Identification of Heat-Tolerant Genes in Non-Reference Sequences in Rice by Integrating Pan-Genome, Transcriptomics, and QTLs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Pan-Genome Assembly and Annotation
2.3. Gene Presence/Absence Variation and Pan-Genome Modeling
2.4. Linking the Known Heat-Resistant QTLs with the Predicted Genes
2.5. Processing of RNA-seq Datasets
2.6. SNP Discovery and Annotation
3. Results
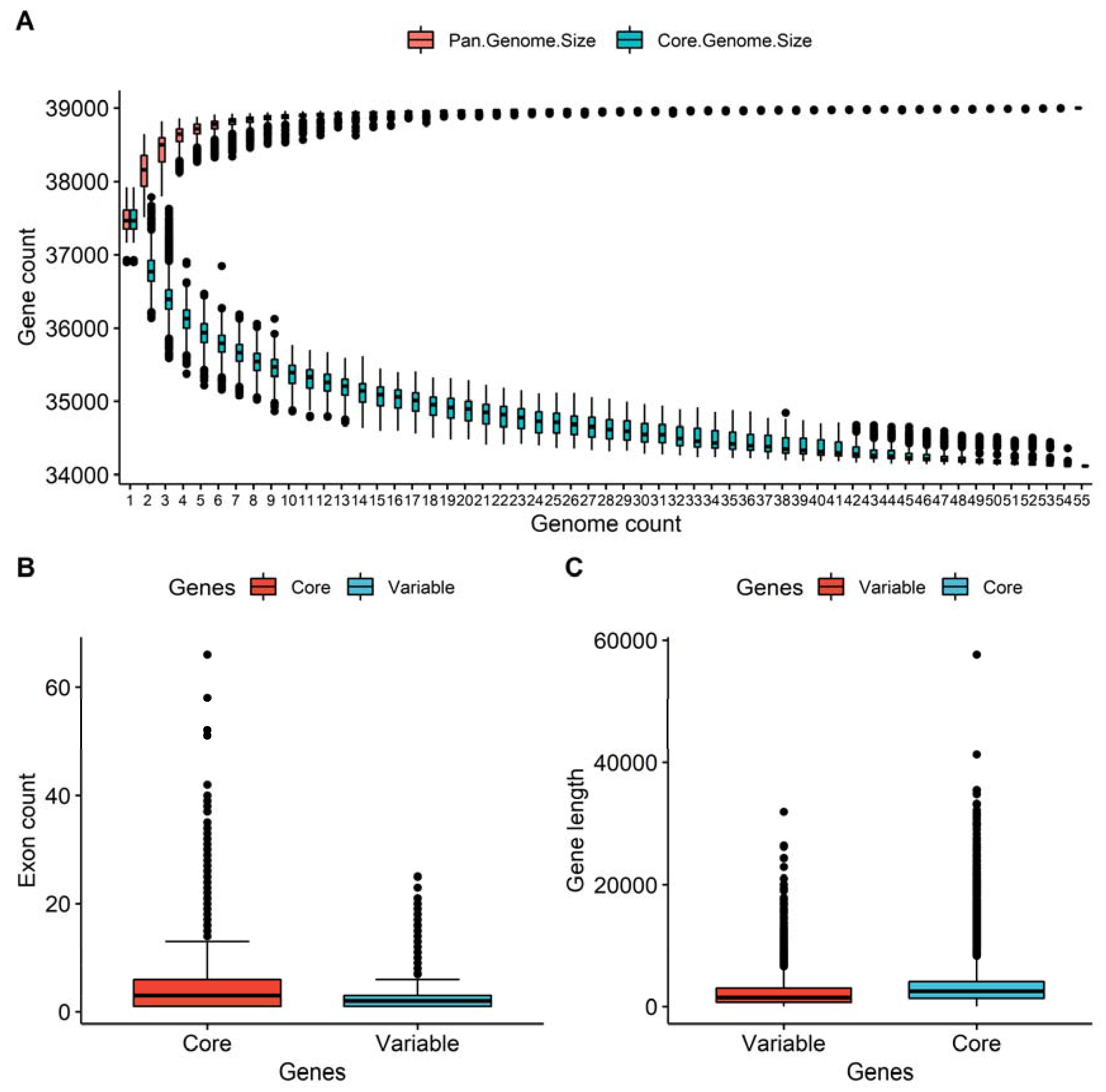
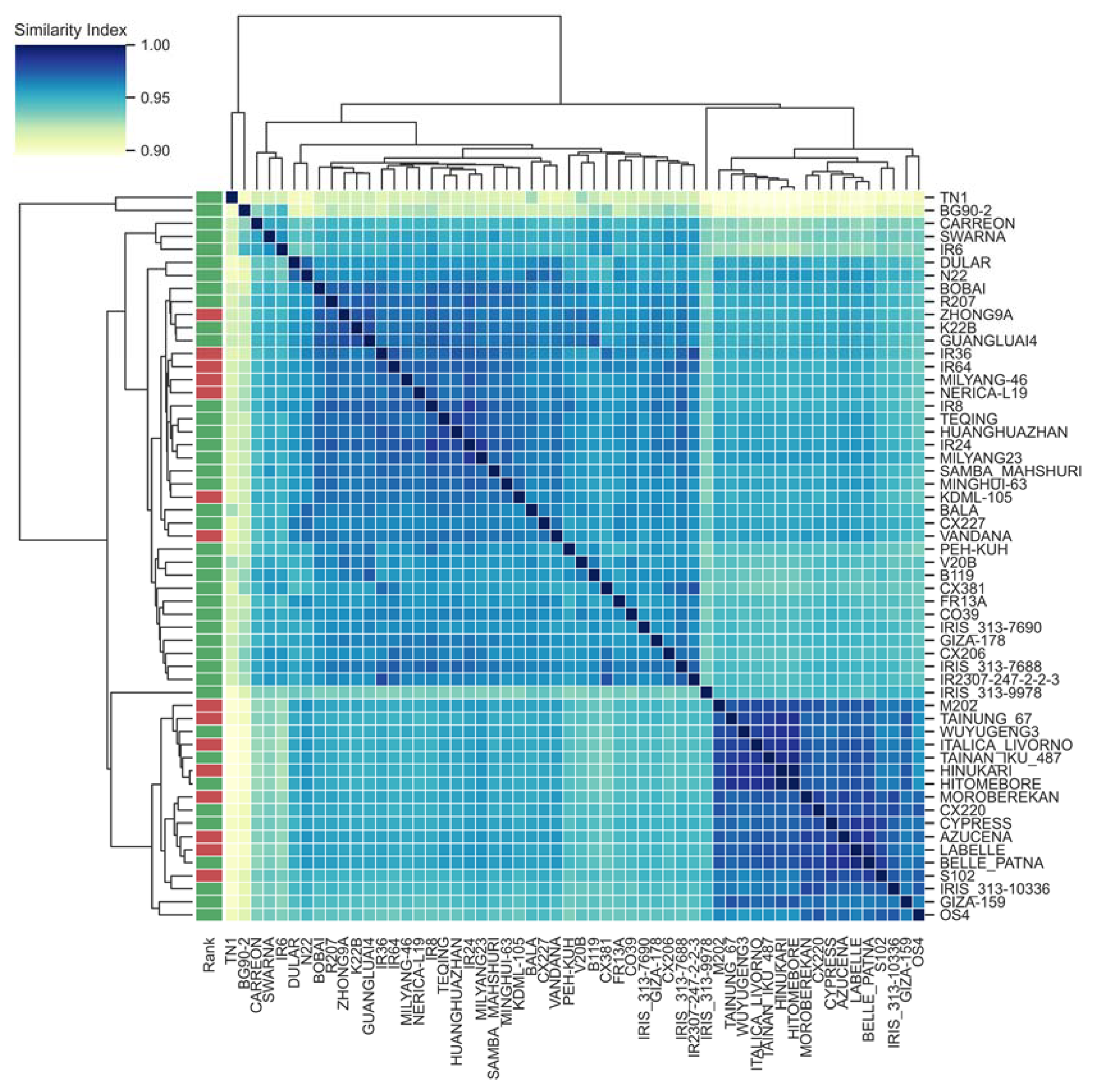
3.1. Pan-Genome Assembly and Annotation
3.2. Core and Variable Genes in the Pan-Genome
3.3. Functional Annotation of Genes
3.4. SNP Analysis
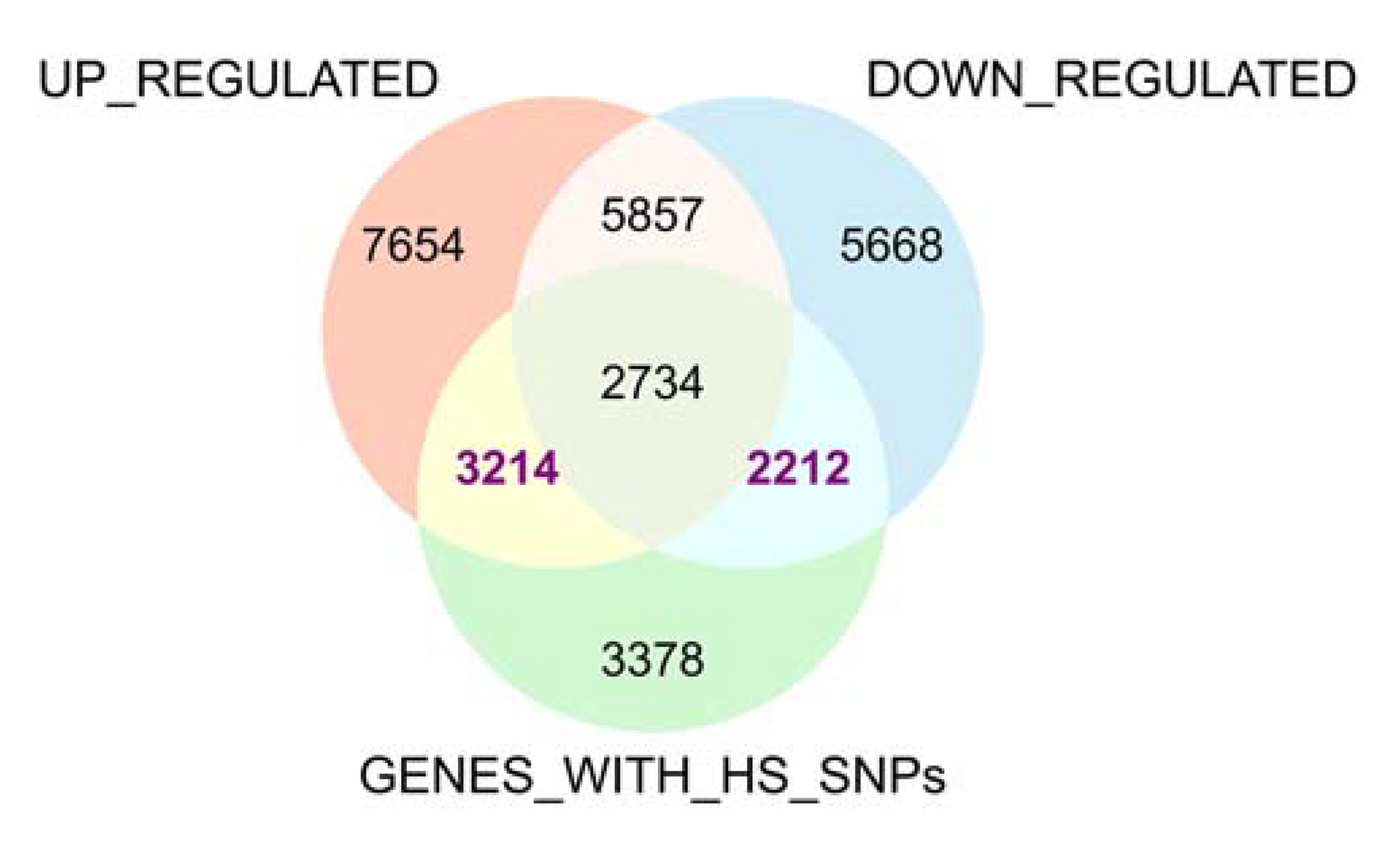
3.5. Identification of Heat Stress Tolerance-Related SNPs and Genes
3.6. Meta-Analysis of Comparative Transcriptomic Data
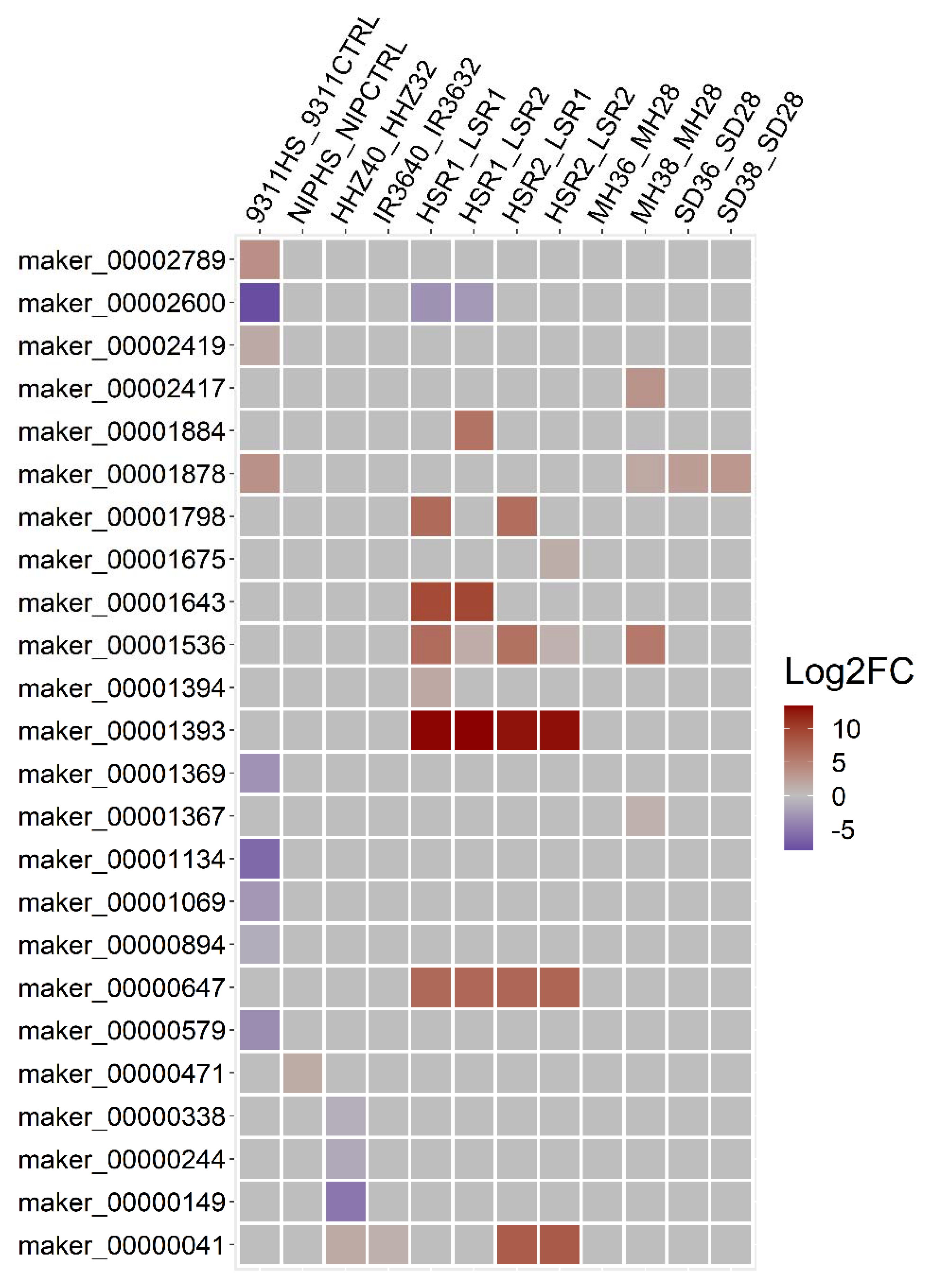
3.7. Mapping DEGs to the Known Heat Stress-Tolerant QTLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tomlinson, I. Doubling Food Production to Feed the 9 Billion: A Critical Perspective on a Key Discourse of Food Security in the UK. J. Rural Stud. 2013, 29, 81–90. [Google Scholar] [CrossRef]
- Challinor, A.; Wheeler, T.; Craufurd, P.; Slingo, J. Simulation of the Impact of High Temperature Stress on Annual Crop Yields. Agric. For. Meteorol. 2005, 135, 180–189. [Google Scholar] [CrossRef] [Green Version]
- Prasad, P.; Boote, K.; Allen, L.; Sheehy, J.; Thomas, J. Species, Ecotype and Cultivar Differences in Spikelet Fertility and Harvest Index of Rice in Response to High Temperature Stress. F. Crop. Res. 2006, 95, 398–411. [Google Scholar] [CrossRef]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive Expression Profiling of Rice Grain Filling-Related Genes under High Temperature Using DNA Microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef] [Green Version]
- Matsui, T.; Omasa, K.; Horie, T. High Temperature Induced Spikelet Sterility of Japonica Rice at Fowering in Relation to Air Humidity and Wind Velocity Conditions. Japan J. Crop Sci. 1997, 66, 449–455. [Google Scholar] [CrossRef] [Green Version]
- Matsui, T.; Omasa, K.; Horie, T. The Difference in Sterility Due to High Temperatures during the Flowering Period among Japonica-Rice Varieties. Plant Prod. Sci. 2001, 4, 90–93. [Google Scholar] [CrossRef]
