Complete Nucleotide Sequence of the Mitogenome of Tapinoma ibericum (Hymenoptera: Formicidae: Dolichoderinae), Gene Organization and Phylogenetics Implications for the Dolichoderinae Subfamily
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Sequencing and Mitogenome Assembly
2.3. Mitogenome Annotation and Sequence Analysis
2.4. Phylogenetic Analysis
3. Results and Discussion
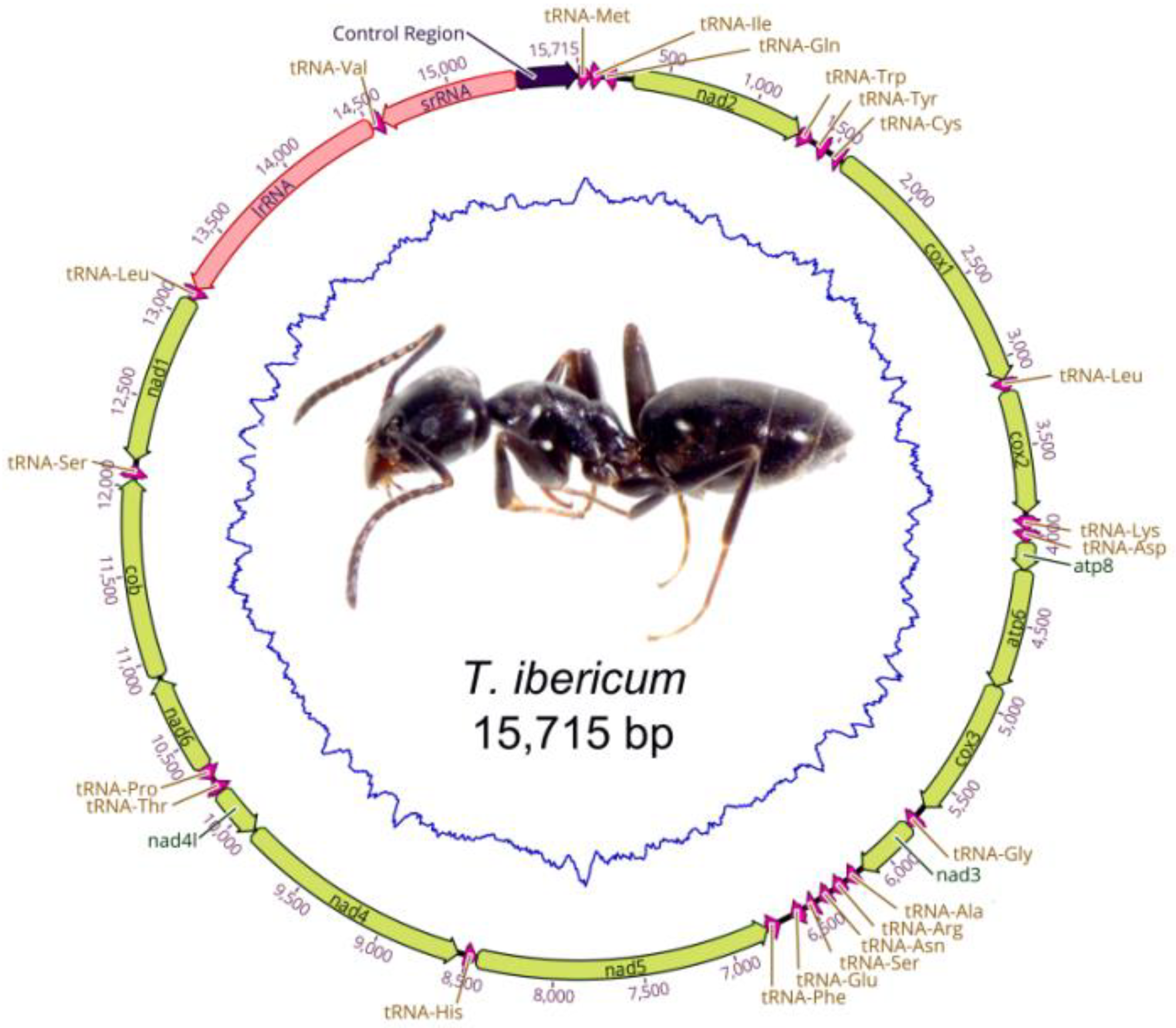
3.1. Gene Organization
3.2. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hölldobler, B.; Wilson, E.O. The Ants; Harvard University Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Bolton, B. An Online Catalog of the Ants of the World. Available online: http://antcat.org (accessed on 1 May 2022).
