Historical Westward Migration Phases of Ovis aries Inferred from the Population Structure and the Phylogeography of Occidental Mediterranean Native Sheep Breeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Design and Genotyping
Population Samples
2.2. Statistical Analysis
3. Results and Discussion
3.1. Microsatellites’ Performance
3.2. Mediterranean Sheep Breeds’ Divergence and Relationships
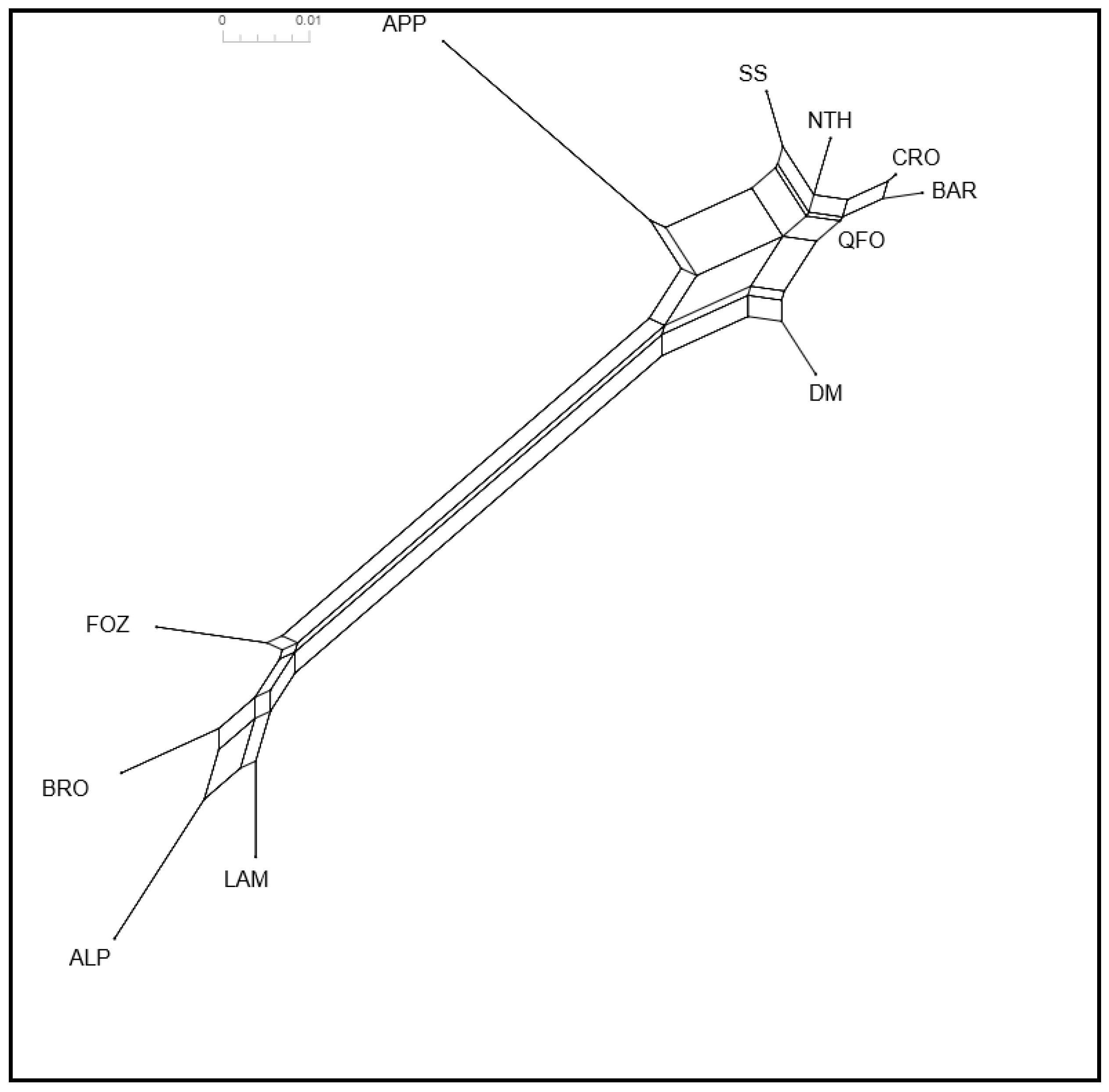
3.2.1. FST Distances and Neighbor Network