- Maruyama, A.; Weerakoon, W.; Wakiyama, Y.; Ohba, K. Effects of Increasing Temperatures on Spikelet Fertility in Different Rice Cultivars Based on Temperature Gradient Chamber Experiments. J. Agron. Crop Sci. 2013, 199, 416–423. [Google Scholar] [CrossRef]
- Tenorio, F.A.; Ye, C.; Redoña, E.; Sierra, S.; Laza, M.; Argayoso, M.A. Screening Rice Genetic Resources for Heat Tolerance. Sabrao J. Breed. Genet. 2013, 45, 371–381. [Google Scholar]
- Cao, Y.; Duan, H.; Yang, L.; Wang, Z.; Zhou, S.; Yang, J. Effect of Heat Stress During Meiosis on Grain Yield of Rice Cultivars Differing in Heat Tolerance and Its Physiological Mechanism. Acta Agron. Sin. 2008, 34, 2134–2142. [Google Scholar] [CrossRef]
- Shi, W.; Ishimaru, T.; Gannaban, R.; Oane, W.; Jagadish, S. Popular Rice (Oryza sativa L.) Cultivars Show Contrasting Responses to Heat Stress at Gametogenesis and Anthesis. Crop Sci. 2015, 55, 589–596. [Google Scholar] [CrossRef]
- Wei, H.; Liu, J.; Wang, Y.; Huang, N.; Zhang, X.; Wang, L.; Zhang, J.; Tu, J.; Zhong, X. A Dominant Major Locus in Chromosome 9 of Rice (Oryza sativa L.) Confers Tolerance to 48 °C High Temperature at Seedling Stage. J. Hered. 2013, 104, 287–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manigbas, N.; Lambio, L.; Madrid, L.; Cardenas, C. Germplasm Innovation of Heat Tolerance in Rice for Irrigated Lowland Conditions in the Philippines. Rice Sci. 2014, 21, 162–169. [Google Scholar] [CrossRef]
- Jagadish, S.; Cairns, J.; Lafitte, R.; Wheeler, T.; Price, A.; Craufurd, P. Genetic Analysis of Heat Tolerance at Anthesis in Rice. Crop Sci. 2010, 50, 1633–1641. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P. Abiotic Stresses: Plant Resistance through Breeding and Molecular Approaches. In Genetic Improvements of Tolerance to High Temperature; Howarth Press Inc.: Binghamton, NY, USA, 2005; pp. 277–300. [Google Scholar]
- Bohnert, H.J.; Gong, Q.; Li, P.; Ma, S. Unraveling Abiotic Stress Tolerance Mechanisms—Getting Genomics Going. Curr. Opin. Plant Biol. 2006, 9, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, L.; Jang, K.; Huang, M.; Zheng, J. QTL Mapping for Heat Tolerance of the Tassel Period of Rice. Mol. Plant Breed. 2008, 6, 867–873. [Google Scholar]
- Qingquan, C.; Sibin, Y.; Chunhai, L. Identification of QTLs for Heat Tolerance at Flowering Stage in Rice. Sci. Agric. Sin. 2009, 41, 315–321. [Google Scholar]