- Schultz, T.R. In search of ant ancestors. Proc. Natl. Acad. Sci. USA 2000, 97, 14028–14029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertelsmeier, C.; Luque, G.M.; Hoffmann, B.D.; Courchamp, F. Worldwide ant invasions under climate change. Biodiver. Conserv. 2015, 24, 117–128. [Google Scholar] [CrossRef]
- Siddiqui, J.A.; Bamisile, B.S.; Khan, M.M.; Islam, W.; Hafeez, M.; Bodlah, I.; Xu, Y. Impact of invasive ant species on native fauna across similar habitats under global environmental changes. Environ. Sci. Pollut. Res. 2021, 28, 54362–54382. [Google Scholar] [CrossRef] [PubMed]
- Klimeš, P.; Okrouhlík, J. Invasive ant Tapinoma melanocephalum (Hymenoptera: Formicidae): A rare guest or increasingly common indoor pest in Europe? Eur. J. Entomol. 2015, 112, 705–712. [Google Scholar] [CrossRef] [Green Version]
- Seifert, B.; D’eustacchio, D.; Kaufmann, B.; Centorame, M.; Lorite, P.; Modica, M.V. Four species within the supercolonial ants of the Tapinoma nigerrimum complex revealed by integrative taxonomy (Hymenoptera: Formicidae). Myrmecol. News 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Smith, M.A.; Fisher, B.L. Invasions, DNA barcodes, and rapid biodiversity assessment using ants of Mauritius. Front. Zool. 2009, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Flucher, S.M.; Krapf, P.; Arthofer, W.; Suarez, A.V.; Crozier, R.H.; Steiner, F.M.; Schlick-Steiner, B.C. Effect of social structure and introduction history on genetic diversity and differentiation. Mol. Ecol. 2021, 30, 2511–2527. [Google Scholar] [CrossRef]
- Simberloff, D. Hybridization between native and introduced wildlife species: Importance for conservation. Wildl. Biol. 1996, 2, 143–150. [Google Scholar] [CrossRef]
- Seifert, B. Hybridization in the European carpenter ants Camponotus herculeanus and C. ligniperda (Hymenoptera: Formicidae). Insect. Soc. 2019, 66, 365–374. [Google Scholar] [CrossRef]
- Song, N.; Yin, X.; Zhao, X.; Chen, J.; Yin, J. Reconstruction of mitogenomes by NGS and phylogenetic implications for leaf beetles. Mitochondrial DNA Part A 2018, 29, 1041–1050. [Google Scholar] [CrossRef]
- Pita, S.; Mora, P.; Rojas-Cortez, M.; Palomeque, T.; Lorite, P.; Panzera, F. The complete nucleotide sequence and gene organization of the mitochondrial genome of Triatoma boliviana (Hemiptera, Reduviidae, Triatominae) and phylogenetic comparisons. Entomology 2022, 1, 2–10. [Google Scholar] [CrossRef]
- Mora, P.; Montiel, E.E.; Palomeque, T.; Lorite, P. Complete mitochondrial genome of the blister beetle Hycleus scutellatus (Coleoptera, Meloidae). Mitochondrial DNA Part B Resour. 2022, 7, 986–988. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Xi, H.; Park, J. Comparative Analysis on Palaearctic Dolichoderus Quadripunctatus Species Group with the Proposal of a Novel Species (Formicidae). In Proceedings of the 2020 Spring International Conference of Korean Society of Applied Entomology, Online Academic Presentation (Society’s Website), 25–29 May 2020. [Google Scholar] [CrossRef]
