Transcriptomic and Metabolomic Analyses Provide Insights into the Formation of the Peach-like Aroma of Fragaria nilgerrensis Schlecht. Fruits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fruit Materials
| Benihoppe (female, 8x) × F. nilgerrensis Schlecht. (male, 2x) |
| ↓ |
| 5x |
| ↓ Chromosome doubling |
| BF (female, 10x) |
| ↓ Seedling selection |
| PA and NA |
2.2. Qualitative and Quantitative Analyses of Extracted Volatiles
2.3. Total RNA Extraction and Sequencing
2.4. Transcriptome Analysis
2.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
2.6. Correlation Analysis of the Transcriptome and Metabolome Data
3. Results
3.1. Metabolomic Profiling
3.2. Identification of DAMs
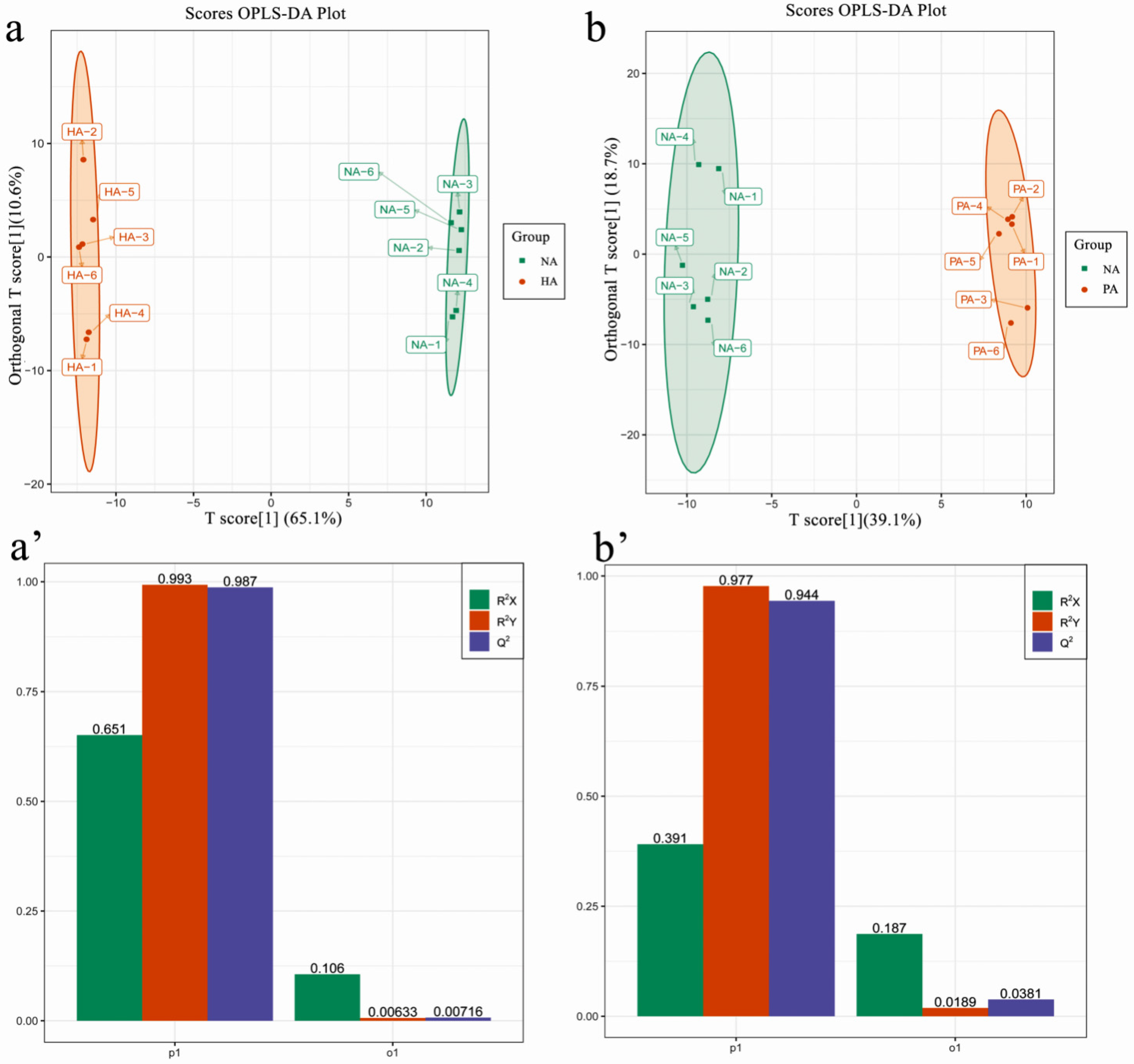
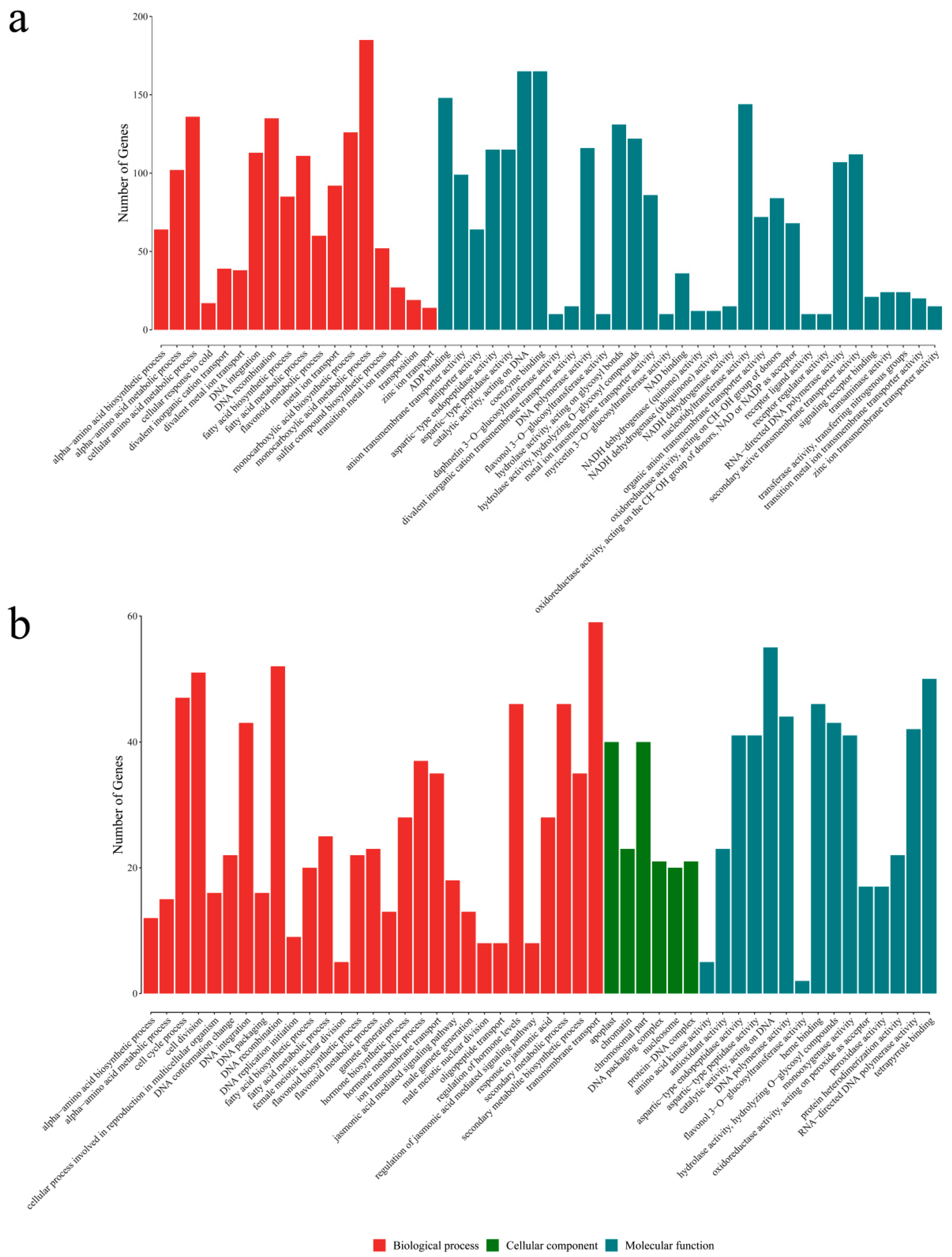
3.3. RNA-seq Analysis and Assembly and Functional Annotation
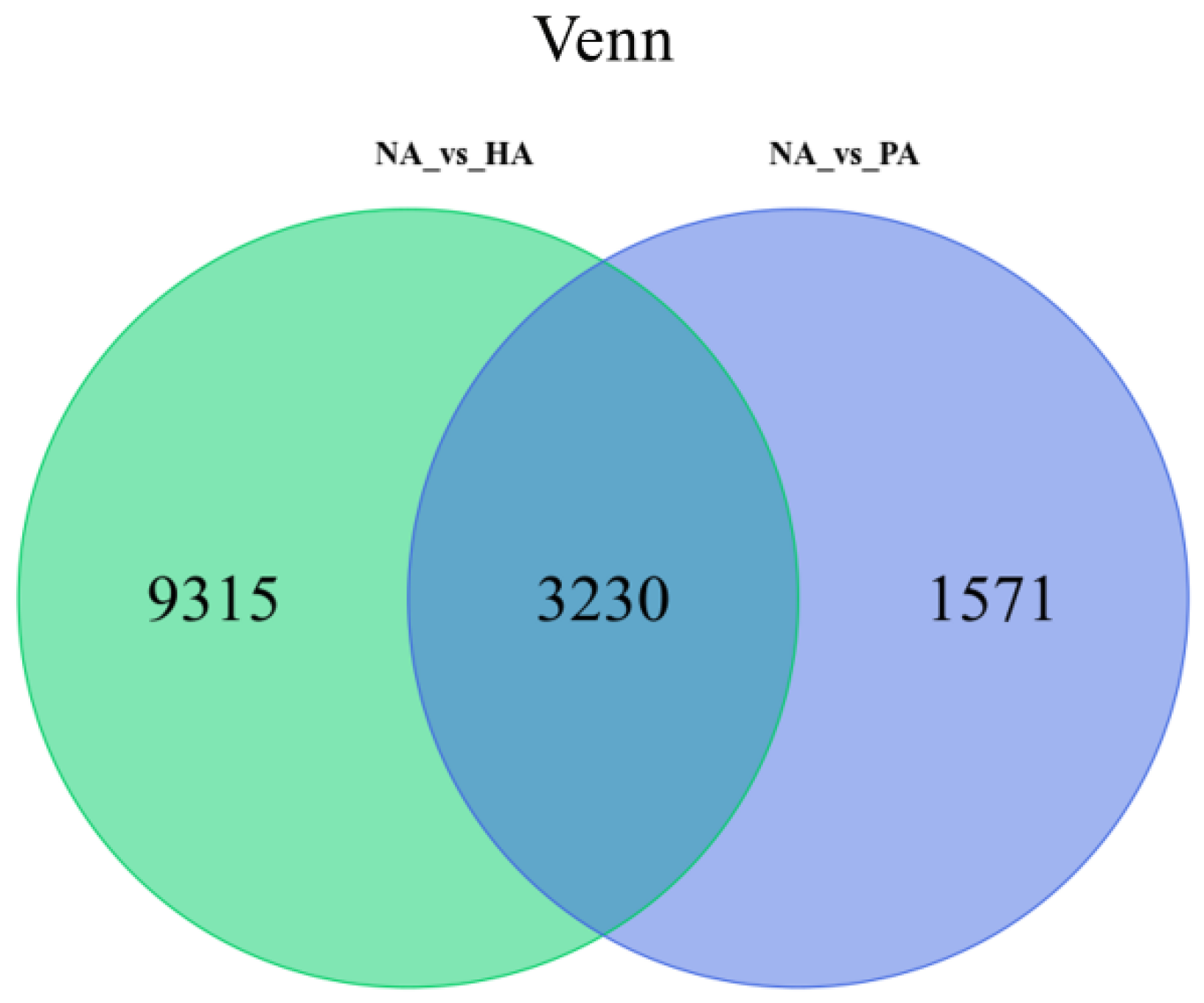
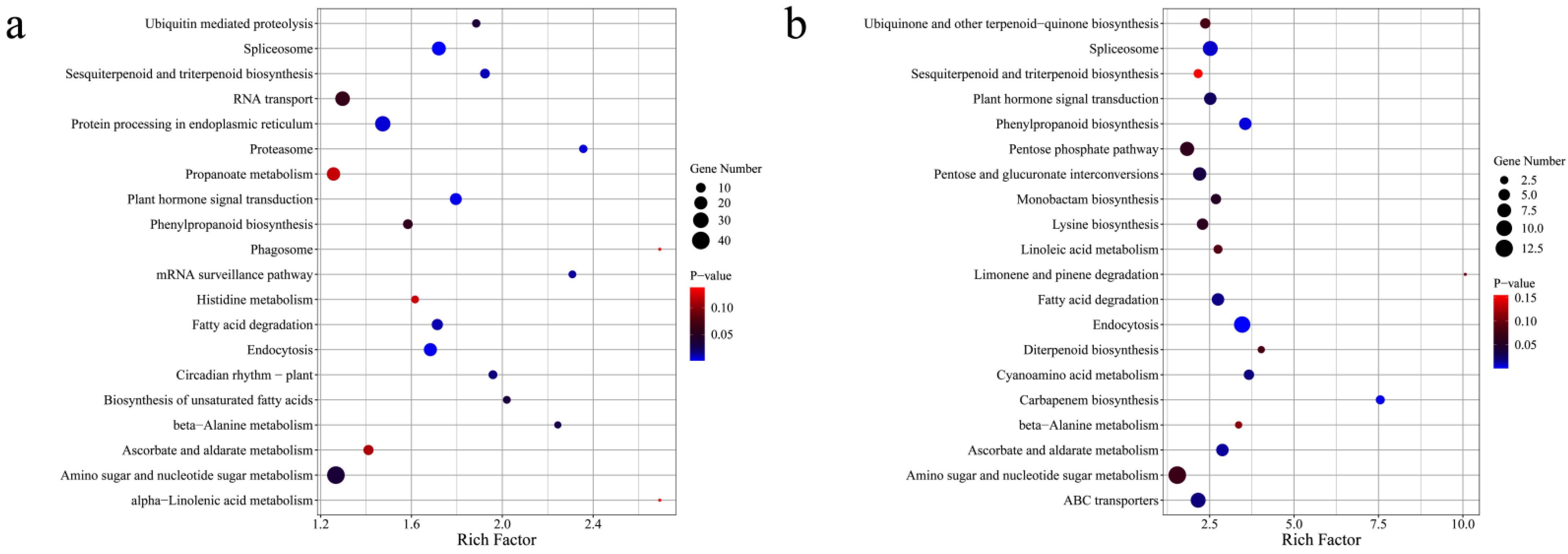
3.4. Identification and Functional Analysis of DEGs
3.5. Expression Patterns of Structural DEGs Related to Ester and Lactone Biosynthesis
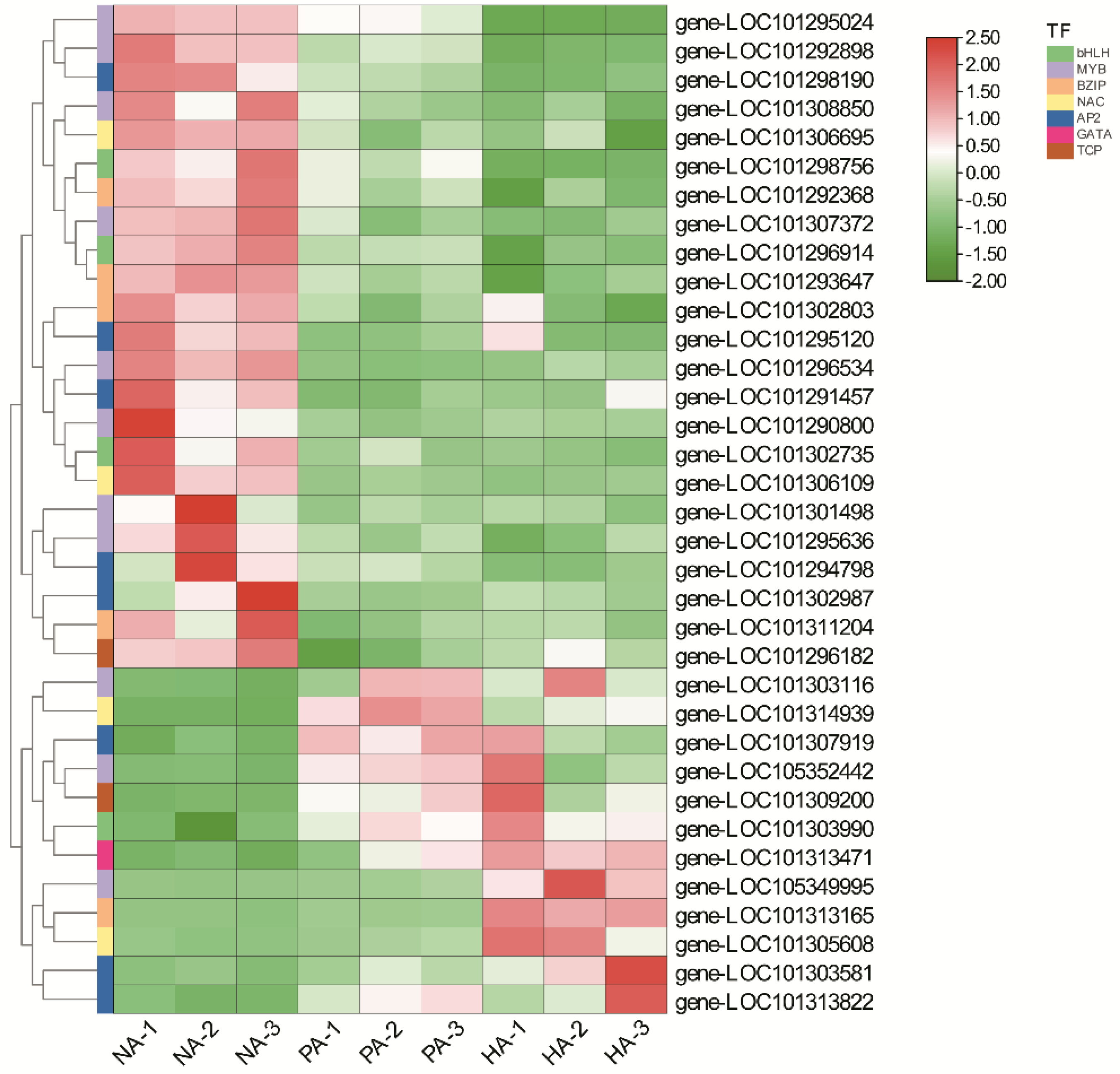
3.6. Expression Patterns of TF Genes Involved in Regulating Aroma Formation
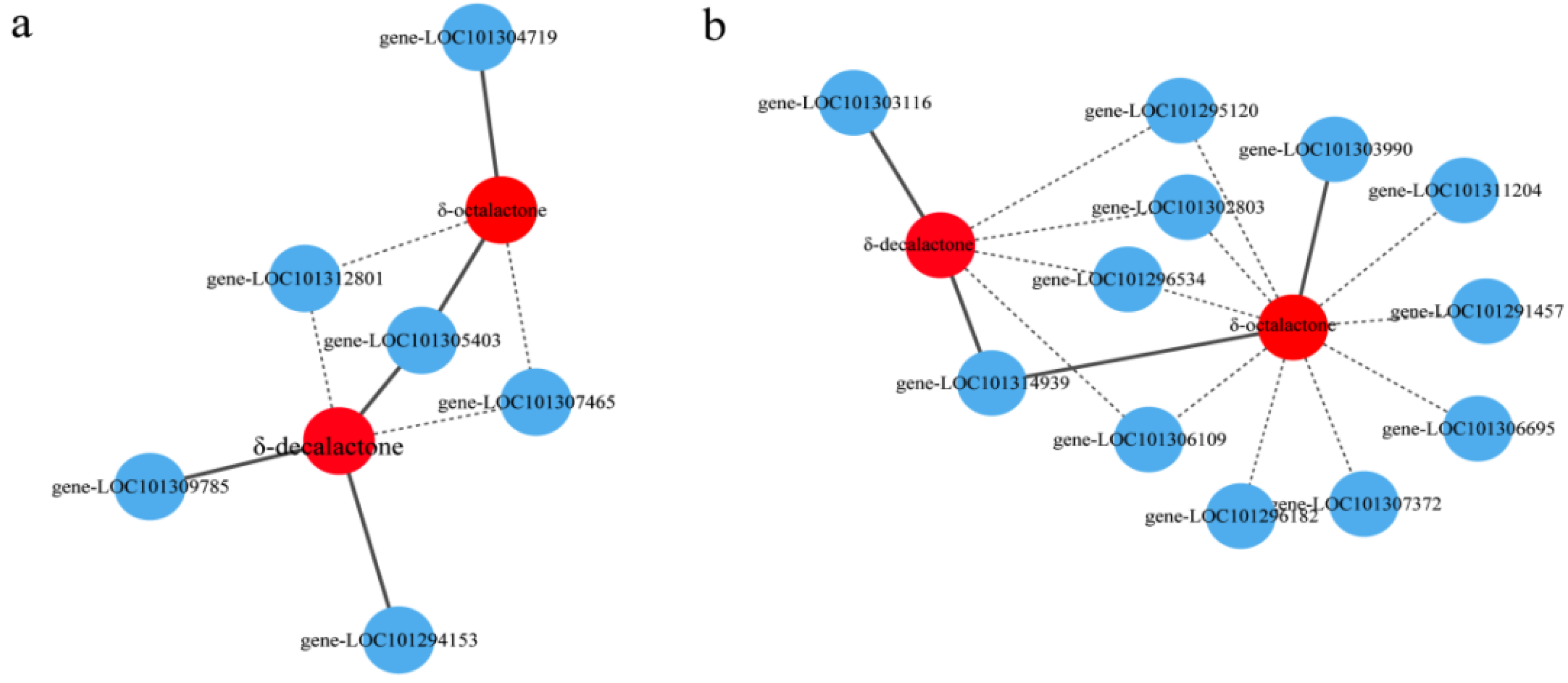
3.7. Correlation between the Transcriptome and Metabolome Data
4. Discussion
4.1. Metabolome Analysis
4.2. Transcriptome Analysis
4.3. Key Metabolites and Candidate Genes Associated with the Peach-like Aroma
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAT | Alcohol acyltransferase |
| LOX | Lipoxygenase |
| AOS | Allene oxide synthase |
| FAD | Fatty acid desaturase |
| EH | Epoxide hydrolase |
| AIM1 | Hydroxyacyl-CoA dehydrogenase |
| FAH | Fatty acid hydroxylase |
| ADH | Alcohol dehydrogenase |
| DEG | Differentially expressed gene |
| DAM | Differentially accumulated metabolite |
References
- Dong, J.; Zhang, Y.; Tang, X.; Jin, W.; Han, Z. Differences in volatile ester composition between Fragaria × ananassa and F. vesca and implications for strawberry aroma patterns. Sci. Hortic. 2013, 150, 47–53. [Google Scholar] [CrossRef]