- Cao, L.; Zhao, J.; Zhan, X.; Li, D.; He, L.; Cheng, S. Mapping QTLs for Heat Tolerance and Correlation between Heat Tolerance and Photosynthetic Rate in Rice. Chin. J. Rice Sci. 2003, 7, 223–227. [Google Scholar]
- Zhang, G.; Chen, L.; Xiao, G.; Xiao, Y.; Chen, X.; Zhang, S. Bulked Segregant Analysis to Detect QTL Related to Heat Tolerance in Rice (Oryza sativa L.) Using SSR Markers. Agric. Sci. China 2009, 8, 482–487. [Google Scholar] [CrossRef]
- Wang, X.; Cai, J.; Jiang, D.; Liu, F.; Dai, T.; Cao, W. Pre-Anthesis High-Temperature Acclimation Alleviates Damage to the Flag Leaf Caused by Post-Anthesis Heat Stress in Wheat. J. Plant Physiol. 2011, 168, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zhu, J.; Zhao, S.; He, L.; Yan, Q. Mapping QTLs for Heat Tolerance in a DH Population from Indica-Japonica Cross of Rice (Oryza sativa). J. Agric. Biotech. 2002, 10, 210–214. [Google Scholar]
- Li, Y.; Dai, Z.; Li, A.; Chen, X.; Wang, B.; Zhao, B.; Liu, G.; Pan, X.; Zhang, H. Role of Rice Main Parent BG90-2 in Breeding of Yangdao Series and Their Bacterial Blight Resistance. Chin. J. Rice Sci. 2011, 4, 439–442. [Google Scholar]
- Zhao, Z.; Jiang, L.; Xiao, Y.; Zhang, W.; Zhai, H.; Wan, J. Identification of QTLs for Heat Tolerance at the Booting Stage in Rice (Oryza sativa L.). Acta Agron. Sin. 2006, 32, 640. [Google Scholar]
- Shanmugavadivel, P.; Amitha, M.; Chandra, P.; Ramkumar, M.; Ratan, T.; Trilochan, M.; Nagendra, K. High Resolution Mapping of QTLs for Heat Tolerance in Rice Using a 5K SNP Array. Rice 2017, 10, 28. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Lei, J.; Huang, Y.; Zhu, S.; Chen, H.; Huang, R.; Peng, Z.; Tu, Q.; Shen, X.; Yan, S. Mapping Quantitative Trait Loci for Heat Tolerance at Anthesis in Rice Using Chromosomal Segment Substitution Lines. Breed. Sci. 2016, 66, 358–366. [Google Scholar] [CrossRef] [Green Version]
- Jagadish, S.; Craufurd, P.; Wheeler, T. Phenotyping Parents of Mapping Populations of Rice for Heat Tolerance during Anthesis. Crop Sci. 2008, 48, 1140–1146. [Google Scholar] [CrossRef]