- Park, J.; Park, C.H.; Park, J. Complete mitochondrial genome of the H3 haplotype Argentine ant Linepithema humile (Mayr, 1868) (Formicidae; Hymenoptera). Mitochondrial DNA Part B Resour. 2021, 6, 786–788. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Varlamov, A.; Vaskin, Y.; Efremov, I.; German Grehov, O.G.; Kandrov, D.; Rasputin, K.; Syabro, M.; et al. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Duan, X.Y.; Peng, X.Y.; Qian, Z.Q. The complete mitochondrial genomes of two globally invasive ants, the Argentine ant Linepithema humile and the little fire ant Wasmannia auropunctata. Conserv. Genet. Resour. 2016, 8, 275–277. [Google Scholar] [CrossRef]
- Cameron, S.L. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef] [Green Version]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Allio, R.; Schomaker-Bastos, A.; Romiguier, J.; Prosdocimi, F.; Nabholz, B.; Delsuc, F. MitoFinder: Efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol. Ecol. Res. 2020, 20, 892–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kwon, W.; Park, J. The complete mitochondrial genome of Siberian odorous ant, Dolichoderus sibiricus Emery, 1889 (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2019, 4, 525–526. [Google Scholar] [CrossRef] [Green Version]
- Berman, M.; Austin, C.M.; Miller, A.D. Characterisation of the complete mitochondrial genome and 13 microsatellite loci through next-generation sequencing for the New Caledonian spider-ant Leptomyrmex pallens. Mol. Biol. Rep. 2014, 41, 1179–1187. [Google Scholar] [CrossRef]
- Zhao, E.; Bi, G.; Yang, J.; Zhang, Z.; Liu, G.; Du, Q.; Shang, E. Complete mitochondrial genome of the argentine ant, Linepithema humile (Hymenoptera: Formicidae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2017, 28, 210–211. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Xi, H.; Park, J. The complete mitochondrial genome of Ochetellus glaber (Mayr, 1862) (Hymenoptera:Formicidae). Mitochondrial DNA Part B Resour. 2020, 5, 147–149. [Google Scholar] [CrossRef] [Green Version]
- Du, Y.; Song, X.; Yu, H.; Lu, Z. Complete mitochondrial genome sequence of Tapinoma melanocephalum (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2019, 4, 3448–3449. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Chiotis, M.; Jermiin, L.S.; Crozier, R.H. A molecular framework for the phylogeny of the ant subfamily Dolichoderinae. Mol. Phylogenet. Evol. 2000, 17, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Gotzek, D.; Clarke, J.; Shoemaker, D. Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae). BMC Evol. Biol. 2010, 10, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodovalho, C.d.M.; Lyra, M.L.; Ferro, M.; Bacci, M., Jr. The mitochondrial genome of the leaf-cutter ant Atta laevigata: A mitogenome with a large number of intergenic spacers. PLoS ONE 2014, 9, e97117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Huang, H.; Liu, Y.; Liu, S.; Xia, J.; Zhang, K.; Geng, J. Organization and phylogenetic relationships of the mitochondrial genomes of Speiredonia retorta and other lepidopteran insects. Sci. Rep. 2021, 11, 2957. [Google Scholar] [CrossRef]
