Comprehensive Analysis of Differentially Expressed mRNAs, lncRNAs and circRNAs Related to Intramuscular Fat Deposition in Laiwu Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Samples Preparation
2.2. RNA Preparation, Library Construction and Sequencing
2.3. Sequencing Data Quality Control, Transcriptome Assembly and Quantification of Gene Expression Level
2.4. lncRNAs Identification and Their Target Genes’s Prediction
2.5. Identification of circRNAs and Analysis of circRNA-miRNA Binding Sites
2.6. Functional Enrichment Analysis of DEGs, Target Genes of DELs and Source Genes of DECs
2.7. Verification of Sequencing Data Using Reverse Transcription Quantitative PCR (RT-qPCR)
3. Results
3.1. IMF Content Detecting and Screening of Samples
3.2. Overview of Sequencing Data
3.3. Predictions and Analysis of mRNAs, lncRNAs and circRNAs
3.4. Differential Expression of mRNAs, lncRNAs and circRNAs
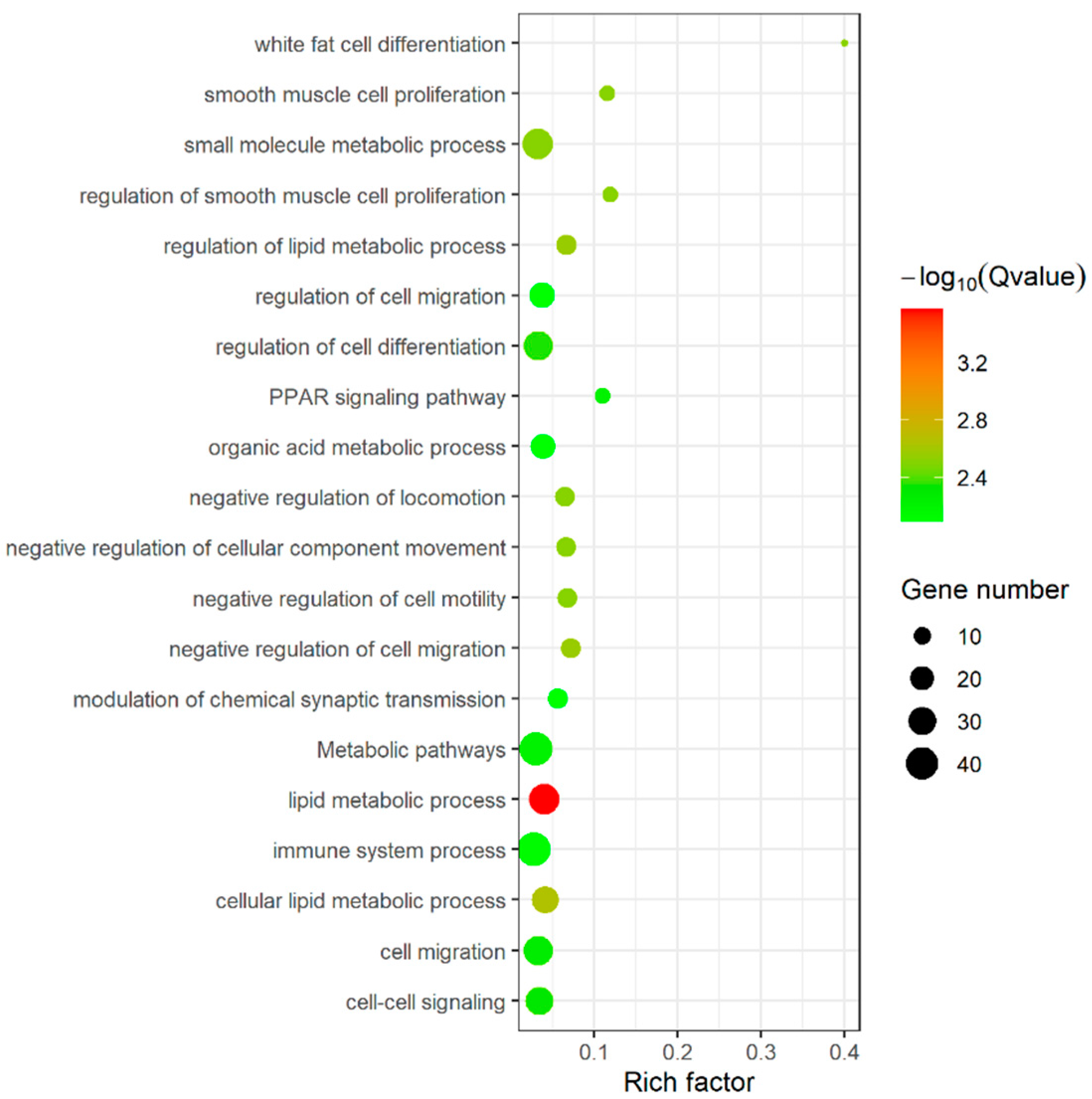
3.5. Function Enrichment Analysis of DEGs, DELs and DECs
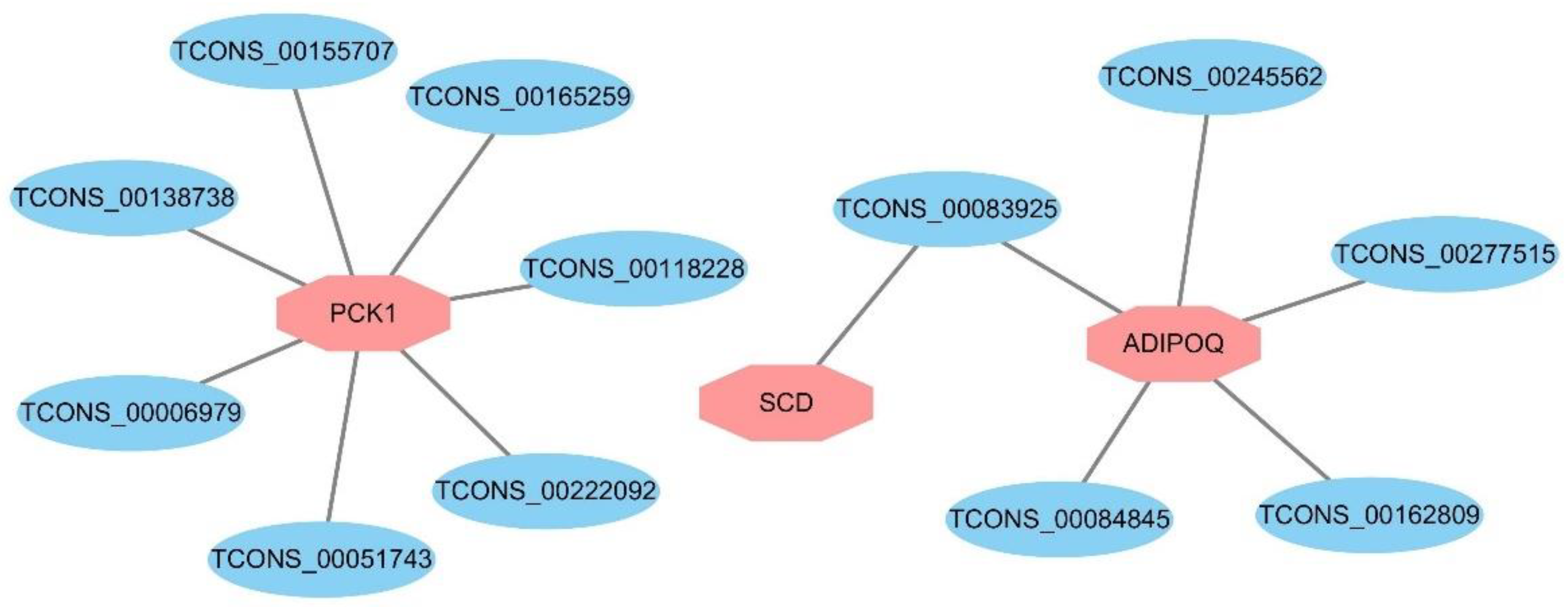
3.6. Expression Regulation Analysis of DELs and Target Genes
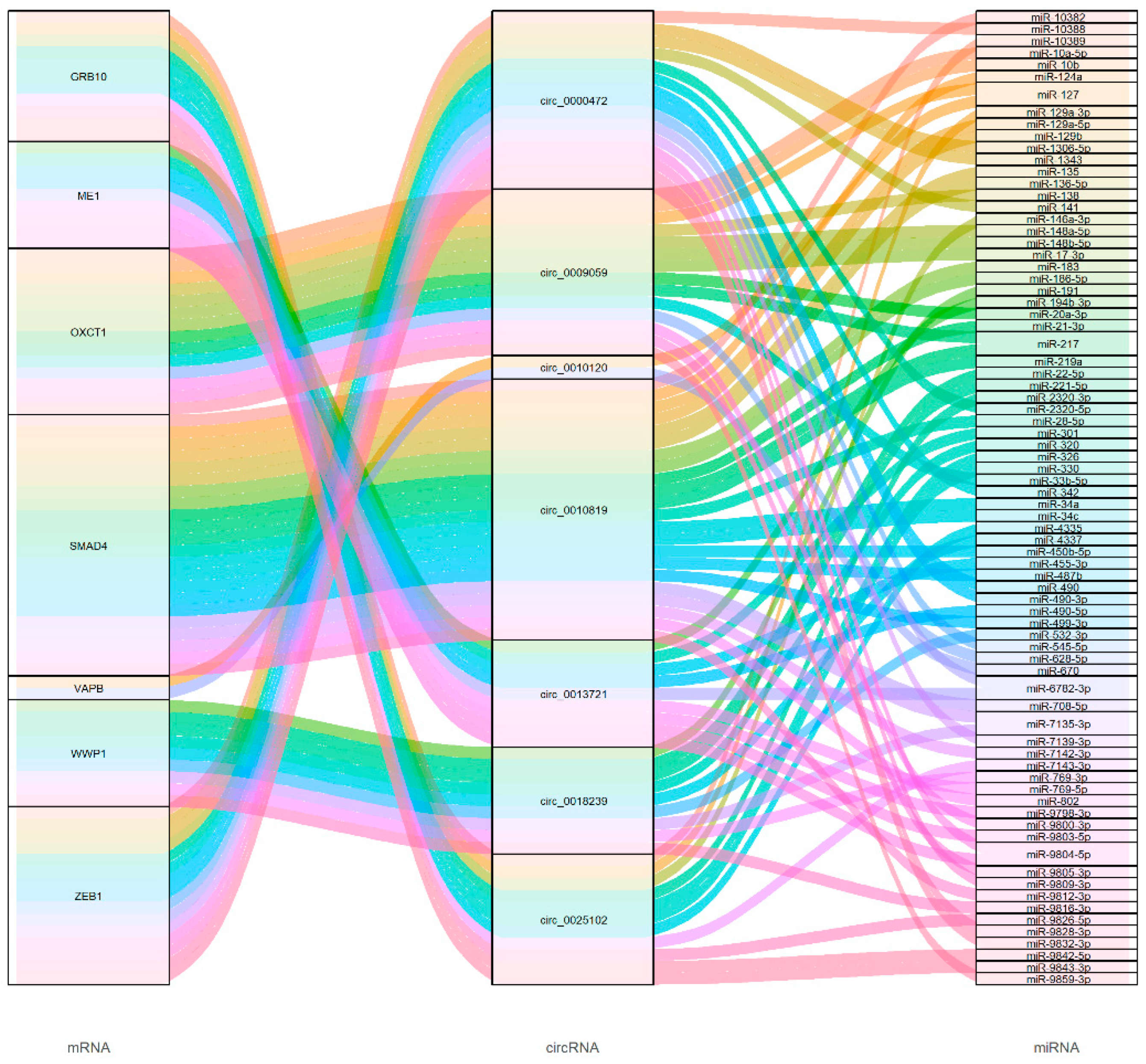
3.7. CircRNA Functional Predictions
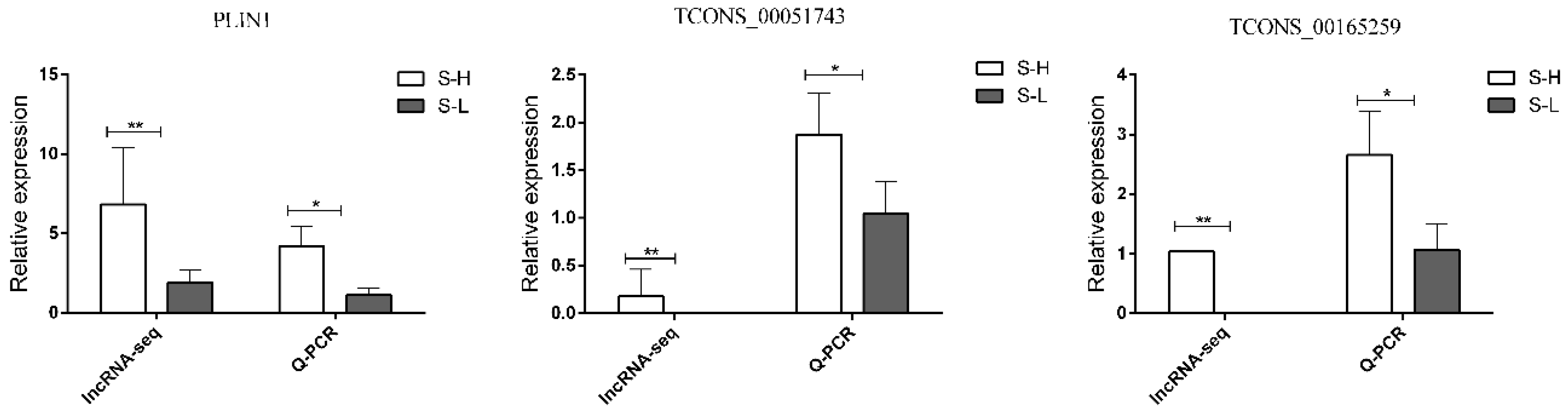
3.8. Validation of DEGs and DELs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| IMF | intramuscular fat |
| lncRNAs | long noncoding RNAs |
| circRNAs | circular RNAs |
| LD | longissimus dorsi |
| DEGs | differentially expressed protein-coding genes |
| DELs | differentially expressed lncRNAs |
| DECs | differentially expressed circRNAs |
| GO | gene ontology |
| KEGG | kyoto encyclopedia of genes and genomes |
| CT | cycle threshold |
| SCD | stearoyl-CoA desaturase |
| PCK1 | phosphoenolpyruvate carboxykinase 1 |
| ADIPOQ | adiponectin |
| FABP4 | fatty acid binding protein 4 |
| ACAA1 | acetyl-coa acyltransferase 1 |
| PLIN1 | perilipin 1 |
| PPARG | peroxisome proliferator activated receptor γ |
| PPAR | peroxisome proliferator-activated receptors |
| AMPK | adenosine 5′-monophosphate (AMP)-activated protein kinase |
| Jak-STAT | janus kinase-signal transducers and activators of transcription |