- Prasanth, V.; Basava, K.; Babu, M.; VGN, V.; Devi, S.; Mangrauthia, S.; Voleti, S.; Sarla, N. Field Level Evaluation of Rice Introgression Lines for Heat Tolerance and Validation of Markers Linked to Spikelet Fertility. Physiol. Mol. Biol. Plants 2016, 22, 179–192. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Pan, Y.; Luo, L.; Deng, H.; Zhang, G.; Tang, W.; Chen, L. Quantitative Trait Loci Associated with Pollen Fertility under High Temperature Stress at Flowering Stage in Rice (Oryza sativa). Rice Sci. 2011, 18, 204–209. [Google Scholar] [CrossRef]
- Ye, C.; Argayoso, M.; Redoña, E.; Sierra, S.; Laza, M.; Dilla, C.; Mo, Y.; Thomson, M.; Chin, J.; Delaviña, C.; et al. Mapping QTL for Heat Tolerance at Flowering Stage in Rice Using SNP Markers. Plant Breed. 2012, 131, 33–41. [Google Scholar] [CrossRef]
- Ye, C.; Tenorio, F.; Argayoso, M.; Laza, M.; Koh, H.; Redoña, E.; Jagadish, K.; Gregorio, G. Identifying and Confirming Quantitative Trait Loci Associated with Heat Tolerance at Flowering Stage in Different Rice Populations. BMC Genet. 2015, 16, 41. [Google Scholar] [CrossRef] [Green Version]
- Nordborg, M.; Tavaré, S. Linkage Disequilibrium: What History Has to Tell Us. Trends Genet. 2002, 18, 83–90. [Google Scholar] [CrossRef]
- Kumar, A.; Gupta, C.; Thomas, J.; Pereira, A. Genetic Dissection of Grain Yield Component Traits Under High Nighttime Temperature Stress in a Rice Diversity Panel. Front. Plant Sci. 2021, 12, 712167. [Google Scholar] [CrossRef] [PubMed]
- Lafarge, T.; Bueno, C.; Frouin, J.; Jacquin, L.; Courtois, B.; Ahmadi, N. Genome-Wide Association Analysis for Heat Tolerance at Flowering Detected a Large Set of Genes Involved in Adaptation to Thermal and Other Stresses. PLoS ONE 2017, 12, e0171254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilasi, N.; Singh, J.; Vallejos, C.; Ye, C.; Jagadish, S.; Kusolwa, P.; Rathinasabapathi, B. Heat Stress Tolerance in Rice (Oryza sativa L.): Identification of Quantitative Trait Loci and Candidate Genes for Seedling Growth under Heat Stress. Front. Plant Sci. 2018, 871, 1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robbins, M.; Sim, S.; Yang, W.; Van Deynze, A.; van der Knaap, E.; Joobeur, T.; Francis, D. Mapping and Linkage Disequilibrium Analysis with a Genome-Wide Collection of SNPs That Detect Polymorphism in Cultivated Tomato. J. Exp. Bot. 2011, 62, 1831–1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNally, K.; Childs, K.; Bohnert, R.; Davidson, R.; Zhao, K.; Ulat, V.; Zeller, G.; Clark, R.; Hoen, D.; Bureau, T.; et al. Genomewide SNP Variation Reveals Relationships among Landraces and Modern Varieties of Rice. Proc. Natl. Acad. Sci. USA. 2009, 106, 12273–12278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, R.; Edwards, D.; Varshney, R. Structural Variations in Plant Genomes. Brief. Funct. Genom. 