3.2.2. Molecular Co-Ancestry and Gene Flow
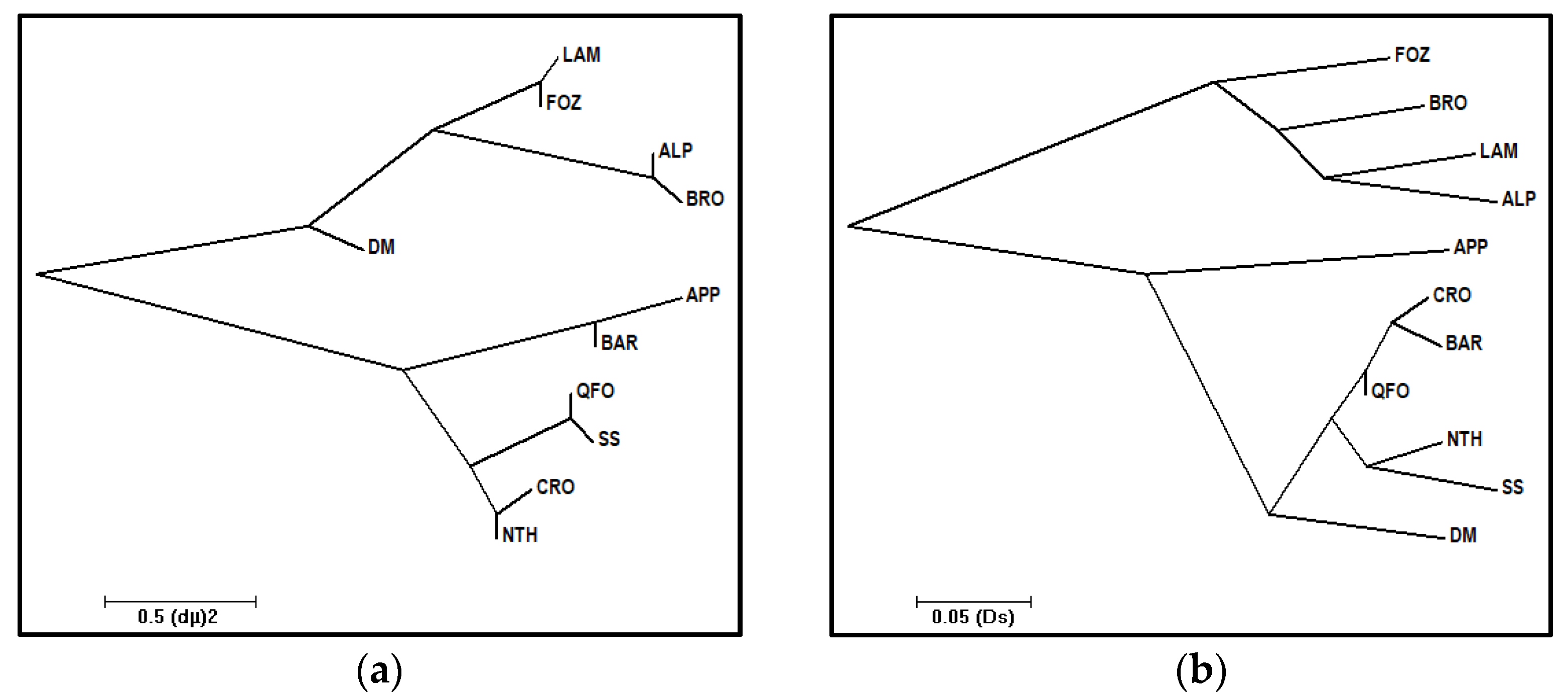
3.2.3. (DS) and (δµ)2 Genetic Distances and Relative Neighbor-Joining Trees
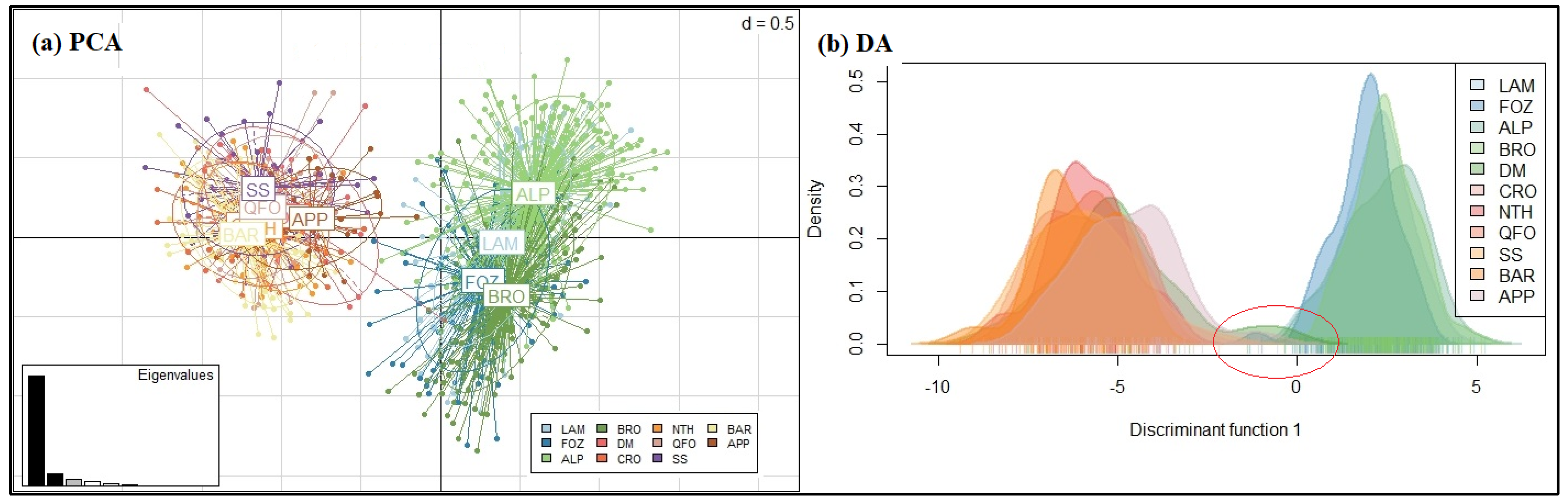
3.2.4. Genetic Structure Analysis
3.2.5. Divergence Time
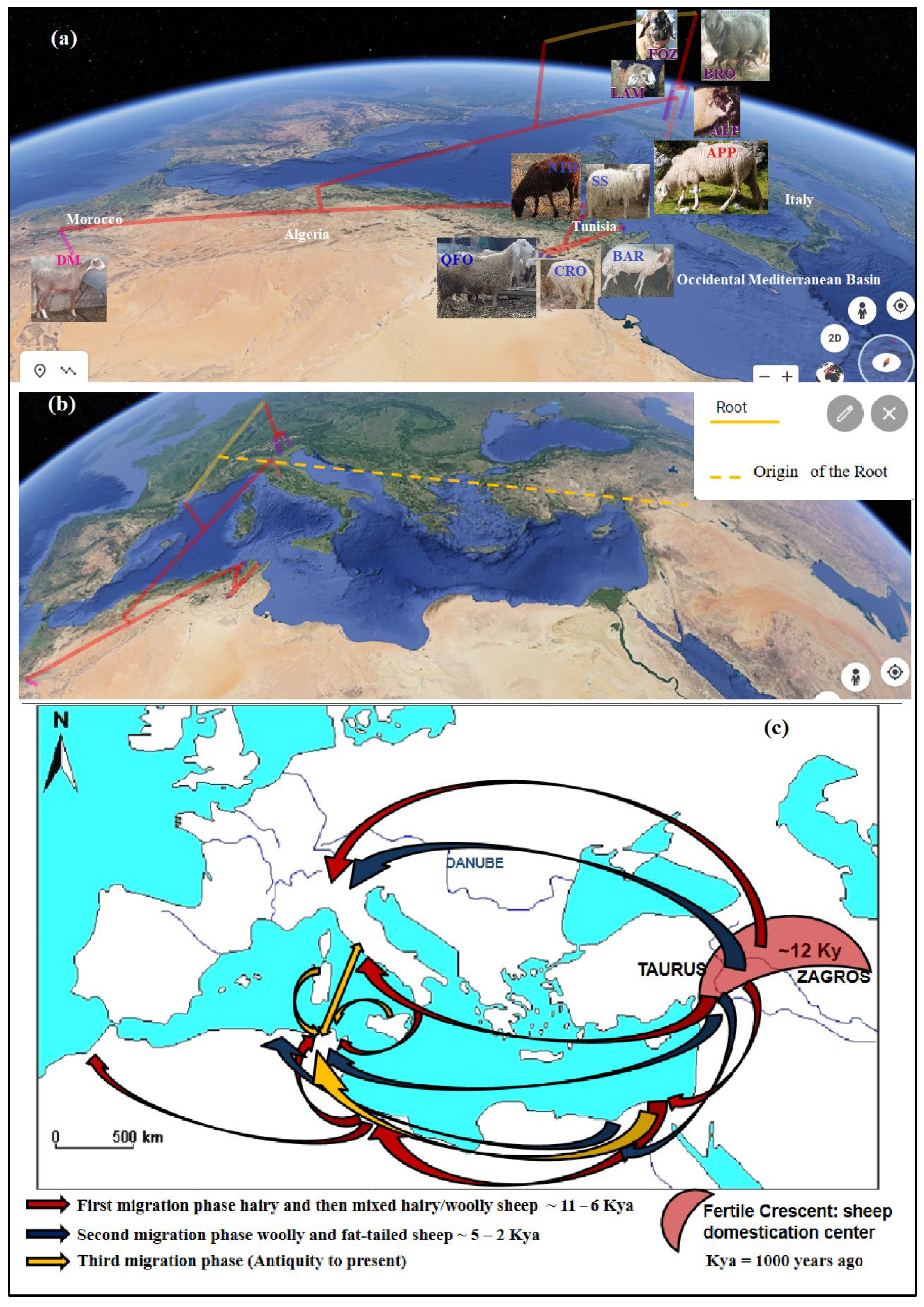
3.3. Phylogeographic Patterns of the Sheep Migration Waves’ Westward Domestication Center Inferred from Genetic Data and Supported by Archeological, Historical and Systematic Data
3.3.1. First Migration Phase
- 1.