- Song, N.; Liang, A. The complete mitochondrial genome sequence of Geisha distinctissima (Hemiptera: Flatidae) and comparison with other hemipteran insects. Acta Biochim. Biophys. Sin. 2009, 41, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Beckenbach, A.T.; Stewart, J.B. Insect mitochondrial genomics 3: The complete mitochondrial genome sequences of representatives from two neuropteroid orders: A dobsonfly (order Megaloptera) and a giant lacewing and an owlfly (order Neuroptera). Genome 2009, 52, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B. The Evolution of Mitochondrial Genome Structure and Function in Insects. Ph.D. Thesis, Simon Fraser University, Burnaby, BC, Canada, 2005. [Google Scholar]
- Yang, S.; Li, X.; Qian, L.G. Characterization of the complete mitochondrial genome of Formica selysi (Insecta: Hymenoptera: Formicidae: Formicinae). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 3378–3380. [Google Scholar] [CrossRef] [PubMed]
- Meza-Lázaro, R.N.; Poteaux, C.; Bayona-Vásquez, N.J.; Branstetter, M.G.; Zaldívar-Riverón, A. Extensive mitochondrial heteroplasmy in the neotropical ants of the Ectatomma ruidum complex (Formicidae: Ectatomminae). Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2018, 29, 1203–1214. [Google Scholar] [CrossRef]
- Wei, S.J.; Tang, P.; Zheng, L.H.; Shi, M.; Chen, X.X. The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A + T content and a long intergenic spacer between atp8 and atp6. Mol. Biol. Rep. 2010, 37, 1931–1942. [Google Scholar] [CrossRef] [Green Version]
- Aydemir, M.N.; Korkmaz, E.M. Comparative mitogenomics of Hymenoptera reveals evolutionary differences in structure and composition. Int. J. Biol. Macromol. 2020, 144, 460–472. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Song, H.; Sheffield, N.C.; Cameron, S.L.; Miller, K.B.; Whiting, M.F. What happens when the phylogenetic assumptions are violated?: The effect of base compositional heterogeneity and among-site rate heterogeneity in beetle mitochondrial phylogenomics. Syst. Entomol. 2010, 35, 429–448. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Wu, Y.; Yang, C.; Gu, X.; Wilson, J.J.; Li, H.; Cai, W.; Yang, H.; Song, F. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: Ichneumonoidea). Int. J. Biol. Macromol. 2020, 164, 540–547. [Google Scholar] [CrossRef]
- Cameron, S.L.; Lambkin, C.L.; Barker, S.C.; Whiting, M.F. A mitochondrial genome phylogeny of Diptera: Whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst. Entomol. 2007, 32, 40–59. [Google Scholar] [CrossRef]
- Taanman, J.W. The mitochondrial genome: Structure, transcription, translation and replication. Biochim. Biophys. Acta 1999, 1410, 103–123. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Phylogenetic approaches for the analysis of mitochondrial genome sequence data in the Hymenoptera—A lineage with both rapidly and slowly evolving mitochondrial genomes. Mol. Phylogenet. Evol. 2009, 52, 512–519. [Google Scholar] [CrossRef]