| PI3K/AKT/mTOR | phosphatidylinositol-3-kinase/protein kinase B/mechanistic target of rapamycin |
References
- Grindflek, E.; Szyda, J.; Liu, Z.; Lien, S. Detection of quantitative trait loci for meat quality in a commercial slaughter pig cross. Mamm. Genome 2001, 12, 299–304. [Google Scholar] [CrossRef]
- Ma, J.; Ren, J.; Guo, Y.; Duan, Y.; Ding, N.; Zhou, L.; Li, L.; Yan, X.; Yang, K.; Huang, L.; et al. Genome-wide identification of quantitative trait loci for carcass composition and meat quality in a large-scale White Duroc x Chinese Erhualian resource population. Anim. Genet. 2009, 40, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hu, H.; Lin, H.; Wang, C.; Wang, Y.; Wang, J. Muscle transcriptome analysis reveals potential candidate genes and pathways affecting intramuscular fat content in pigs. Front. Genet. 2020, 11, 877. [Google Scholar] [CrossRef]
- Zhen, Q.; Gao, L.N.; Wang, R.F.; Chu, W.W.; Zhang, Y.X.; Zhao, X.J.; Lv, B.L.; Liu, J.B. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control 2018, 25, 1073274818769849. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Y.; Zhang, Y.; Xia, Y.; Zhao, H.; Liu, A.; Chen, Y. LncRNA MEG3 targeting miR-424-5p via MAPK signaling pathway mediates neuronal apoptosis in ischemic stroke. Aging 2020, 12, 3156–3174. [Google Scholar] [CrossRef]
- Luo, H.; Xu, C.; Le, W.; Ge, B.; Wang, T. lncRNA CASC11 promotes cancer cell proliferation in bladder cancer through miRNA-150. J. Cell. Biochem. 2019, 120, 13487–13493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Shan, G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 2021, 505, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Zhang, X.; Odame, E.; Xu, X.; Chen, Y.; Ye, J.; Zhou, H.; Dai, D.; Kyei, B.; Zhan, S.; et al. CircRNA-protein interactions in muscle development and diseases. Int. J. Mol. Sci. 2021, 22, 3262. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lian, C.; Chen, G.; Zou, P.; Qin, B.G. CircRNA FUT10 regulates the regenerative potential of aged skeletal muscle stem cells by targeting HOXA9. Aging 2021, 13, 17428–17441. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Yi, X.; He, Z.; Tian, T.; Kou, Z.; Pang, W. LncIMF2 promotes adipogenesis in porcine intramuscular preadipocyte through sponging MiR-217. Anim. Biotechnol. 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhou, Z.Y.; Zhang, T.; Zhang, L.; Hou, X.; Yan, H.; Wang, L. IRLnc: A novel functional noncoding RNA contributes to intramuscular fat deposition. BMC Genom. 2021, 22, 95. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Tang, G.; Zhang, Q.; Yong, W.; Zhang, W.; Xiao, J.; Wei, C.; He, C.; Yang, G.; Pang, W. A novel lnc-RNA, named lnc-ORA, is identified by RNA-Seq analysis, and its knockdown inhibits adipogenesis by regulating the PI3K/AKT/mTOR signaling pathway. Cells 2019, 8, 477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, H.; Li, Y.; Mao, R.; Yang, H.; Zhang, Y.; Zhang, Y.; Guo, P.; Zhan, D.; Zhang, T. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 2020, 10, 4705–4719. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Tang, J.; Ru, W.; Zhang, X.; Huang, Y.; Lei, C.; Cao, H.; Lan, X.; Chen, H. CircINSR regulates fetal bovine muscle and fat development. Front. Cell Dev. Biol. 2020, 8, 615638. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, S.; Jiang, E.; Wang, X.; Wang, Z.; Chen, H.; Lan, X. circFLT1 and lncCCPG1 sponges miR-93 to regulate the proliferation and differentiation of adipocytes by promoting lncSLC30A9 expression. Mol. Ther. Nucleic Acids 2020, 22, 484–499. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, X.; Li, A.; Xie, L.; Miao, X. Differential regulation of mRNAs and lncRNAs related to lipid metabolism in two pig breeds. Oncotarget 2017, 8, 87539–87553. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zhou, L.; Zhang, J.; Liu, X.; Zhang, Y.; Cai, L.; Zhang, W.; Cui, L.; Yang, J.; Ji, J.; et al. A large-scale comparison of meat quality and intramuscular fatty acid composition among three Chinese indigenous pig breeds. Meat Sci. 2020, 168, 108182. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq data using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar] [PubMed]