2014, 13, 296–307. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-Genome Analysis Highlights the Extent of Genomic Variation in Cultivated and Wild Rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhou, G.; Ma, J.; Jiang, W.; Jin, L.; Zhang, Z.; Guo, Y.; Zhang, J.; Sui, Y.; Zheng, L.; et al. De Novo Assembly of Soybean Wild Relatives for Pan-Genome Analysis of Diversity and Agronomic Traits. Nat. Biotechnol. 2014, 32, 1045–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golicz, A.; Bayer, P.; Barker, G.; Edger, P.; Kim, H.; Martinez, P.; Chan, C.K.K.; Severn-Ellis, A.; McCombie, W.; Parkin, I.; et al. The Pangenome of an Agronomically Important Crop Plant Brassica Oleracea. Nat. Commun. 2016, 7, 13390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.; Burzynski-Chang, E.; Fish, T.; Stromberg, K.; Sacks, G.; et al. The Tomato Pan-Genome Uncovers New Genes and a Rare Allele Regulating Fruit Flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Ruperao, P.; Thirunavukkarasu, N.; Gandham, P.; Selvanayagam, S.; Govindaraj, M.; Nebie, B.; Manyasa, E.; Gupta, R.; Das, R.R.; Odeny, D.A.; et al. Sorghum Pan-Genome Explores the Functional Utility for Genomic-Assisted Breeding to Accelerate the Genetic Gain. Front. Plant Sci. 2021, 12, 963. [Google Scholar] [CrossRef]
- Zhao, J.; Bayer, P.; Ruperao, P.; Saxena, R.; Khan, A.; Golicz, A.; Nguyen, H.; Batley, J.; Edwards, D.; Varshney, R. Trait Associations in the Pangenome of Pigeon Pea (Cajanus cajan). Plant Biotechnol. J. 2020, 18, 1946–1954. [Google Scholar] [CrossRef] [Green Version]
- Cook, D.; Lee, T.; Guo, X.; Melito, S.; Wang, K.; Bayless, A.; Wang, J.; Hughes, T.; Willis, D.; Clemente, T.; et al. Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. Science 2012, 338, 1206–1209. [Google Scholar] [CrossRef] [Green Version]
- Maron, L.; Guimarães, C.; Kirst, M.; Albert, P.; Birchler, J.; Bradbury, P.; Buckler, E.; Coluccio, A.; Danilova, T.; Kudrna, D.; et al. Aluminum Tolerance in Maize Is Associated with Higher MATE1 Gene Copy Number. Proc. Natl. Acad. Sci. USA. 2013, 110, 5241–5246. [Google Scholar] [CrossRef] [Green Version]