- The first wave of this earliest migration phase was not highlighted by the present study. It would have occurred in the precedent Millennia, since the earliest divergence time was revealed in the present study between the analyzed Mediterranean breeds remote to 11,000 years ago. This first wave could correspond to the earliest form of the first domesticated sheep in the Fertile Crescent in the early Holocene before 12,000 years ago, which was very close to the wild ancestor (O. g.). This earliest domesticated form migrated via the Mediterranean Sea and occupied Cyprus since 12,000 [87], and later on, the Tyrrhenian area (Corsica and Sardinia), where a return to the feral form occurred, leading to, respectively, the Cyprus mouflon (O. g. ophion) and the European mouflon (O. g. musimon) of Corsica and Sardinia [87,88]. The presence of a second post glacial human occupation was highlighted in the Tyrrhenian islands and in the south-western Europe in the early 12th millennium B.P., described as the Holocene human re-expansion [86,89] induced by climate warming, which favored the early Holocene seasonality just preceding the Younger Dryas and the high fire frequency events [90]. Recent studies demonstrated that sheep domestication was pushed back to the late Epipaleolithic (13th–12th millennium B.P.), allowing for the first domesticated sheep with wild morphology [2,91]. This sheep was a hairy and short-tailed sheep, such as the wild ancestor and the remnant mouflon. The findings of Vigne et al. [83], highlighting the early Mediterranean transportation of mammals by boats starting in the 13th millennium and later specialized by human voyagers, supported this earliest migration wave of domesticated sheep with wild morphology, which led to the establishment of the Mediterranean mouflon. This first wave seems to concern only the northern Mediterranean rim, since:
- No evidence of mouflon and short-tailed sheep was noted in North Africa;
- Waves of human population from the Middle East to North Africa occurred 15,000 and 9000 years ago [85], before the initiation of sheep domestication and after the first appearance of the morphologically domestic sheep with a long tail, out of the domestication center in the Mediterranean Cyprus, around 10,500 years ago [2,83,95];
- The climate conditions in the 12th millennium’s North African Paleolithic noted a relatively dry episode [96,97,98] that could have prevented the introduction of this westward Middle Eastern Human wave, with their first domesticated wild sheep (mouflon), where the Northern Mediterranean basin conditions were more favorable.
- 2.
- The second migration wave, perfectly highlighted by the present sheep genetic analysis, is considered as the first Neolithic westward introduction of phenotypically domesticated sheep. This migration wave could have been represented by the three contemporaneous migrant sheep that are the ancestors of the center Italian APP sheep, the Venetian sheep and the DM sheep of the Maghreb. The very ancient relatedness between these sheep was prove by the (fij), and the (δµ)2 distances and the shared ancestral gene pool, especially between the DM and Venetian breeds, depicted by the structure analysis, and mainly by their divergence, approximately between 11,000 to 9000 years ago (Table 6). These three ancestors, as depicted by the phylogenetic trees (Figure 2), diverged from a common ancestor (first node), which would be the earliest domestic-like sheep and the descendant of the morphologically wild sheep domesticated in the Fertile Crescent and described above in the first wave. Our revealed genetic divergence faithfully coincided with the management step of the morphologically domesticated sheep, which replaced the first domesticated sheep with a wild morphological form in the domestication center mentioned by Zeder [2,95] and Vigne et al. [83]. In fact, these scholars and Zeder [91] mentioned that the most important phenotypic changes—mostly in horn and body sizes and in shape, such as changes in rachis morphology influencing the tail length—had been noted over a 1000 years ago, following the 12th millennium, which reflected a change in the management practices of animals. It is worth mentioning that actual Mediterranean sheep breeds belong to the long-tailed group and migrated westward from the Fertile Crescent during this second wave of the first migration phase after these mentioned changes, especially the rachis morphology change. In fact, tail length, which is a strongly inherited trait and academically interesting feature in sheep classifications [102], is based on the number of coccygeal vertebrae. The mouflon presented a short-tailed vertebrae (no more than 11); the North European short-tailed primitive sheep group, such as the Soay breed, had 8 to 10 vertebrae; and the long tailed sheep group had 16 to 18, reaching to 24 vertebrae in the tail [101,102]. This progression in the tail length reflects the sheep evolution morphology after the domestication effect. Otherwise, recent structure analysis, including mouflon, primitive breeds and modern Mediterranean breeds, demonstrated a clear distinction (at K = 2) between the two ancestral populations of mouflon and domesticated sheep [21,70,82]. Barbato et al. [82] and Ciani et al. [70] highlighted the first differentiation of the primitive short-tailed Soay sheep from the morphologically long-tailed domestic sheep, which faithfully argues our hypothesis of two distinct waves in the first phase of the westward migration of sheep, in where the short-tailed Soay breed would occupied an intermediate position between these two waves. Consequently, the three contemporaneous ancestors of this second wave should have been derived from a common long-tailed domesticated sheep ancestor, which was already evolved in the domestication center and then migrated westward, leading to its recent descendants: The Venetian breeds, the central Italian APP and the Maghrebian DM breed.
- The first ancestor, from which derived the central Italian APP sheep diverged from the Maghrebian DM breed since 11,000 years ago, would be the first Neolithic sheep migrated by maritime road from the domestication center, reaching South Europe, going through Cyprus, and then the Greece islands to reach the Apennine peninsula—the cradle of the APP breed—where the spread of the earliest Impressa Neolithic group characterized by the sheep breeding dominance at the 9th millennium was revealed [16,86,95]. This early Neolithic sheep presence in the Italian peninsula was achieved or reduced probably due to the 8.2 cal ka climatic cooling event and the maximum fire activity noted in this region [86,95,97]. The APP’s ancestor would have migrated, after these unfavorable conditions, to the southern Mediterranean coasts, where the humid climate of the green North African begins [97], as evidenced by the high molecular co-ancestry and gene flow values between APP and the Maghrebian breeds—greater than the values between APP and the Venetian breeds—and their shared common ancestral populations of up to nine assumed ancestral populations (K = 9). This was depicted by the structure analysis and the phylogenetic trees (Figure 2a,b) clustering together the APP and Maghrebian breeds. The genetic relatedness between the Maghrebian and the south-western European breeds was highlighted by Kandoussi et al. [32], who revealed the closeness of the Moroccan and the Italian sheep groups and suggested a south Italian–Tunisian introduction route of Neolithic sheep from the Middle East domestication center. The hypothesis that the APP’s ancestor has contemporarily directly reached the southern Mediterranean rim might not have to be excluded, since a strong molecular co-ancestry relatedness had been revealed between the APP and Maghrebian breeds, in addition to the next gene flow reported between them. However, the absence of a sheep trace in North Africa in the 9th millennium can be explained by the geoarchaeological data revealing a dry North African climate in this period [97] and, thus, avoiding the early Neolithic expansion in the southern Mediterranean area, and/or probably due to the absence or the scarcity of zooarcheological research highlighting this presence in the southern rim comparatively to those of the northern Mediterranean, as proposed by Zeder [95], explaining the lack of evidence of the earliest Neolithic trace in North Africa before the 8th millennium B.P. The two remaining ancestors, from which the Venetian breeds and Maghrebian DM breed were derived, diverged between the 11th and the 10th millennium (Table 6) from a common ancestor, as revealed by the molecular co-ancestry (fij) relatedness and the (δµ)2 distances-based tree, and was migrated by two different land roads;
- The second ancestor is the Venetian breeds’ ancestor. It was diverged from the Fertile Crescent ancestor since the 11th millennium (10,736 years ago, Table 6), and went to South Europe by the northwestern overland road known as the Danubian route, considered as a gateway of the Europe Neolithization [103]. The very ancient remote relatedness between the Venetian breeds before their recent differentiation revealed by the (fij) parameter strongly supported this idea and was well proven by the two phylogenetic trees (Figure 2a,b), as well as the PCA, DAPC and Bayesian structure analysis. This ancient genetic relatedness was supported by the historic data revealing that by the Danubian, immigrant farmers—Danilo groups from Balkans—introduced domestic animals into Istria and northeastern Italy. This northern land introduction to the actual Venetian region was mentioned in addition to the earliest and speeder migration by the Impressa group, which introduced agriculture to the southern and center-west of Italy, probably introducing the ancestor of APP. These two different introduction roads of agriculture into Italy explained the genetic differentiation of the two ancestral populations of Venetian breeds, and APP was described as the first and the second ancestors of the westward second sheep wave. The two different sheep introductions to the Italian Peninsula, the Mediterranean maritime route and the land route via the Balkans were highlighted by Ciani et al. [21].