- Sharma, D.; Sehgal, P.; Hariprakash, J.; Sivasubbu, S.; Scaria, V. Methods for annotation and validation of circular RNAs from RNAseq data. Methods Mol. Biol. 2019, 1912, 55–76. [Google Scholar] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Ren, Q.; Xu, Z.L.; Wang, X.W.; Zhao, X.F.; Wang, J.X. Clip domain serine protease and its homolog respond to Vibrio challenge in Chinese white shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. 2009, 26, 787–798. [Google Scholar] [CrossRef]
- Richard, A.J.; Stephens, J.M. Emerging roles of JAK-STAT signaling pathways in adipocytes. Trends Endocrinol. Metab. 2011, 22, 325–332. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Xue, W.; Jin, B.; Zhang, X.; Ma, F.; Xu, X. Candidate gene expression affects intramuscular fat content and fatty acid composition in pigs. J. Appl. Genet. 2013, 54, 113–118. [Google Scholar] [CrossRef]
- Reardon, W.; Mullen, A.M.; Sweeney, T.; Hamill, R.M. Association of polymorphisms in candidate genes with colour, water-holding capacity, and composition traits in bovine M. longissimus and M. semimembranosus. Meat Sci. 2010, 86, 270–275. [Google Scholar] [CrossRef]
- Fortin, A.; Robertson, W.M.; Tong, A.K. The eating quality of Canadian pork and its relationship with intramuscular fat. Meat Sci. 2005, 69, 297–305. [Google Scholar] [CrossRef]
- Huff-Lonergan, E.; Baas, T.J.; Malek, M.; Dekkers, J.C.; Prusa, K.; Rothschild, M.F. Correlations among selected pork quality traits. J. Anim. Sci. 2002, 80, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasturi, R.; Joshi, V.C. Hormonal regulation of stearoyl coenzyme A desaturase activity and lipogenesis during adipose conversion of 3T3-L1 cells. J. Biol. Chem. 1982, 257, 12224–12230. [Google Scholar] [CrossRef]
- Tan, Z.; Du, J.; Shen, L.; Liu, C.; Ma, J.; Bai, L.; Jiang, Y.; Tang, G.; Li, M.; Li, X.; et al. miR-199a-3p affects adipocytes differentiation and fatty acid composition through targeting SCD. Biochem. Biophys. Res. Commun. 2017, 492, 82–88. [Google Scholar] [CrossRef]