- Urrutia, M.; Rambla, J.L.; Alexiou, K.G.; Granell, A.; Monfort, A. Genetic analysis of the wild strawberry (Fragaria vesca) volatile composition. Plant Physiol. Biochem. 2017, 121, 99–117. [Google Scholar] [CrossRef] [PubMed]
- Schwieterman, M.L.; Colquhoun, T.A.; Jaworski, E.A.; Bartoshuk, L.M.; Gilbert, J.L.; Tieman, D.M.; Odabasi, A.Z.; Moskowitz, H.R.; Folta, K.M.; Klee, H.J.; et al. Strawberry flavor: Diverse chemical compositions, a seasonal influence, and effects on sensory perception. PLoS ONE 2014, 9, e88446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negri, A.S.; Allegra, D.; Simoni, L.; Rusconi, F.; Tonelli, C.; Espen, L.; Galbiati, M. Comparative analysis of fruit aroma patterns in the domesticated wild strawberries “Profumata di Tortona” (F. moschata) and “Regina delle Valli” (F. vesca). Front. Plant Sci. 2015, 11, 56. [Google Scholar]
- Zhang, J.; Lei, Y.; Wang, B.; Li, S.; Yu, S.; Wang, Y.; Li, H.; Liu, Y.; Ma, Y.; Dai, H.; et al. The high-quality genome of diploid strawberry (Fragaria nilgerrensis) provides new insights into anthocyanin accumulation. Plant Biotechnol. J. 2020, 18, 1908–1924. [Google Scholar] [CrossRef] [Green Version]
- Sheng, L.; Ni, Y.; Wang, J.; Chen, Y.; Gao, H. Characteristic-aroma-component-based evaluation and classification of strawberry varieties by aroma type. Molecules 2021, 26, 6219. [Google Scholar] [CrossRef]
- Jetti, R.R.; Yang, E.; Kurnianta, A.; Finn, C.; Qian, M.C. Quantification of selected aroma-active compounds in strawberries by headspace solid-phase microextraction gas chromatography and correlation with sensory descriptive analysis. J. Food Sci. 2007, 72, S487–S496. [Google Scholar] [CrossRef]
- Ulrich, D.; Komes, D.; Olbricht, K.; Hoberg, E. Diversity of aroma patterns in wild and cultivated Fragaria accessions. Genet. Resour. Crop Evol. 2007, 54, 1185. [Google Scholar] [CrossRef]