- Exploring the systematic data of the Venetian sheep, we discovered, unexpectedly, the systematic classification “O. a. sudanica Sanson” of the Alpine sheep group, from which the Venetian and Bergamesca breeds were derived [104,105]. This classification is relative to an ancient supposed Sudanese origin of the Alpine sheep based on the phenotypic similarity between old Alpine sheep and Sudanese native sheep [105,106,107]. This classification would explain the noticeable level of the molecular ancestry identity between the Venetian sheep and the North African DM sheep or its ancestor, described as the third ancestor of this westward second sheep wave;
- The third ancestor is the direct ancestor of the DM breed due to the high conservation level of its non-miscegenated ancestral gene pool. This ancestor would have diverged in the Fertile Crescent since the 11th millennium. It would have taken the southwestern overland road, via the Suez Ishim, Egypt and Libya, in order to reach Tunisia, Algeria and Morocco. The DM seems to be the legacy of this ancestor, and is perfectly conserved after its genetic drift and isolation in the northern Sahara oasis. Our hypothesis based on phylogenetic inference is strongly supported by historical and archeological data. In fact, the first ancestral sheep reaching North Africa was depicted in an old Egyptian monument and in the Maghrebian Cavern, most often apprehended through engraved or painted representations as a hairy sheep with long tail and legs [108,109]. Basing on sheep representations found in Egyptian, Libyan and Atlas monuments (Saharan Algeria), this ancestry was respectively identified as Ovis longipes aegyptiaca for Egyptian ancestry sheep and, for Maghrebian ancestry sheep, as Ovis longipes libyca Fitzinger (1860) or “Fezzan sheep“ defined by Fitzinger [110] using the first representation of this ancestral sheep found in Fezzan in Libya (Figure 6a). As mentioned by Joleaud [111] and Camps [108,109], these subspecies were regrouped by Fitzinger (1860) under a unique systematic classification as O. a. longipes Fitzinger (1860), in relation to the very long legs and tail of this group of the first ancestors of hairy domestic sheep. The gap of the archeological evidence of this ancestral sheep in Tunisia was filled after the recent findings of Ben Naser [112,113] discovering the sheep (O. a. longipes Fitzinger) representations, in the Central Tunisian caves of Jbel Ouesslat, considered as the northernmost currently known engravings on the rocks of the Maghreb, dating from the early to middle Holocene and illustrating both Capsian and Neolithic periods (8200 to 6000 years ago). Until our findings, it was assumed that this ancestry was extremely extinct from North Africa (Egypt, Libya, Algeria and Morocco) and that its descendants still existed in the Sahel and Sub Saharan region from Mauritania to Chad, as the Tuareg sheep of the Saharan Algeria (Figure 6b) and West African sheep as the Dwarf of Nigeria and Cameroon [108,109,111]. The phylogenetic data from our genetic analysis led us to investigate the archeological, historical and morphological data to confirm our hypothesis, considering the actual native DM breed of the Maghreb as the last representative of the first ancestral sheep reaching North Africa in the first Neolithic immigration wave. As mentioned by Tourte [114], in 1068, the Arab historian El Bekri described the sheep of the western Sahara as a hairy sheep with long legs and tail named “Dammanian sheep”; this nomenclature is considered as the Arabic name of the subspecies O. a. longipes Fitzinger. In fact, the actual name “D’man” or “Damman” (DM) of the Maghrebian breed was derived from this Arabic nomenclature of the Sub Saharan hairy sheep. Consequently, the DM is the last representative of this subspecies that is supposed to be extinct from the Maghreb. At a morphological level, we discovered a perfect congruence between the current DM breed and the first ancestor of the North African sheep, faithfully reproduced on the engraved or painted representations in Maghrebian caves of the Saharan Atlas dated from the early Neolithic. In 1860, Fitzinger [110] finely reproduced the features of this ancestral sheep of North Africa dated from the earlier Neolithic period in a figure entitled “Das Fezzan-Schaf. O. a. longipes libyca” (Figure 6a). We illustrated the fine compatibility—big size, very long legs and tail, presence of man, roman nose (a slightly domed chamfer) and mixed hairy and woolly coat between Fitzinger’s lithography of “Fezzan sheep” (Figure 6a) and the current DM sheep of the Maghreb (Figure 6c).