- Garin-Shkolnik, T.; Rudich, A.; Hotamisligil, G.S.; Rubinstein, M. FABP4 attenuates PPARgamma and adipogenesis and is inversely correlated with PPARgamma in adipose tissues. Diabetes 2014, 63, 900–911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Calvo, R.; Girona, J.; Rodriguez, M.; Samino, S.; Barroso, E.; de Gonzalo-Calvo, D.; Guaita-Esteruelas, S.; Heras, M.; van der Meer, R.W.; Lamb, H.J.; et al. Fatty acid binding protein 4 (FABP4) as a potential biomarker reflecting myocardial lipid storage in type 2 diabetes. Metabolism 2019, 96, 12–21. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Cao, Y.; Xiao, C.; Liu, Y.; Jin, H.; Cao, Y. Effect of the ACAA1 gene on preadipocyte differentiation in sheep. Front. Genet. 2021, 12, 649140. [Google Scholar] [CrossRef]
- Li, B.; Weng, Q.; Dong, C.; Zhang, Z.; Li, R.; Liu, J.; Jiang, A.; Li, Q.; Jia, C.; Wu, W.; et al. A key gene, PLIN1, can affect porcine intramuscular fat content based on transcriptome analysis. Genes 2018, 9, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Li, F.; Sun, J.W.; Li, D.H.; Li, W.T.; Jiang, R.R.; Li, Z.J.; Liu, X.J.; Han, R.L.; Li, G.X.; et al. LncRNA IMFNCR promotes intramuscular adipocyte differentiation by sponging miR-128-3p and miR-27b-3p. Front. Genet. 2019, 10, 42. [Google Scholar] [CrossRef]
- Liu, W.; Ma, C.; Yang, B.; Yin, C.; Zhang, B.; Xiao, Y. LncRNA Gm15290 sponges miR-27b to promote PPARgamma-induced fat deposition and contribute to body weight gain in mice. Biochem. Biophys. Res. Commun. 2017, 493, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [Green Version]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Mu, Y.; Ma, L.; Wang, C.; Tang, Z.; Yang, S.; Zhou, R.; Hu, X.; Li, M.H.; Li, K. Systematic identification and characterization of long intergenic non-coding RNAs in fetal porcine skeletal muscle development. Sci. Rep. 2015, 5, 8957. [Google Scholar] [CrossRef] [Green Version]
- Bertone, P.; Stolc, V.; Royce, T.E.; Rozowsky, J.S.; Urban, A.E.; Zhu, X.; Rinn, J.L.; Tongprasit, W.; Samanta, M.; Weissman, S.; et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004, 306, 2242–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponjavic, J.; Ponting, C.P.; Lunter, G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007, 17, 556–565. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.Q.; Zhu, L.; Ma, M.L.; Chen, X.C.; Gao, Y.; Yu, T.Y.; Yang, G.S.; Pang, W.J. Mulberry 1-deoxynojirimycin inhibits adipogenesis by repression of the ERK/PPARgamma signaling pathway in porcine intramuscular adipocytes. J. Agric. Food Chem. 2015, 63, 6212–6220. [Google Scholar] [CrossRef]