- Larsen, M.; Poll, L. Odour thresholds of some important aroma compounds in strawberries. Z. Lebensm. Unters. Forsch. 1992, 195, 120–123. [Google Scholar] [CrossRef]
- Schieberle, P.; Hofmann, T. Evaluation of the character impact odorants in fresh strawberry juice by quantitative measurements and sensory studies on model mixtures. J. Agric. Food Chem. 1997, 45, 227–232. [Google Scholar] [CrossRef]
- Staudt, G.; Drawert, F.; Tressl, R. Gas chromatographic-mass spectrometric differentiation of aroma compounds from strawberry varieties. II. Fragaria Nilgerrensis. Z. Fuer Pflanzenzuechtung. 1975, 75, 36–42. [Google Scholar]
- Sánchez-Sevilla, J.F.; Cruz-Rus, E.; Valpuesta, V.; Botella, M.A.; Amaya, I. Deciphering γ-decalactone biosynthesis in strawberry fruit using a combination of genetic mapping, RNA-Seq and eQTL analyses. BMC Genom. 2014, 15, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, M.; Ulrich, D. Characterisation of the aroma pattern of the wild strawberry Fragaria nilgerrensis (Schltdl.) as a precondition for the use in strawberry breeding. Mitt. Aus Dem Jul. Kühn-Inst. 2009, 419, 36. [Google Scholar]
- Gong, C.; Diao, W.; Zhu, H.; Umer, M.J.; Zhao, S.; He, N.; Lu, X.; Yuan, P.; Anees, M.; Yang, D.; et al. Metabolome and transcriptome integration reveals insights into flavor formation of ‘Crimson’ watermelon flesh during fruit development. Front. Plant Sci. 2021, 12, 629361. [Google Scholar] [CrossRef] [PubMed]
- El Hadi, M.A.; Zhang, F.J.; Wu, F.F.; Zhou, C.H.; Tao, J. Advances in fruit aroma volatile research. Molecules 2013, 18, 8200–8229. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Barbey, C.R.; Chandra, S.; Bai, J.; Fan, Z.; Plotto, A.; Pillet, J.; Folta, K.M.; Whitaker, V.M.; Lee, S. Genomic characterization of the fruity aroma gene, FaFAD1, reveals a gene dosage effect on γ-decalactone production in strawberry (Fragaria × ananassa). Front. Plant Sci. 2021, 12, 639345. [Google Scholar] [CrossRef]