- Knox, A.; Dhillon, T.; Cheng, H.; Tondelli, A.; Pecchioni, N.; Stockinger, E. CBF Gene Copy Number Variation at Frost Resistance-2 Is Associated with Levels of Freezing Tolerance in Temperate-Climate Cereals. Theor. Appl. Genet. 2010, 121, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Zimin, A.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.; Yorke, J. The MaSuRCA Genome Assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef] [Green Version]
- Holt, C.; Yandell, M. MAKER2: An Annotation Pipeline and Genome-Database Management Tool for Second-Generation Genome Projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korf, I. Gene Finding in Novel Genomes. BMC Bioinform. 2004, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Keller, O.; Gunduz, I.; Hayes, A.; Waack, S.; Morgenstern, B. AUGUSTUS: Ab Initio Prediction of Alternative Transcripts. Nucleic Acids Res. 2006, 34, W435–W439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Cantalapiedra, C.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Golicz, A.; Martinez, P.; Zander, M.; Patel, D.; Van De Wouw, A.; Visendi, P.; Fitzgerald, T.; Edwards, D.; Batley, J. Gene Loss in the Fungal Canola Pathogen Leptosphaeria Maculans. Funct. Integr. Genomics. 2015, 15, 189–196. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Eisen, J.A.; Peterson, S.; Wessels, M.R.; Paulsen, I.T.; Nelson, K.E.; Margarit, I.; Read, T.D.; et al. Complete Genome Sequence and Comparative Genomic Analysis of an Emerging Human Pathogen, Serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 2002, 99, 12391–12396. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Chebotarov, D.; Kudrna, D.; Llaca, V.; Lee, S.; Rajasekar, S.; Mohammed, N.; Al-Bader, N.; Sobel-Sorenson, C.; Parakkal, P.; et al. A Platinum Standard Pan-Genome Resource That Represents the Population Structure of Asian Rice. Sci. Data 2020, 7, 113. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Van der Auwera, G.; Carneiro, M.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinforma. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. bioRxiv 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Broad Institute Picard Toolkit. Available online: http://broadinstitute.github.io/picard (accessed on 26 October 2020).