- Huang, J.; Feng, X.; Zhu, R.; Guo, D.; Wei, Y.; Cao, X.; Ma, Y.; Shi, D. Comparative transcriptome analysis reveals that PCK1 is a potential gene affecting IMF deposition in buffalo. BMC Genom. 2020, 21, 710. [Google Scholar] [CrossRef]
- Qiao, L.; Yoo, H.S.; Madon, A.; Kinney, B.; Hay, W.W., Jr.; Shao, J. Adiponectin enhances mouse fetal fat deposition. Diabetes 2012, 61, 3199–3207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Xie, M.; Huang, Z.; Li, H.; Chen, T.; Sun, R.; Wang, J.; Xi, Q.; Wu, T.; Zhang, Y. Integrated analysis of non-coding RNA and mRNA expression profiles of 2 pig breeds differing in muscle traits. J. Anim. Sci 2017, 95, 1092–1103. [Google Scholar] [CrossRef]
- Liu, X.; Wei, S.; Deng, S.; Li, D.; Liu, K.; Shan, B.; Shao, Y.; Wei, W.; Chen, J.; Zhang, L. Genome-wide identification and comparison of mRNAs, lncRNAs and circRNAs in porcine intramuscular, subcutaneous, retroperitoneal and mesenteric adipose tissues. Anim. Genet. 2019, 50, 228–241. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Liang, W.; Wang, S.; Wang, Z.; Bai, H.; Jiang, Y.; Bi, Y.; Chen, G.; Chang, G. Circular RNA expression profiling reveals that circ-PLXNA1 functions in duck adipocyte differentiation. PLoS ONE 2020, 15, e0236069. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bai, Y.; Cui, R.; He, S.; Zhao, X.; Wu, K.; Fang, M. Sus_circPAPPA2 regulates fat deposition in castrated pigs through the miR-2366/GK pathway. Biomolecules 2022, 12, 753. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, Y.; Chen, X.; Peng, Y.; Chen, F.; He, Y.; Pang, W.; Yang, G.; Yu, T. MiR-127 attenuates adipogenesis by targeting MAPK4 and HOXC6 in porcine adipocytes. J. Cell. Physiol. 2019, 234, 21838–21850. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, H.; Li, J.; Chen, Y.; Cao, J.; Zhong, T.; Wang, L.; Guo, J.; Li, L.; Zhang, H. MiR-183 promotes preadipocyte differentiation by suppressing Smad4 in goats. Gene 2018, 666, 158–164. [Google Scholar] [CrossRef] [PubMed]