- Zhang, D.X.; Szymura, J.M.; Hewitt, G.M. Evolution and structural conservation of the control region of insect mitochondrial DNA. J. Mol. Evol. 1995, 40, 382–391. [Google Scholar] [CrossRef]
- Ward, P.S.; Brady, S.G.; Fisher, B.F.; Schultz, T. Phylogeny and biogeography of Dolichoderine ants: Effects of data partitioning and relict taxa on historical inference. Syst. Biol. 2010, 59, 342–362. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.C.; Wang, J.; Matsuura, K.; Yang, C.C.S. The complete mitochondrial genome of yellow crazy ant, Anoplolepis gracilipes (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2018, 3, 622–623. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Park, J. Complete mitochondrial genome of the gate-keeper ant Colobopsis nipponica (Wheeler, W.M., 1928) (Formicidae: Hymenoptera). Mitochondrial DNA Part B Resour. 2021, 6, 86–88. [Google Scholar] [CrossRef] [PubMed]
- Babbucci, M.; Basso, A.; Scupola, A.; Patarnello, T.; Negrisolo, E. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol. Evol. 2014, 6, 3326–3343. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J. Complete mitochondrial genome of the jet ant Lasius spathepus Wheeler, W.M., 1910 (Formicidae; Hymenoptera). Mitochondrial DNA Part B Resour. 2021, 6, 505–507. [Google Scholar] [CrossRef]
- Liu, J.H.; Jia, P.F.; Fu, J.Q.; Dan, W.L.; Yang, L.Y.; Wang, Q.M.; Li, Z.N. Characterization of mitochondrial genome and phylogenetic implications for Chinese black ant, Polyrhachis dives (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2017, 2, 679–680. [Google Scholar] [CrossRef]
- Park, J.; Xi, H.; Park, J. The complete mitochondrial genome of Aphaenogaster famelica (Smith, 1874) (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2020, 5, 492–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suen, G.; Teiling, C.; Li, L.; Holt, C.; Abouheif, E.; Bornberg-Bauer, E.; Bouffard, P.; Caldera, E.J.; Cash, E.; Cavanaugh, A.; et al. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 2011, 7, e1002007. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, J.T.V.; Barbosa, M.S.; Morais, S.; Santana, A.E.G.; Almeida, C. Mitochondrial genomes of genus Atta (Formicidae: Myrmicinae) reveal high gene organization and giant intergenic spacers. Genet. Mol. Biol. 2020, 42, e20180055. [Google Scholar] [CrossRef]
- Liu, L.; Chen, F.; Wang, Q.X.; Zhang, Z.Y.; Tang, Y.; Li, F.; Qian, Z.Q. Characterization of the complete mitochondrial genome of the invasive tramp ant Cardiocondyla obscurior (Hymenoptera: Formicidae: Myrmicinae). Mitochondrial DNA Part B Resour. 2019, 4, 1496–1498. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Xi, H.; Park, J. Complete mitochondrial genome of the acrobat ant Crematogaster teranishii Santschi, 1930 (Formicidae; Hymenoptera). Mitochondrial DNA Part B Resour. 2021, 6, 593–595. [Google Scholar] [CrossRef]
- Zhang, X.M.; Han, X.; Liu, X.; Xu, Z.H. Characterization of the complete mitochondrial genome of a harvesting ant Messor structor (Hymenoptera: Formicidae: Myrmicinae). Mitochondrial DNA Part B Resour. 2022, 7, 933–935. [Google Scholar] [CrossRef]