3.3.2. Second Migration Phase
3.3.3. Third Migration Phase
- The westward migration wave aspect, which corresponded to the second migration wave reintroducing fat-tailed sheep in the western Mediterranean basin, caused the final divergence between the fat-tailed BAR and the thin-tailed QFO 1440 years ago (Table 6). This divergence time is contemporary to the Islamic and Arab conquest started in the 7th century [139], about 1300 years ago, which concentrated for five centuries mostly in Tunisia, named Ifrikiya in this periode, the capital of the North African and Maghrebian part of the Islamic period [85]. This recent fat-tailed migration wave explained the regeneration of the fat-tailed phenotype only in the southern Mediterranean rim, and its concentration in Eastern North Africa (Egypt, Libya and Tunisia), despite its precedented presence in the western North African thin-tailed sheep genome (Algerian and Moroccan) [25,140], and in the southern European thin-tailed sheep genome, as revealed in the recent genetic structure investigation of Mediterranean breeds [21,33]. These findings justify the dominance of the thin-tailed character [138];
- The local western Mediterranean gene flow aspect between already installed and more or less evolved and selected woolly Mediterranean breeds. This west Mediterranean breeds’ gene flow finalized the genetic makeup of the present day northern and southern Mediterranean shores’ sheep breeds. This intensive gene flow is strongly related to the wide exchange and trades of finer woolly sheep starting in the Roman period and continues into the medieval period under the merinization process [70,123]. The highest gene flow value revealed between the Algerian-originated QFO breed and Italian APP seems to be an earlier evidence of this process. In fact, the participation of North African sheep in shaping the merino genome was mentioned by Ciani et al. [70]. Unfortunately, the present study did not include merino breeds to highlight the genetic relationship between North African and merino or merino-derived breeds. However, in the clustering analysis result elaborated by Ben Jemaa et al. [33], we revealed the shared genome between thin-tailed Tunisian breeds and merino breeds, with the only difference in the clusters’ proportions. This last study highlighted the strong gen flow and the past introgression, interpreted as unexpected, between the western Mediterranean breeds.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Childe, V.G. Man Makes Himself; Watts and Co.: London, UK, 1936; p. 275. [Google Scholar]
- Zeder, M.A. The origins of agriculture in the Near East. Curr. Anthropol. 2011, 52, S221–S235. [Google Scholar] [CrossRef]
- Zeder, M.A.; Emshwiller, E.; Smith, B.D.; Bradley, D.G. Documenting domestication: The intersection of genetics and archaeology. Trends Genet. 2006, 22, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Vigne, J.-D.; Helmer, D.; Peters, J. First Steps of Animal Domestication: New Archaeozoological Approaches; Dir.; Oxbow Books: Oxford, UK, 2005. [Google Scholar]
- McDonald, M.M.A. The pattern of Neolithization in Dakhleh Oasis in the Eastern Sahara. Quat. Int. 2016, 410, 181–197. [Google Scholar] [CrossRef]
- Pereira, F.; Davis, S.J.M.; Pereira, L.; McEvoy, B.; Bradley, D.G.; Amorim, A. Genetic signatures of a Mediterranean influence in Iberian Peninsula sheep husbandry. Mol. Biol. Evol. 2006, 23, 1420–1426. [Google Scholar] [CrossRef]
- Pereira, F.; Queiros, S.; Gusmao, L.; Nijman, I.J.; Cuppen, E.; Lenstra, J.A.; Consortium, E.; Davis, S.J.M.; Nejmeddine, F.; Amorim, A. Tracing the History of Goat Pastoralism: New Clues from Mitochondrial and Y Chromosome DNA in North Africa. Mol. Biol. Evol. 2009, 26, 2765–2773. [Google Scholar] [CrossRef]
- Vigne, J.-D. The large “true” Mediterranean islands as a model for the Holocene human impact on the European vertebrate fauna ? Recent data and new reflections. In The Holocene History of European Vertebrate Fauna. Modern Aspects and Research; Deutsches Archaologisches Institut Eurasien-Abteilung; Verlag Marie Leidorf GmbH Rahden/Westf.: Rahden, Germany, 1999; pp. 295–322. [Google Scholar]
- Rezaei, H.R.; Naderi, S.; Chintauan-Marquier, I.C.; Taberlet, P.; Virk, A.T.; Naghash, H.R.; Rioux, D.; Kaboli, M.; Pompanon, F. Evolution and taxonomy of the wild species of the genus Ovis (Mammalia, Artiodactyla, Bovidae). Mol. Phylogenet. Evol. 2010, 54, 315–326. [Google Scholar] [CrossRef]
- Martínez, A.; Manunza, A.; Delgado, J.V.; Landi, V.; Adebambo, A.; Ismaila, M.; Capote, J.; El Ouni, M.; Elbeltagy, A.; Abushady, A.M.; et al. Detecting the existence of gene flow between Spanish and North African goats through a coalescent approach. Sci. Rep. 2016, 6, 38935. [Google Scholar] [CrossRef]
- Dolfini, A. From the Neolithic to the Bronze Age in Central Italy: Settlement, Burial, and Social Change at the Dawn of Metal Production. J. Archaeol. Res. 2020, 28, 503–556. [Google Scholar] [CrossRef]
- Ryder, M.L. Sheep. In Evolution of domesticated Animals; Mason, I.L., Ed.; Longman: London, UK; New York, NY, USA, 1984. [Google Scholar]
- Manning, K.; Stopp, B.; Colledge, S.; Downey, S.; Conolly, J.; Dobney, K.; Shennan, S. Animal exploitation in the early Neolithic of the Balkans and Central Europe. In Origins and Spread of Domestic Animals in Southwest Asia and Europe; Colledge, S., Conolly, J., Dobney, K., Manning, K., Shennan, S., Eds.; Left Coast Press: Walnut Creek, CA, USA, 2013; pp. 237–251. [Google Scholar]
- Arbuckle, B.; Kansa, S.; Kansa, E.; Orton, D.; Çakırlar, C. Data Sharing Reveals Complexity in the Westward Spread of Domestic Animals across Neolithic Turkey. PLoS ONE 2014, 9, e99845. [Google Scholar] [CrossRef]
- Lv, F.-H.; Peng, W.-F.; Yang, J.; Zhao, Y.-X.; Li, W.-R.; Liu, M.-J.; Ma, Y.-H.; Zhao, Q.-J.; Yang, G.-L.; Wang, F.; et al. Mitogenomic Meta-Analysis Identifies Two Phases of Migration in the History of Eastern Eurasian Sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef]
- Machová, K.; Málková, A.; Vostrý, L. Sheep Post-Domestication Expansion in the Context of Mitochondrial and Y Chromosome Haplogroups and Haplotypes. Genes 2022, 13, 613. [Google Scholar] [CrossRef]
- Muigai, A.W.T.; Hanotte, O. The Origin of African Sheep: Archaeological and Genetic Perspectives. Afr. Archaeol. Rev. 2013, 30, 39–50. [Google Scholar] [CrossRef]
- Tapio, M.; Marzanov, N.; Ozerov, M.; Cinkulov, M.; Gonzarenko, G.; Kiselyova, T.; Murawski, M.; Viinalass, H.; Kantanen, J. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol. Biol. Evol. 2006, 23, 1776–1783. [Google Scholar] [CrossRef]