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.; Banks, E.; DePristo, M.; Handsaker, R.; Lunter, G.; Marth, G.; Sherry, S.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Sun, C.; Hu, Z.; Zheng, T.; Lu, K.; Zhao, Y.; Wang, W.; Shi, J.; Wang, C.; Lu, J.; Zhang, D.; et al. RPAN: Rice Pan-Genome Browser for ∼3000 Rice Genomes. Nucleic Acids Res. 2016, 45, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza Sativa Nipponbare Reference Genome Using next Generation Sequence and Optical Map Data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Dolatabadian, A.; Bayer, P.E.; Tirnaz, S.; Hurgobin, B.; Edwards, D.; Batley, J. Characterization of Disease Resistance Genes in the Brassica Napus Pangenome Reveals Significant Structural Variation. Plant Biotechnol. J. 2020, 18, 969–982. [Google Scholar] [CrossRef] [Green Version]
- Gabur, I.; Chawla, H.S.; Lopisso, D.T.; von Tiedemann, A.; Snowdon, R.J.; Obermeier, C. Gene Presence-Absence Variation Associates with Quantitative Verticillium Longisporum Disease Resistance in Brassica Napus. Sci. Rep. 2020, 10, 4131. [Google Scholar] [CrossRef] [Green Version]
- Weisweiler, M.; de Montaigu, A.; Ries, D.; Pfeifer, M.; Stich, B. Transcriptomic and Presence/Absence Variation in the Barley Genome Assessed from Multi-Tissue MRNA Sequencing and Their Power to Predict Phenotypic Traits. BMC Genom. 2019, 20, 787. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, L.; Ou, S.; Wang, R.; Wang, Y.; Chu, C.; Yao, S. Natural Variations of SLG1 Confer High-Temperature Tolerance in Indica Rice. Nat. Commun. 2020, 11, 5441. [Google Scholar] [CrossRef]
- Xu, Y.; Chu, C.; Yao, S. The Impact of High-Temperature Stress on Rice: Challenges and Solutions. Crop J. 2021, 9, 963–976. [Google Scholar] [CrossRef]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.; Scheffler, B.; Sanabria, Y.; de Guzman, C.; Galam, D.; Farmer, A.; Woodward, J.; May, G.; Oard, J. Identification of Candidate Genes in Rice for Resistance to Sheath Blight Disease by Whole Genome Sequencing. Theor. Appl. Genet. 2012, 124, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yuan, Y.; Xu, Y.; Zhang, G.; Guo, X.; Wu, F.; Wang, Q.; Rong, T.; Pan, G.; Cao, M.; et al. Identification of Candidate Genes for Drought Tolerance by Whole-Genome Resequencing in Maize. BMC Plant Biol. 2014, 14, 83. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.S.; Ali, G.S.; Reddy, A.S.N. Genes Encoding Calmodulin-Binding Proteins in the Arabidopsis Genome. J. Biol. Chem. 2002, 277, 9840–9852. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-T.; Li, G.-L.; Chang, H.; Sun, D.-Y.; Zhou, R.-G.; Li, B. Calmodulin-Binding Protein Phosphatase PP7 Is Involved in Thermotolerance in Arabidopsis. Plant. Cell Environ. 2007, 30, 156–164. [Google Scholar] [CrossRef]
- Liu, H.-T.; Gao, F.; Li, G.-L.; Han, J.-L.; Liu, D.-L.; Sun, D.-Y.; Zhou, R.-G. The Calmodulin-Binding Protein Kinase 3 Is Part of Heat-Shock Signal Transduction in Arabidopsis thaliana. Plant J. 2008, 55, 760–773. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When Defense Pathways Collide. The Response of Arabidopsis to a Combination of Drought and Heat Stress 1[W]. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Jiang, Y.; Dong, J.; Zhang, X.; Xiao, H.; Xu, Z.; Gao, X. Genome-Wide Analysis and Environmental Response Profiling of SOT Family Genes in Rice (Oryza sativa). Genes Genom. 2012, 34, 549–560. [Google Scholar] [CrossRef]
- Zhang, X.; Rerksiri, W.; Liu, A.; Zhou, X.; Xiong, H.; Xiang, J.; Chen, X.; Xiong, X. Transcriptome Profile Reveals Heat Response Mechanism at Molecular and Metabolic Levels in Rice Flag Leaf. Gene 2013, 530, 185–192. [Google Scholar] [CrossRef]
- Hu, T.; Sun, X.; Zhang, X.; Nevo, E.; Fu, J. An RNA Sequencing Transcriptome Analysis of the High-Temperature Stressed Tall Fescue Reveals Novel Insights into Plant Thermotolerance. BMC Genom. 2014, 15, 1147. [Google Scholar] [CrossRef] [Green Version]
- Mani, B.; Agarwal, M.; Katiyar-Agarwal, S. Comprehensive Expression Profiling of Rice Tetraspanin Genes Reveals Diverse Roles during Development and Abiotic Stress. Front. Plant Sci. 2015, 6, 1088. [Google Scholar] [CrossRef] [Green Version]
- Idänheimo, N.; Gauthier, A.; Salojärvi, J.; Siligato, R.; Brosché, M.; Kollist, H.; Mähönen, A.P.; Kangasjärvi, J.; Wrzaczek, M. The Arabidopsis Thaliana Cysteine-Rich Receptor-like Kinases CRK6 and CRK7 Protect against Apoplastic Oxidative Stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef]