- Pillet, J.; Chambers, A.H.; Barbey, C.; Bao, Z.; Plotto, A.; Bai, J.; Schwieterman, M.; Johnson, T.; Harrison, B.; Whitaker, V.M.; et al. Identification of a methyltransferase catalyzing the final step of methyl anthranilate synthesis in cultivated strawberry. BMC Plant Biol. 2017, 17, 147. [Google Scholar] [CrossRef]
- Lyzhin, A.S.; Luk’yanchuk, I.V.; Zhbanova, E.V. Polymorphism of the FaOMT and FaFAD1 genes for fruit flavor volatiles in strawberry varieties and wild species from the genetic collection of the Michurin Federal Research Center. Vavilovskii. Zhurnal. Genet. Selektsii. 2020, 24, 5–11. [Google Scholar]
- Aharoni, A.; Giri, A.P.; Verstappen, F.W.; Bertea, C.M.; Sevenier, R.; Sun, Z.; Jongsma, M.A.; Schwab, W.; Bouwmeester, H.J. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 2004, 16, 3110–3131. [Google Scholar] [CrossRef] [Green Version]
- Lu, H.; Luo, Z.; Wang, L.; Liu, W.; Li, D.; Belwal, T.; Xu, Y.; Li, L. FaMYB9 is involved in the regulation of C6 volatile biosynthesis in strawberry. Plant Sci. 2020, 293, 110422. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Feng, C.; Yang, L.; Edger, P.P.; Kang, M. Genomic population structure and local adaptation of the wild strawberry Fragaria nilgerrensis. Hortic. Res. 2022, 9, uhab059. [Google Scholar] [CrossRef]
- Noguchi, Y. “Tokun”: A new decaploid interspecific hybrid strawberry having the aroma of the wild strawberry. J. Jpn. Assoc. Odor Environ. 2011, 42, 122–128. [Google Scholar]
- Ménager, I.; Jost, M.; Aubert, C. Changes in physicochemical characteristics and volatile constituents of strawberry (Cv. Cigaline) during maturation. J. Agric. Food Chem. 2004, 52, 1248–1254. [Google Scholar] [CrossRef]
- Zhao, M.Z.; Wang, J.; Wang, Z.W.; Qian, Y.M. GC-MS analysis of volatile components in Chinese wild strawberry (F. nilgerrensis Schlecht.). Acta Hortic. 2014, 1049, 467–469. [Google Scholar] [CrossRef]