- Tapio, M.; Ozerov, M.; Tapio, I.; Toro, M.A.; Marzanov, N.; Cinkulov, M.; Goncharenko, G.; Kiselyova, T.; Murawski, M.; Kantanen, J. Microsatellite-based genetic diversity and population structure of domestic sheep in northern Eurasia. BMC Genet. 2010, 11, 76. [Google Scholar] [CrossRef]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Delvento, C.; et al. On the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools. Genet. Sel. Evol. 2020, 52, 25. [Google Scholar] [CrossRef]
- Blackburn, H.D.; Toishibekov, Y.; Toishibekov, M.; Welsh, C.S.; Spiller, S.F.; Brown, M.; Paiva, S.R. Genetic diversity of Ovis aries populations near domestication centers and in the New World. Genetica 2011, 139, 1169–1178. [Google Scholar] [CrossRef]
- Ciani, E.; Ciampolini, R.; D’Andrea, M.; Castellana, E.; Cecchi, F.; Incoronato, C.; d’Angelo, F.; Albenzio, M.; Pilla, F.; Matassino, D.; et al. Analysis of genetic variability within and among Italian sheep breeds reveals population stratification and suggests the presence of a phylogeographic gradient. Small Rumin. Res. 2013, 112, 21–27. [Google Scholar] [CrossRef]
- Ben Sassi-Zaidy, Y.; Maretto, F.; Charfi-Cheikrouha, F.; Cassandro, M. Genetic diversity, structure, and breed relationships in Tunisian sheep. Small Rumin. Res. 2014, 119, 52–56. [Google Scholar] [CrossRef]
- Gaouar, S.B.S.; Da Silva, A.; Ciani, E.; Kdidi, S.; Aouissat, M.; Dhimi, L.; Lafri, M.; Maftah, A.; Mehtar, N. Admixture and Local Breed Marginalization Threaten Algerian Sheep Diversity. PLoS ONE 2015, 10, e0122667. [Google Scholar] [CrossRef]
- Harkat, S.; Laoun, A.; Belabdi, I.; Benali, R.; Outayeb, D.; Payet-Duprat, N.; Blanquet, V.; Lafri, M.; Da Silva, A. Assessing patterns of genetic admixture between sheep breeds: Case study in Algeria. Ecol. Evol. 2017, 7, 6404–6412. [Google Scholar] [CrossRef]
- Gaouar, S.B.S.; Kdidi, S.; Tabet Aouel, N.; Aït-Yahia, R.; Boushaba, N.; Aouissat, M.; Dhimi, L.; Yahyaoui, M.; Saidi-Mehtar, N. Genetic admixture of North-African ovine breeds as revealed by microsatellite loci. Livest. Res. Rural Dev. 2015, 26, 118. [Google Scholar]
- Leroy, G.; Danchin-Burge, C.; Palhière, I.; Sancristobal, M.; Nédélec, Y.; Verrier, E.; Rognon, X. How do introgression events shape the partitioning of diversity among breeds: A case study in sheep. Genet. Sel. Evol. 2015, 47, 48. [Google Scholar] [CrossRef]
- Ben Sassi-Zaidy, Y.; Maretto, F.; Charfi-cheikhrouha, F.; Mohamed-Brahmi, A.; Cassandro, M. Contribution of microsatellites markers in the clarification of the origin, genetic risk factors, and implications for conservation of Tunisian native sheep breeds. Genet. Mol. Res. 2016, 15, 15017059. [Google Scholar] [CrossRef] [PubMed]
- Gaouar, S.B.S.; Kdidi, S.; Ouragh, L. Estimating population structure and genetic diversity of five Moroccan sheep breeds by microsatellite markers. Small Rumin. Res. 2016, 144, 23–27. [Google Scholar] [CrossRef]
- Landi, V.; Lasagna, E.; Ceccobelli, S.; Martinez, A.; Santos-Silva, F.; Vega-Pla, J.L.; Panella, F.; Allain, D.; Palhiere, I.; Murawski, M.; et al. An historical and biogeographical assessment of European Merino sheep breeds by microsatellite markers. Small Rumin. Res. 2019, 177, 76–81. [Google Scholar] [CrossRef]
- Kandoussi, A.; Boujenane, I.; Auger, C.; Serranito, B.; Germot, A.; Piro, M.; Maftah, A.; Badaoui, B.; Petit, D. The origin of sheep settlement in Western Mediterranean. Sci. Rep. 2020, 10, 10225. [Google Scholar] [CrossRef]
- Ben Jemaa, S.; Kdidi, S.; Gdura, A.M.; Dayhum, A.S.; Eldaghayes, I.M.; Boussaha, M.; Rebours, E.; Yahyaoui, M.H. Inferring the population structure of the Maghreb sheep breeds using a medium-density SNP chip. Anim. Genet. 2019, 50, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Ghernouti, N.; Bodinier, M.; Ranebi, D.; Maftah, A.; Petit, D.; Gaouar, S.B.S. Control Region of mtDNA identifies three migration events of sheep breeds in Algeria. Small Rumin. Res. 2017, 155, 66–71. [Google Scholar] [CrossRef]
- Rekik, M.; Aloulou, R.; Ben Hamouda, M. Small ruminant breeds of Tunisia. In Characterisation of Small Ruminant Breeds in West Asia and North Africa; Iniguez, L., Ed.; North Africa; International Centre for Agricultural Research in the Dry Areas (ICARDA): Aleppo, Syria, 2005; pp. 91–140. [Google Scholar]
- Khaldi, Z.; Haddad, B.; Souid, S.; Rouissi, H.; Ben Gara, A.; Rekik, B. Caracterisation Phenotypique de la Population Ovine du Sud Ouest de la Tunisie. Anim. Genet. Resour. Génétiques Anim. Genéticos Anim. 2011, 49, 1–8. [Google Scholar] [CrossRef]
- Bondesan, V.; Tormen, N.; Bittante, G.; Ribeca, C.; Pellettero, E.; Stelletta, C.; Vencato, Y.; Schiavon, E.; Mutinelli, F.; Granato, A.; et al. Conservazione e Caratterizzazione delle Razze Ovine Venete—Programma Bionet. Rete Regionale per la Conservazione e Caratterizzazione della Biodiversità di Interesse Agrario; Gruppo di Lavoro Ovini, Veneto Agricoltura: Legnaro, Italy, 2014. [Google Scholar]
- ISAG Standing Committee. Applied Genetics in Sheep and Goats. Workshop Report, ISAG Conference 2014, Xi’an, China. 2014. Available online: https://www.isag.us/Docs/AppGenSheepGoat2014.pdf (accessed on 16 February 2016).
- Baumung, R.; Schwend, K.; Achmann, R.; So, J. Genetic characterisation and breed assignment in Austrian sheep breeds using microsatellite marker information. J. Anim. Breed. Genet. 2006, 123, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; De Marchi, M.; Zanetti, E.; Cassandro, M. Genetic variation and population structure of Italian native sheep breeds undergoing in situ conservation. J. Anim. Sci. 2009, 87, 3837–3844. [Google Scholar] [CrossRef] [PubMed]
- Ben Sassi-Zaidy, Y.; Mohamed-Brahmi, A.; Nouairia, G.; Charfi-Cheikhrouha, F.; Djemali, M.N.; Cassandro, M. Genetic Variability and Population Structure of the Tunisian Sicilo-Sarde Dairy Sheep Breed Inferred from Microsatellites Analysis. Genes 2022, 13, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.; Wills, D.; Shipley, P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlötterer, C. Microsatellite analyser (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel Sous WindowsTM pour la Génétique des Populations. Laboratoire Génome, Populations, Interaction. CNRS UMR 5000, Université de Montpellier II: Montpellier, France; Available online: http://www.genetix.univ-montp2.fr/genetix/intro.htm (accessed on 19 April 2022).