| Group | Samples | Carcass Weight (kg) | Average (kg) | p Value of t-Test | IMF Content (%) | Average | p Value of t-Test |
|---|---|---|---|---|---|---|---|
| High IMF content group | LW63 | 77.7 | 74.6 | 0.1428 | 18.85 | 16.66 | 0.0032 |
| LW70 | 71.6 | 14.38 | |||||
| LW119 | 74.6 | 16.74 | |||||
| Low IMF content group | LW82 | 66.4 | 70.0 | 5.90 | 4.59 | ||
| LW88 | 72.5 | 3.39 | |||||
| LW90 | 71.0 | 4.49 |
| Samples | Raw Reads | Clean Reads | Error Rate (%) | Total Mapped (%) | Uniquely Mapped (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|---|
| LW119 | 99403460 | 97533328 | 0.02 | 95.11% | 86.22% | 98.11 | 94.66 | 51.26 |
| W63 | 86027762 | 84512732 | 0.03 | 94.65% | 83.13% | 97.97 | 94.31 | 53.01 |
| W70 | 83537954 | 82059898 | 0.02 | 94.67% | 84.25% | 98.05 | 94.51 | 53.27 |
| LW82 | 89852982 | 87838194 | 0.03 | 94.26% | 83.94% | 97.88 | 94.11 | 51.55 |
| LW88 | 78555518 | 77268288 | 0.02 | 92.08% | 80.88% | 97.96 | 94.39 | 53.68 |
| LW90 | 99854922 | 98262148 | 0.02 | 94.72% | 84.39% | 98.07 | 94.58 | 52.47 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhao, X.; Wang, Y.; Wang, J. Comprehensive Analysis of Differentially Expressed mRNAs, lncRNAs and circRNAs Related to Intramuscular Fat Deposition in Laiwu Pigs. Genes 2022, 13, 1349. https://doi.org/10.3390/genes13081349
Li J, Zhao X, Wang Y, Wang J. Comprehensive Analysis of Differentially Expressed mRNAs, lncRNAs and circRNAs Related to Intramuscular Fat Deposition in Laiwu Pigs. Genes. 2022; 13(8):1349. https://doi.org/10.3390/genes13081349
Chicago/Turabian StyleLi, Jingxuan, Xueyan Zhao, Yanping Wang, and Jiying Wang. 2022. "Comprehensive Analysis of Differentially Expressed mRNAs, lncRNAs and circRNAs Related to Intramuscular Fat Deposition in Laiwu Pigs" Genes 13, no. 8: 1349. https://doi.org/10.3390/genes13081349
APA StyleLi, J., Zhao, X., Wang, Y., & Wang, J. (2022). Comprehensive Analysis of Differentially Expressed mRNAs, lncRNAs and circRNAs Related to Intramuscular Fat Deposition in Laiwu Pigs. Genes, 13(8), 1349. https://doi.org/10.3390/genes13081349