- Zeliou, K.; Papasotiropoulos, V.; Manoussopoulos, Y.; Lamari, F.N. Physical and chemical quality characteristics and antioxidant properties of strawberry cultivars (Fragaria × ananassa Duch.) in Greece: Assessment of their sensory impact. J. Sci. Food Agric. 2018, 9811, 4065–4073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shen, J.Y.; Wei, W.W.; Xi, W.P.; Xu, C.J.; Ferguson, I.; Chen, K. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J. Agric. Food Chem. 2010, 58, 6157–6165. [Google Scholar] [CrossRef]
- Braga, A.; Belo, I. Biotechnological production of γ-decalactone, a peach like aroma, by Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2016, 32, 169. [Google Scholar] [CrossRef] [Green Version]
- Budak, N.H.; Özdemir, N.; Gökırmaklı, Ç. The changes of physicochemical properties, antioxidants, organic, and key volatile compounds associated with the flavor of peach (Prunus cerasus L. Batsch) vinegar during the fermentation process. J. Food Biochem. 2022, 46, e13978. [Google Scholar] [CrossRef]
- Zabetakis, I.; Holden, M.A. Strawberry flavor: Analysis and biosynthesis. J. Sci. Food Agric. 1997, 74, 421–434. [Google Scholar] [CrossRef]
- Ulrich, D.; Kecke, S.; Olbricht, K. What do we know about the chemistry of strawberry aroma? J. Agric. Food Chem. 2018, 66, 3291–3301. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Li, G.; Zhao, X.; Zhao, F.; Li, L.; Zhou, H. Transcriptome profiling by RNA-Seq reveals differentially expressed genes related to fruit development and ripening characteristics in strawberries (Fragaria × ananassa). PeerJ 2018, 27, e4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbey, C.R.; Hogshead, M.H.; Harrison, B.; Schwartz, A.E.; Verma, S.; Oh, Y.; Lee, S.; Folta, K.M.; Whitaker, V.M. Genetic Analysis of Methyl Anthranilate, Mesifurane, Linalool, and Other Flavor Compounds in Cultivated Strawberry (Fragaria × ananassa). Front. Plant Sci. 2021, 19, 615749. [Google Scholar] [CrossRef]