- Idogawa, N.; Lee, C.C.; Yang, C.S.; Dobata, S. The complete mitochondrial genome of a parthenogenetic ant Monomorium triviale (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2021, 6, 2793–2795. [Google Scholar] [CrossRef] [PubMed]
- Ströher, P.R.; Zarza, E.; Tsai, W.L.E.; McCormack, J.E.; Feitosa, R.M.; Pie, M.R. The mitochondrial genome of Octostruma stenognatha and its phylogenetic implications. Insectes Sociaux 2017, 64, 149–154. [Google Scholar] [CrossRef]
- Sang, Y.; Yin, R.Y.; Luo, Y.; Zhou, Z.M. The complete mitochondrial genome of Pheidole nodus (Smith, 1874) (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2022, 7, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, E.; Kobayashi, K.; Yagi, N.; Tsuji, K. Complete mitochondrial genomes of normal and cheater morphs in the parthenogenetic ant Pristomyrmex punctatus (Hymenoptera: Formicidae). Myrmecol. News 2011, 15, 85–90. [Google Scholar]
- Yin, R.Y.; Luo, Y.; Zhou, Z.M. The complete mitochondrial genome of Tetramorium tsushimae (Emery, 1925) (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2021, 7, 40–42. [Google Scholar] [CrossRef]
- Liu, N.; Duan, X.Y.; Qian, Z.Q.; Wang, X.Y.; Li, X.L.; Ding, M.Y. Characterization of the complete mitochondrial genome of the myrmicine ant Vollenhovia emeryi (Insecta: Hymenoptera: Formicidae). Conserv. Genet. Resour. 2016, 8, 211–214. [Google Scholar] [CrossRef]
- McKenzie, S.K.; Kronauer, D.J.C. The genomic architecture and molecular evolution of ant odorant receptors. Genome Res. 2018, 28, 1757–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kwon, W.; Park, J. The complete mitochondrial genome of Cryptopone sauteri Wheeler, W.M., 1906 (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2019, 4, 614–615. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kwon, W.; Park, J. The complete mitochondrial genome of Ectomomyrmex javanus Mayr, 1867 (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2019, 4, 1636–1637. [Google Scholar] [CrossRef]
- Vieira, G.A.; Prosdocimi, F. Accessible molecular phylogenomics at no cost: Obtaining 14 new mitogenomes for the ant subfamily Pseudomyrmecinae from public data. PeerJ 2019, 7, e6271. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kwon, W.; Park, J. The complete mitochondrial genome of Camponotus concavus Kim & Kim, 1994 (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2019, 4, 1243–1244. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Hong, E.J.; Kim, I. Complete mitochondrial genome of Camponotus atrox (Hymenoptera: Formicidae): A new tRNA arrangement in Hymenoptera. Genome 2016, 59, 59–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Xi, H.; Park, J. The complete mitochondrial genome of Nylanderia flavipes (Smith, 1874) (Hymenoptera: Formicidae). Mitochondrial DNA Part B Resour. 2020, 5, 420–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Species | Genome Size (bp) | Origen Country | Accession Number | Reference |
|---|---|---|---|---|