- Gutierrez, J.P.; Royo, L.J.; Alvarez, I.; Goyache, F. MolKin v2.0: A Computer Program for Genetic Analysis of Populations Using Molecular Coancestry Information. J. Hered. 2005, 96, 718–721. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Zhivotovsky, L.A. Estimating Divergence Time with the Use of Microsatellite Genetic Distances: Impacts of Population Growth and Gene Flow. Mol. Biol. Evol. 2001, 18, 700–709. [Google Scholar] [CrossRef]
- Alvarez, I.; Gutiérrez, J.P.; Royo, L.J.; Fernandez, I.; Gomez, E.; Arranz, J.J.; Goyache, F. Testing the usefulness of the molecular coancestry information to assess genetic relationships in livestock using a set of Spanish sheep breeds. J. Anim. Sci. 2005, 83, 737–744. [Google Scholar] [CrossRef]
- Richard, M.; Thorpe, R.S. Can Microsatellites Be Used to Infer Phylogenies? Evidence from Population Affinities of the Western Canary Island Lizard (Gallotia galloti). Mol. Phylogenet. Evol. 2001, 20, 351–360. [Google Scholar] [CrossRef]
- Ramakrishnan, U.; Mountain, J.L. Precision and Accuracy of Divergence Time Estimates from STR and SNPSTR Variation. Mol. Biol. Evol. 2004, 21, 1960–1971. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogeny inference package (PHYLIP). In Genomes Sciences; Software; Departement of Genetics, University of Washington: Seattle, WA, USA, 1989; Available online: http://evolution.gs.washington.edu/phylip.html (accessed on 27 April 2022).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Puigbo, P.; Major, J.M. GPT: A web-server to map phylogenetic trees on a virtual globe. PeerJ Prepr. 2015, 3, e840v1. [Google Scholar] [CrossRef]
- Sheriff, O.; Alemayehu, K. Genetic diversity studies using microsatellite markers and their contribution in supporting sustainable sheep breeding programs: A review. Cogent Food Agric. 2018, 4, 1459062. [Google Scholar] [CrossRef]
- Olschewsky, A.; Hinrichs, D. An overview of the use of genotyping techniques for assessing genetic diversity in local farm animal breeds. Animals 2021, 11, 2016. [Google Scholar] [CrossRef]
- Trouette, G. L’élevage Indigène en Algérie; Pub Sociét: Paris, France, 1931. [Google Scholar]
- Djamaï, A.; Zebiri, M.E. L’activité Sexuelle de la Brebis. Ph.D. Thesis, Université Mentouri de Constantine, Constantine, Algeria, 2007. [Google Scholar]
- Edea, Z.; Dessie, T.; Dadi, H.; Do, K.-T.; Kim, K.-S. Genetic Diversity and Population Structure of Ethiopian Sheep Populations Revealed by High-Density SNP Markers. Front. Genet. 2017, 8, 218. [Google Scholar] [CrossRef]
- Lawson Handley, L.-J.; Byrne, K.; Santucci, F.; Townsend, S.; Taylor, M.; Bruford, M.W.; Hewitt, G.M. Genetic structure of European sheep breeds. Heredity 2007, 99, 620–631. [Google Scholar] [CrossRef]
- Goldstein, D.B.; Ruiz Linares, A.; Cavalli-Sforza, L.L.; Feldman, M.W. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. USA 1995, 92, 6723–6727. [Google Scholar] [CrossRef]
- Pérez, T.; Albornoz, J.; Domínguez, A. Phylogeography of chamois (Rupicapra spp.) inferred from microsatellites. Mol. Phylogenet. Evol. 2002, 25, 524–534. [Google Scholar] [CrossRef]
- Ciani, E.; Lasagna, E.; D’Andrea, M.; Alloggio, I.; Marroni, F.; Ceccobelli, S.; Delgado Bermejo, J.V.; Sarti, F.M.; Kijas, J.; Lenstra, J.A.; et al. Merino and Merino-derived sheep breeds: A genome-wide intercontinental study. Genet. Sel. Evol. 2015, 47, 64. [Google Scholar] [CrossRef]
- Ciani, E.; Crepaldi, P.; Nicoloso, L.; Lasagna, E.; Sarti, F.M.; Moioli, B.; Napolitano, F.; Carta, A.; Usai, G.; D’Andrea, M.; et al. Genome-wide analysis of Italian sheep diversity reveals a strong geographic pattern and cryptic relationships between breeds. Anim. Genet. 2014, 45, 256–266. [Google Scholar] [CrossRef]
- Pastore, E. Le Razze Ovine Autoctone del Veneto; Isabella Lavezzo, M.M., Ed.; Veneto Agricoltura—Settore Ricerca e Sperimentazione Agraria ed Ittica, Viale dell’Università: Legnago, Italy, 2005. [Google Scholar]
- Creech, T.G.; Epps, C.W.; Wehausen, J.D.; Crowhurst, R.S.; Jaeger, J.R.; Longshore, K.; Holton, B.; Sloan, W.B.; Monello, R.J. Genetic and Environmental Indicators of Climate Change Vulnerability for Desert Bighorn Sheep. Front. Ecol. Evol. 2020, 8, 279. [Google Scholar] [CrossRef]
- Crawford, A.M.; Cuthbertson, R.P. Mutations in sheep microsatellites. Genome Res. 1996, 6, 876–879. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat. Genet. 2000, 24, 400–402. [Google Scholar] [CrossRef]
- Li, B.; Kimmel, M. Factors influencing ascertainment bias of microsatellite allele sizes: Impact on estimates of mutation rates. Genetics 2013, 195, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Elfawal, M.A.; Galal, S.; Abdelsalam, A.Z.E.; Mona, A. Osman4 Hassanane, M.S. Microsatellite polymorphism in three Egyptian sheep breeds. Egypt. J. Anim. Prod. 2008, 45, 1–14. [Google Scholar]
- Esmaeilkhanian, S.; Banabazi, M.H. Genetic Variation Within and Between Five Iranian Sheep Populations Using Microsatellites Markers. Pakistan J. Biol. Sci. 2006, 9, 2488–2492. [Google Scholar] [CrossRef]
- Rochus, C.M.; Johansson, A.M. Estimation of genetic diversity in Gute sheep: Pedigree and microsatellite analyses of an ancient Swedish breed. Hereditas 2017, 154, 4. [Google Scholar] [CrossRef] [PubMed]
- Ben Gara, A. Définition des objectifs de la sélection des ovins de race Barbarine en Tunisie. In Analysis and Definition of the Objectives in Genetic Improvement Programmes in Sheep and Goats. An Economic Approach to Increase Their Profitability; Options Méditerranéennes: Série A. Séminaires Méditerranéens; n. 43; CIHEAM: Zaragoza, Spain, 2000; pp. 111–116. [Google Scholar]
- Chessa, B.; Pereira, F.; Arnaud, F.; Amorim, A.; Goyache, F.; Mainland, I.; Kao, R.R.; Pemberton, J.M.; Beraldi, D.; Stear, M.J.; et al. Revealing the history of sheep domestication using retrovirus integrations. Science 2009, 324, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Barbato, M.; Hailer, F.; Orozco-Terwengel, P.; Kijas, J.; Mereu, P.; Cabras, P.; Mazza, R.; Pirastru, M.; Bruford, M.W. Genomic signatures of adaptive introgression from European mouflon into domestic sheep. Sci. Rep. 2017, 7, 7623. [Google Scholar] [CrossRef]
- Vigne, J.-D.; Zazzo, A.; Cucchi, T.; Briois, F.; Guilaine, J. The transportation of mammals to Cyprus shed light on early voyaging and boats in the mediterranean sea. Eurasian Prehistory 2014, 10, 157–176. [Google Scholar]
- Moradi, M.H.; Nejati-Javaremi, A.; Moradi-Shahrbabak, M.; Dodds, K.G.; McEwan, J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genet. 2012, 13, 10. [Google Scholar] [CrossRef]