- Pandian, B.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Derakhshani, B.; Jafary, H.; Zanjani, B.M.; Hasanpur, K.; Mishina, K.; Tanaka, T.; Kawahara, Y.; Oono, Y. Combined QTL Mapping and RNA-Seq Profiling Reveals Candidate Genes Associated with Cadmium Tolerance in Barley. PLoS ONE 2020, 15, e0230820. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Jiang, F.; Weng, Y.; Sun, M.; Shi, X.; Zhou, Y.; Yu, L.; Wu, Z. Identification of Heat-Tolerance QTLs and High-Temperature Stress-Responsive Genes through Conventional QTL Mapping, QTL-Seq and RNA-Seq in Tomato. BMC Plant Biol. 2019, 19, 398. [Google Scholar] [CrossRef]
- Li, J.; Yuan, D.; Wang, P.; Wang, Q.; Sun, M.; Liu, Z.; Si, H.; Xu, Z.; Ma, Y.; Zhang, B.; et al. Cotton Pan-Genome Retrieves the Lost Sequences and Genes during Domestication and Selection. Genome Biol. 2021, 22, 119. [Google Scholar] [CrossRef]
- Qian, D.; Tian, L.; Qu, L. Proteomic Analysis of Endoplasmic Reticulum Stress Responses in Rice Seeds. Sci. Rep. 2015, 5, 14255. [Google Scholar] [CrossRef] [Green Version]


| Variant Type | Pan-Genome | Reference | Non-Reference | Cluster 2 | Cluster 1 |
|---|---|---|---|---|---|
| Bi-allele SNP | 5,059,798 | 4,868,611 | 191,187 | 4,763,997 | 2,373,085 |
| Splicing | 15,986 | 15,881 | 105 | 15,210 | 7773 |
| Exonic | 284,016 | 281,185 | 2831 | 269,738 | 140,517 |
| Intronic | 578,887 | 576,534 | 2353 | 548,909 | 277,418 |
| UTR | 224,599 | 224,127 | 472 | 213,375 | 105,933 |
| Upstream | 1,229,370 | 1,224,403 | 4967 | 2,790,074 | 1,351,465 |
| Downstream | 1,100,851 | 1,095,937 | 4914 | 2,595,947 | 1,262,776 |
| Missense | 143,819 | 142,071 | 1748 | 136,135 | 70,833 |
| Stop gained | 1936 | 1900 | 36 | 1816 | 840 |
| Annotation | SNPs in Resistant Cultivars | SNPs in Highly Resistant Cultivars |
|---|---|---|
| Downstream | 29,082 | 14,759 |
| Exon | 358 | 185 |
| Intron | 18,846 | 9541 |
| Non_Synonymous | 10,194 | 5046 |
| Splice site acceptor | 28 | 16 |
| Splice Site donor | 38 | 19 |
| Start gained | 769 | 366 |
| Start lost | 25 | 11 |
| Stop gained | 225 | 116 |
| Stop lost | 26 | 14 |
| Synonymous | 7827 | 4090 |
| Upstream | 66,718 | 35,970 |
| UTR_3 | 8820 | 4422 |
| UTR_5 | 3817 | 1880 |
| Total | 146,773 | 76,435 |
| ProjectID * | Test Cultivars | Comparisons | Pan-Genome Upregulated Genes | Reference Upregulated Genes | Non-Reference Upregulated Genes | Pan-Genome Downregulated Genes | Reference Upregulated Genes | Non-Reference Downregulated Genes |
|---|---|---|---|---|---|---|---|---|
| PRJNA604026 | 9311 | 9311HS_9311CTRL | 8248 | 8202 | 46 | 9691 | 9616 | 75 |
| Nipponbare | NIPHS_NIPCTRL | 4914 | 4909 | 5 | 6504 | 6495 | 9 | |
| PRJNA508820 | Huanghuazhan | HHZ40_HHZ32 | 2091 | 2064 | 27 | 1819 | 1800 | 19 |
| IR36 | IR3640_IR3632 | 1395 | 1364 | 31 | 1503 | 1486 | 17 | |
| PRJNA610667 | HSR1 | HSR1_LSR1 | 2143 | 2058 | 85 | 1650 | 1605 | 45 |
| HSR2 | HSR1_LSR2 | 1704 | 1645 | 59 | 1048 | 1020 | 28 | |
| LSR1 | HSR2_LSR1 | 1617 | 1560 | 57 | 2232 | 2180 | 52 | |
| LSR2 | HSR2_LSR2 | 1596 | 1532 | 64 | 2117 | 2060 | 57 | |
| PRJNA633211 | MH101 | MH36_MH28 | 1825 | 1809 | 16 | 1361 | 1354 | 7 |
| MH38_MH28 | 5181 | 5145 | 36 | 2898 | 2881 | 17 | ||
| SDW005 | SD36_SD28 | 1380 | 1375 | 5 | 110 | 103 | 7 | |
| SD38_SD28 | 4618 | 4599 | 19 | 923 | 916 | 7 |
| BioSample ID * | Number of Genes |
|---|---|
| SAMN08217222 | 37 |
| SAMN10564385 | 60 |
| SAMN12715984 | 49 |
| SAMN12721963 | 46 |
| SAMN12672924 | 55 |
| SAMN12718029 | 49 |
| SAMN12748569 | 38 |
| SAMN12748589 | 42 |
| SAMN12748590 | 55 |
| SAMN12748600 | 41 |
| SAMN12748601 | 39 |
| SAMN13021815 | 51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woldegiorgis, S.T.; Wu, T.; Gao, L.; Huang, Y.; Zheng, Y.; Qiu, F.; Xu, S.; Tao, H.; Harrison, A.; Liu, W.; et al. Identification of Heat-Tolerant Genes in Non-Reference Sequences in Rice by Integrating Pan-Genome, Transcriptomics, and QTLs. Genes 2022, 13, 1353. https://doi.org/10.3390/genes13081353
Woldegiorgis ST, Wu T, Gao L, Huang Y, Zheng Y, Qiu F, Xu S, Tao H, Harrison A, Liu W, et al. Identification of Heat-Tolerant Genes in Non-Reference Sequences in Rice by Integrating Pan-Genome, Transcriptomics, and QTLs. Genes. 2022; 13(8):1353. https://doi.org/10.3390/genes13081353
Chicago/Turabian StyleWoldegiorgis, Samuel Tareke, Ti Wu, Linghui Gao, Yunxia Huang, Yingjie Zheng, Fuxiang Qiu, Shichang Xu, Huan Tao, Andrew Harrison, Wei Liu, and et al. 2022. "Identification of Heat-Tolerant Genes in Non-Reference Sequences in Rice by Integrating Pan-Genome, Transcriptomics, and QTLs" Genes 13, no. 8: 1353. https://doi.org/10.3390/genes13081353