- Sheng, L.; Ma, C.; Chen, Y.; Gao, H.; Wang, J. Genome-Wide screening of AP2 transcription factors involving in fruit color and aroma regulation of cultivated strawberry. Genes 2021, 12, 530. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, G.; Venegas-Calerón, M.; Salas, J.J.; Monforte, A.; Badenes, M.L.; Granell, A. An integrative “omics” approach identifies new candidate genes to impact aroma volatiles in peach fruit. BMC Genom. 2013, 23, 343. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.J.; Liu, H.R.; Gao, J.; Huang, Y.J.; Zhang, B.; Chen, K.S. Two ω-3 FADs are associated with peach fruit volatile formation. Int. J. Mol. Sci. 2016, 29, 464. [Google Scholar] [CrossRef] [Green Version]
- Peng, B.; Yu, M.; Zhang, B.; Xu, J.; Ma, R. Differences in PpAAT1 Activity in High- and Low-Aroma Peach Varieties Affect γ-Decalactone Production. Plant Physiol. 2020, 182, 2065–2080. [Google Scholar] [CrossRef]
- Song, Z.Z.; Peng, B.; Gu, Z.X.; Tang, M.L.; Li, B.; Liang, M.X.; Wang, L.M.; Guo, X.T.; Wang, J.P.; Sha, Y.F.; et al. Site-directed mutagenesis identified the key active site residues of alcohol acyltransferase PpAAT1 responsible for aroma biosynthesis in peach fruits. Hortic. Res. 2021, 8, 32. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Gao, L.; Qi, Y.; Fu, W.; Li, X.; Zhou, X.; Gao, Q.; Gao, Z.; Jia, H. Acyl-CoA oxidase 1 is involved in γ-decalactone release from peach (Prunus persica) fruit. Plant Cell Rep. 2017, 36, 829–842. [Google Scholar] [CrossRef]
- Deshpande, A.B.; Chidley, H.G.; Oak, P.S.; Pujari, K.H.; Giri, A.P.; Gupta, V.S. Isolation and characterization of 9-lipoxygenase and epoxide hydrolase 2 genes: Insight into lactone biosynthesis in mango fruit (Mangifera indica L.). Phytochemistry 2017, 138, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Wei, C.; Duan, W.; Gao, Y.; Kuang, J.; Liu, M.; Chen, K.; Klee, H.; Zhang, B. Transcriptional and epigenetic analysis reveals that NAC transcription factors regulate fruit flavor ester biosynthesis. Plant J. 2021, 106, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Li, M.; Cao, X.; Jin, Z.; Zhang, C.; Xu, M.; Chen, K.; Zhang, B. Linalool synthesis related PpTPS1 and PpTPS3 are activated by transcription factor PpERF61 whose expression is associated with DNA methylation during peach fruit ripening. Plant Sci. 2022, 317, 111200. [Google Scholar] [CrossRef]