| Dolichoderus lamellosus | 16,234 | Costa Rica | BK012125 | [24] |
| Dolichoderus pustulatus | 16,224 | Canada | BK012668 | [24] |
| Dolichoderus quadripunctatus | 16,017 | Poland | MT178447 | [15] |
| Dolichoderus sibiricus | 16,086 | South Korea | MH719017 | [25] |
| 16,044 | South Korea | MK801110 | [15] | |
| 16,067 | Russia | MT919976 | unpublished | |
| 16,110 | Taiwan | MW160468 | unpublished | |
| Dorymyrmex brunneus | 15,848 | MG253267 | unpublished | |
| Leptomyrmex erythrocephalus | 15,546 | Australia | BK012481 | [24] |
| Leptomyrmex pallens | 15,591 | New Calcedonia | KC160533 | [26] |
| Linepithema humile | 16,098 | USA | KT428891 | [27] |
| 15,929 | KX146468 | [20] | ||
| 15,934 | South Korea | MT890564 | [16] | |
| Ochetellus glaber | 16,259 | South Korea | MN044390 | [28] |
| Tapinoma ibericum | 15,715 | Spain | ON746721 | This study |
| Tapinoma melanocephalum | 15,499 | China | MN397938 | [29] |
| Tapinoma sessile | 15,287 | USA | BK012786 | [24] |
| Gene | Anticodon | Strand | Nucleotide Number | Start Codon | Stop Codon | IGN |
|---|---|---|---|---|---|---|
| (M) tRNA-Met | CAU | H | 1–68 | 3 | ||
| (I) tRNA-Ile | GAU | H | 72–137 | −3 | ||
| (Q) tRNA-Gln | UUG | L | 135–203 | 104 | ||
| nad2 | H | 308–1291 | ATA | TAA | 1 | |
| (W) tRNA-Trp | UCA | H | 1293–1366 | 32 | ||
| (Y) tRNA-Tyr | GUA | L | 1399–1467 | 56 | ||
| (C) tRNA-Cys | GCA | L | 1501–1567 | 21 | ||
| cox1 | H | 1589–3118 | ATG | TAA | 0 | |
| (L1) tRNA-Leu | UAA | H | 3119–3184 | 0 | ||
| cox2 | H | 3185–3874 | ATT | TAA | 24 | |
| (K) tRNA-Lys | UUU | H | 3899–3971 | 0 | ||
| (D) tRNA-Asp | GUC | H | 3972–4040 | 69 | ||
| atp8 | H | 4041–4202 | ATC | TAA | −7 | |
| atp6 | H | 4196–4864 | ATG | TAA | 6 | |
| cox3 | H | 4871–5653 | ATG | TAA | 74 | |
| (G) tRNA-Gly | UCC | H | 5728–5795 | 0 | ||
| nad3 | H | 5796–6146 | ATA | TAA | 37 | |
| (A) tRNA-Ala | UGC | H | 6184–6256 | 22 | ||
| (R) tRNA-Arg | UCG | H | 6279–6354 | 16 | ||
| (N) tRNA-Asn | GUU | H | 6371–6439 | 31 | ||
| (S1) tRNA-Ser | UCU | H | 6471–6533 | 20 | ||
| (E) tRNA-Glu | UUC | H | 6554–6624 | 76 | ||
| (F) tRNA-Phe | GAA | L | 6701–6772 | 0 | ||
| nad5 | L | 6773–8440 | ATA | TAA | 0 | |
| (H) tRNA-His | GUG | L | 8441–8509 | 37 | ||
| nad4 | L | 8547–9887 | ATG | TAA | 5 | |
| nad4l | L | 9893–10,180 | ATT | TAA | 10 | |
| (T) tRNA-Thr | UGU | H | 10,191–10,261 | 8 | ||
| (P) tRNA-Pro | UGG | L | 10,270–10,341 | 6 | ||
| nad6 | H | 10,348–10,893 | ATG | TAA | 23 | |
| cob | H | 10,917–12,038 | ATG | TAA | 10 | |
| (S2) tRNA-Ser | UGA | H | 12,049–12,115 | 31 | ||
| nad1 | L | 12,147–13,094 | ATT | TAA | 0 | |
| (L2) tRNA-Leu | UAG | L | 13,095–13,166 | 0 | ||
| lrRNA | L | 13,167–14,511 | 0 | |||
| (V) tRNA-Val | UAC | L | 14,512–14,583 | 0 | ||
| srRNA | L | 14,584–15,374 | 0 | |||
| Control Region | 15,375–15,715 |
| Codon | n | % | RSCU | Codon | n | % | RSCU |
|---|---|---|---|---|---|---|---|
| UUU(F) | 368 | 1.91 | 9.96 | UAU(Y) | 198 | 5.36 | 1.82 |
| UUC(F) | 18 | 0.09 | 0.49 | UAC(Y) | 20 | 0.54 | 0.18 |
| UUA(L) | 443 | 5.13 | 11.99 | UAA(*) | 13 | 0.35 | 2 |
