar] [CrossRef]
- Stroop, S.D.; Beavo, J.A. Structure and function studies of the cGMP-stimulated phosphodiesterase. J. Biol. Chem. 1991, 266, 23802–23809. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manjunath, M.; Swaroop, S.; Pradhan, S.S.; Rao K, R.; Mahadeva, R.; Sivaramakrishnan, V.; Choudhary, B. Integrated Transcriptome and Metabolomic Analysis Reveal Anti-Angiogenic Properties of Disarib, a Novel Bcl2-Specific Inhibitor. Genes 2022, 13, 1208. https://doi.org/10.3390/genes13071208
Manjunath M, Swaroop S, Pradhan SS, Rao K R, Mahadeva R, Sivaramakrishnan V, Choudhary B. Integrated Transcriptome and Metabolomic Analysis Reveal Anti-Angiogenic Properties of Disarib, a Novel Bcl2-Specific Inhibitor. Genes. 2022; 13(7):1208. https://doi.org/10.3390/genes13071208
Chicago/Turabian StyleManjunath, Meghana, Sai Swaroop, Sai Sanwid Pradhan, Raksha Rao K, Raghunandan Mahadeva, Venketesh Sivaramakrishnan, and Bibha Choudhary. 2022. "Integrated Transcriptome and Metabolomic Analysis Reveal Anti-Angiogenic Properties of Disarib, a Novel Bcl2-Specific Inhibitor" Genes 13, no. 7: 1208. https://doi.org/10.3390/genes13071208
APA StyleManjunath, M., Swaroop, S., Pradhan, S. S., Rao K, R., Mahadeva, R., Sivaramakrishnan, V., & Choudhary, B. (2022). Integrated Transcriptome and Metabolomic Analysis Reveal Anti-Angiogenic Properties of Disarib, a Novel Bcl2-Specific Inhibitor. Genes, 13(7), 1208. https://doi.org/10.3390/genes13071208