- Jian, W.; Cao, H.; Yuan, S.; Liu, Y.; Lu, J.; Lu, W.; Li, N.; Wang, J.; Zou, J.; Tang, N.; et al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 2019, 6, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, X.; Wan, S.; Lin, C.; Zhou, C.; Hu, L.; Deng, C.; Zhang, L. Integration of metabolome and transcriptome reveals the relationship of benzenoid-phenylpropanoid pigment and aroma in purple tea flowers. Front. Plant Sci. 2021, 12, 762330. [Google Scholar] [CrossRef]
- Miao, L.; Di, Q.; Sun, T.; Li, Y.; Duan, Y.; Wang, J.; Yan, Y.; He, C.; Wang, C.; Yu, X. Integrated metabolome and transcriptome analysis provide insights into the effects of grafting on fruit flavor of cucumber with different rootstocks. Int. J. Mol. Sci. 2019, 20, 3592. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Huang, L.; Chen, M.; Zeng, W.; Feng, Z.; Huang, S.; Liu, T. Integrated analysis of the transcriptome and metabolome reveals genes involved in terpenoid and flavonoid biosynthesis in the loblolly pine (Pinus taeda L.). Front. Plant Sci. 2021, 12, 729161. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Shi, T.; Chen, M.; Li, Y.; Du, J.; Jiang, H.; Yang, X.; Hu, H.; Wang, L. Integrating transcriptomic and GC-MS metabolomic analysis to characterize color and aroma formation during tepal development in Lycoris longituba. Plants 2019, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Fei, X.; Qi, Y.; Lei, Y.; Wang, S.; Hu, H.; Wei, A. Transcriptome and metabolome dynamics explain aroma differences between green and red prickly ash fruit. Foods 2021, 10, 391. [Google Scholar] [CrossRef]