| UUG(L) | 13 | 0.15 | 0.35 | UAG(*) | 0 | 0 | 0 |
| CUU(L) | 31 | 0.36 | 0.84 | CAU(H) | 47 | 1.27 | 1.52 |
| CUC(L) | 2 | 0.02 | 0.05 | CAC(H) | 15 | 0.41 | 0.48 |
| CUA(L) | 29 | 0.34 | 0.78 | CAA(Q) | 42 | 1.14 | 1.91 |
| CUG(L) | 0 | 0 | 0 | CAG(Q) | 2 | 0.05 | 0.09 |
| AUU(I) | 490 | 1.91 | 13.26 | AAU(N) | 220 | 5.95 | 1.86 |
| AUC(I) | 22 | 0.09 | 0.60 | AAC(N) | 17 | 0.46 | 0.14 |
| AUA(M) | 347 | 1.88 | 9.39 | AAA(K) | 112 | 3.03 | 1.9 |
| AUG(M) | 23 | 0.12 | 0.62 | AAG(K) | 6 | 0.16 | 0.1 |
| GUU(V) | 63 | 2.12 | 1.71 | GAU(D) | 48 | 1.30 | 1.6 |
| GUC(V) | 7 | 0.24 | 0.19 | GAC(D) | 12 | 0.32 | 0.4 |
| GUA(V) | 45 | 1.51 | 1.22 | GAA(E) | 62 | 1.68 | 1.8 |
| GUG(V) | 4 | 0.13 | 0.11 | GAG(E) | 7 | 0.19 | 0.2 |
| UCU(S) | 116 | 3.14 | 2.75 | UGU(C) | 31 | 0.84 | 1.82 |
| UCC(S) | 11 | 0.30 | 0.26 | UGC(C) | 3 | 0.08 | 0.18 |
| UCA(S) | 98 | 2.65 | 2.32 | UGA(W) | 80 | 2.17 | 1.95 |
| UCG(S) | 1 | 0.03 | 0.02 | UGG(W) | 2 | 0.05 | 0.05 |
| CCU(P) | 54 | 1.46 | 1.79 | CGU(R) | 15 | 0.41 | 1.4 |
| CCC(P) | 12 | 0.32 | 0.4 | CGC(R) | 0 | 0 | 0 |
| CCA(P) | 51 | 1.38 | 1.69 | CGA(R) | 25 | 0.68 | 2.33 |
| CCG(P) | 4 | 0.11 | 0.13 | CGG(R) | 3 | 0.08 | 0.28 |
| ACU(T) | 64 | 1.73 | 2.02 | AGU(S) | 20 | 0.54 | 0.47 |
| ACC(T) | 4 | 0.11 | 0.13 | AGC(S) | 4 | 0.11 | 0.09 |
| ACA(T) | 58 | 1.57 | 1.83 | AGA(S) | 75 | 2.03 | 1.78 |
| ACG(T) | 1 | 0.03 | 0.03 | AGG(S) | 13 | 0.35 | 0.31 |
| GCU(A) | 50 | 1.35 | 2.67 | GGU(G) | 25 | 0.68 | 0.67 |
| GCC(A) | 3 | 0.08 | 0.16 | GGC(G) | 8 | 0.22 | 0.21 |
| GCA(A) | 21 | 0.57 | 1.12 | GGA(G) | 89 | 2.41 | 2.39 |
| GCG(A) | 1 | 0.03 | 0.05 | GGG(G) | 27 | 0.73 | 0.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Mena, A.; Mora, P.; Montiel, E.E.; Palomeque, T.; Lorite, P. Complete Nucleotide Sequence of the Mitogenome of Tapinoma ibericum (Hymenoptera: Formicidae: Dolichoderinae), Gene Organization and Phylogenetics Implications for the Dolichoderinae Subfamily. Genes 2022, 13, 1325. https://doi.org/10.3390/genes13081325
Ruiz-Mena A, Mora P, Montiel EE, Palomeque T, Lorite P. Complete Nucleotide Sequence of the Mitogenome of Tapinoma ibericum (Hymenoptera: Formicidae: Dolichoderinae), Gene Organization and Phylogenetics Implications for the Dolichoderinae Subfamily. Genes. 2022; 13(8):1325. https://doi.org/10.3390/genes13081325
Chicago/Turabian StyleRuiz-Mena, Areli, Pablo Mora, Eugenia E. Montiel, Teresa Palomeque, and Pedro Lorite. 2022. "Complete Nucleotide Sequence of the Mitogenome of Tapinoma ibericum (Hymenoptera: Formicidae: Dolichoderinae), Gene Organization and Phylogenetics Implications for the Dolichoderinae Subfamily" Genes 13, no. 8: 1325. https://doi.org/10.3390/genes13081325
APA StyleRuiz-Mena, A., Mora, P., Montiel, E. E., Palomeque, T., & Lorite, P. (2022). Complete Nucleotide Sequence of the Mitogenome of Tapinoma ibericum (Hymenoptera: Formicidae: Dolichoderinae), Gene Organization and Phylogenetics Implications for the Dolichoderinae Subfamily. Genes, 13(8), 1325. https://doi.org/10.3390/genes13081325