- Chen, X.; Cai, W.; Xia, J.; Yu, H.; Wang, Q.; Pang, F.; Zhao, M. Metabolomic and transcriptomic analyses reveal that blue light promotes chlorogenic acid synthesis in strawberry. J. Agric. Food Chem. 2020, 68, 12485–12492. [Google Scholar] [CrossRef]
- Xin, M.; Li, C.; Khoo, H.E.; Li, L.; He, X.; Yi, P.; Tang, Y.; Sun, J. Dynamic analyses of transcriptome and metabolic rrofiling: Revealing molecular insight of aroma synthesis of mango (Mangifera indica L. Var. Tainong). Front. Plant Sci. 2021, 12, 666805. [Google Scholar] [CrossRef] [PubMed]




| Sample | Raw Reads (M) | Clean Reads (M) | Clean Base (G) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| HA-1 | 47.30 | 44.37 | 6.66 | 98.17 | 94.54 | 45.75 |
| HA-2 | 45.76 | 42.85 | 6.43 | 97.98 | 94.27 | 46.44 |
| HA-3 | 47.59 | 44.11 | 6.62 | 97.49 | 92.99 | 46.14 |
| NA-1 | 43.43 | 42.00 | 6.3 | 98.06 | 94.32 | 46.30 |
| NA-2 | 48.95 | 47.23 | 7.09 | 97.95 | 94.14 | 46.54 |
| NA-3 | 48.01 | 46.93 | 7.04 | 98.00 | 94.13 | 46.35 |
| PA-1 | 45.89 | 43.92 | 6.59 | 98.09 | 94.39 | 46.08 |
| PA-2 | 46.13 | 43.45 | 6.52 | 98.07 | 94.34 | 45.45 |
| PA-3 | 47.79 | 45.60 | 6.84 | 98.05 | 94.27 | 45.97 |
| Sum | 420.86 | 400.45 | 60.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.-H.; Ma, H.-Y.; Zhang, B.-H.; Mo, C.-Y.; Li, E.-H.; Li, F. Transcriptomic and Metabolomic Analyses Provide Insights into the Formation of the Peach-like Aroma of Fragaria nilgerrensis Schlecht. Fruits. Genes 2022, 13, 1285. https://doi.org/10.3390/genes13071285
Wang A-H, Ma H-Y, Zhang B-H, Mo C-Y, Li E-H, Li F. Transcriptomic and Metabolomic Analyses Provide Insights into the Formation of the Peach-like Aroma of Fragaria nilgerrensis Schlecht. Fruits. Genes. 2022; 13(7):1285. https://doi.org/10.3390/genes13071285
Chicago/Turabian StyleWang, Ai-Hua, Hong-Ye Ma, Bao-Hui Zhang, Chuan-Yuan Mo, En-Hong Li, and Fei Li. 2022. "Transcriptomic and Metabolomic Analyses Provide Insights into the Formation of the Peach-like Aroma of Fragaria nilgerrensis Schlecht. Fruits" Genes 13, no. 7: 1285. https://doi.org/10.3390/genes13071285