- Elkamel, S.; Boussetta, S.; Khodjet-El-Khil, H.; Benammar Elgaaied, A.; Cherni, L. Ancient and recent Middle Eastern maternal genetic contribution to North Africa as viewed by mtDNA diversity in Tunisian Arab populations. Am. J. Hum. Biol. 2018, 30, e23100. [Google Scholar] [CrossRef]
- Rowley-Conwy, P.; Gourichon, L.; Helmer, D.; Vigne, J.-D. Early domestic animals in Italy, Istria, the Tyrrhenian Islands and Southern France. In The Origins and Spread of Domestic Animals in Southwest Asia and Europe; Routledge: New York, NY, USA, 2013; pp. 161–194. [Google Scholar]
- Demirci, S.; Koban Baştanlar, E.; Dağtaş, N.D.; Pişkin, E.; Engin, A.; Özer, F.; Yüncü, E.; Doğan, Ş.A.; Togan, İ. Mitochondrial DNA Diversity of Modern, Ancient and Wild Sheep (Ovis gmelinii anatolica) from Turkey: New Insights on the Evolutionary History of Sheep. PLoS ONE 2013, 8, e81952. [Google Scholar] [CrossRef]
- Sanna, D.; Barbato, M.; Hadjisterkotis, E.; Cossu, P.; Decandia, L.; Trova, S.; Pirastru, M.; Leoni, G.G.; Naitana, S.; Francalacci, P.; et al. The First Mitogenome of the Cyprus Mouflon (Ovis gmelini ophion): New Insights into the Phylogeny of the Genus Ovis. PLoS ONE 2015, 10, e0144257. [Google Scholar] [CrossRef]
- Cruciani, F.; La Fratta, R.; Trombetta, B.; Santolamazza, P.; Sellitto, D.; Colomb, E.B.; Dugoujon, J.-M.; Crivellaro, F.; Benincasa, T.; Pascone, R.; et al. Tracing past human male movements in northern/eastern Africa and western Eurasia: New clues from Y-chromosomal haplogroups E-M78 and J-M12. Mol. Biol. Evol. 2007, 24, 1300–1311. [Google Scholar] [CrossRef]
- Finsinger, W.; Tinner, W.; Van Der Knaap, W.O.; Ammann, B. The expansion of hazel (Corylus avellana L.) in the southern Alps: A key for understanding its early Holocene history in Europe? Quat. Sci. Rev. 2006, 25, 612–631. [Google Scholar] [CrossRef]
- Zeder, M.A. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc. Natl. Acad. Sci. USA 2008, 105, 11597–11604. [Google Scholar] [CrossRef]
- Francalacci, P.; Sanna, D. History and geography of human Y-chromosome in Europe: A SNP perspective. JASS Invit. Rev. J. Anthropol. Sci. 2008, 86, 59–89. [Google Scholar]
- Battaglia, V.; Fornarino, S.; Al-Zahery, N.; Olivieri, A.; Pala, M.; Myres, N.M.; King, R.J.; Rootsi, S.; Marjanovic, D.; Primorac, D.; et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur. J. Hum. Genet. 2009, 17, 820–830. [Google Scholar] [CrossRef]
- Szécsényi-Nagy, A.; Brandt, G.; Haak, W.; Keerl, V.; Jakucs, J.; Möller-Rieker, S.; Köhler, K.; Mende, B.G.; Oross, K.; Marton, T.; et al. Tracing the genetic origin of Europe’s first farmers reveals insights into their social organization. Proc. Biol. Sci. 2015, 282, 20150339. [Google Scholar] [CrossRef]
- Zeder, M.A. Out of the Fertile Crescent: The dispersal of domestic livestock through Europe and Africa. In Human Dispersal and Species Movement, from Prehistory to the Present; Boivin, N., Petraglia, M., Crassard, R., Eds.; Cambridge University Press: Cambridge, UK, 2017; pp. 261–303. [Google Scholar]
- Cheddadi, R.; Carré, M.; Nourelbait, M.; François, L.; Rhoujjati, A.; Manay, R.; Ochoa, D.; Schefuß, E. Early Holocene greening of the Sahara requires Mediterranean winter rainfall. Proc. Natl. Acad. Sci. USA 2021, 118, e2024898118. [Google Scholar] [CrossRef]
- Berger, J.F. Geoarchaeological and Paleo-Hydrological Overview of the Central-Western Mediterranean Early Neolithic Human-Environment Interactions. Open Archaeol. 2021, 7, 1371–1397. [Google Scholar] [CrossRef]
- Hassan, F.A. Holocene Palaeoclimates of Africa. Afr. Archaeol. Rev. 1997, 14, 213–230. [Google Scholar] [CrossRef]
- Ryder, M.L. The History of Sheep Breeds in Britain. Agric. Hist. Rev. 1964, 12, 65–82. [Google Scholar]
- Fleming, A. Soay Sheep: The Back-story. Camb. Archaeol. J. 2021, 31, 419–436. [Google Scholar] [CrossRef]
- Dýrmundsson, Ó.R.; Niżnikowski, R. North European short-tailed breeds of sheep: A review. Animal 2010, 4, 1275–1282. [Google Scholar] [CrossRef]
- Scobie, D.R.; O’connell, D. Genetic reduction of tail length in New Zealand sheep. Proc. N. Zeal. Soc. Anim. Prod. 2002, 62, 195–198. [Google Scholar]
- Kovacevic, L.; Tambets, K.; Ilumäe, A.-M.; Kushniarevich, A.; Yunusbayev, B.; Solnik, A.; Bego, T.; Primorac, D.; Skaro, V.; Leskovac, A.; et al. Standing at the Gateway to Europe—The Genetic Structure of Western Balkan Populations Based on Autosomal and Haploid Markers. PLoS ONE 2014, 9, e105090. [Google Scholar] [CrossRef]
- Sanson, A. Traité de zootechnie. Tome 5: Zoologie et Zootechnie Spéciales, Ovidés Ariétins et Caprins, Suidés et Porcins; Bibliothèque Agricole; Librairie Agricole de La Maison Rustique: Paris, France, 1886. [Google Scholar]
- Corti, M. La Pecora Bergamasca. Storia e Presente di una Razza Ovina; Ed.: Provin, France, 1999. [Google Scholar]
- Bonacini, I.; Lauvergne, J.-J.; Succi, G.; Rognoni, G. Etude du profil génétique des ovins de l’Arc Alpin italien à l’aide de marqueurs à effets visibles. Genet. Sel. Evol. 1982, 14, 417–434. [Google Scholar] [CrossRef]
- Corti, M.; Foppa, G. La Pecora Bergamasca (The Bergamasca Sheep), 1st ed.; FAO: Bergamo, Italy, 1999; p. 156. [Google Scholar]
- Camps, G. Le mouton au Néolithique ancien dans les pays de la Méditerranée occidentale. In Premières Communautés Paysannes en Méditerranée Occidentale: Actes du Colloque International du CNRS, Montpellier, France, 26–29 Avril 1983; Guilaine, J., Courtin, J., Roudil, J., Vernet, J., Eds.; CNRS: Paris, France, 1987; pp. 209–214. [Google Scholar]
- Camps, G. Bélier à sphéroïde. Encycl. Berbère 1991, 9, 1417–1433. [Google Scholar] [CrossRef]
- Fitzinger, L.J.F.J. Über die Racen des Zahmen Schafes. Sitz. Akad. Wiss. Math. Kl. 1860, 41, 151–246. [Google Scholar]
- Joleaud, L. Bœufs, Moutons et Chèvres sauvages de Berbérie aux temps préhistoriques. Terre Vie 1933, 10, 150–174. [Google Scholar]
- Ben Nasr, J. Des gravures rupestres de la Tunisie Centrale. Sahara 2012, 23, 113. [Google Scholar]
- Ben Nasr, J.; Walsh, K.J. Environment and Rock Art in the Jebel Ousselat, Atlas Mountains, Tunisia. J. Mediterr. Archaeol. 2020, 33, 3–28. [Google Scholar] [CrossRef]
- Tourte, R. Aux sources de l’agriculture africaine: De la préhistoire au moyen âge. In Histoire de la Recherche Agricole en Afrique Tropicale Farncophone; FAO: Rome, Italy, 2005. [Google Scholar]
- Wilson, R.T. Small Ruminant Production and the Small Ruminant Genetic Resource in Tropical Africa; Animal Production and Health Paper N° 88; FAO ANIMAL; Food and Agriculture Organization of the United Nations: Rome, Italy, 1991; p. 231. ISBN 9251029989. [Google Scholar]
- Kröpelin, S.; Verschuren, D.; Lézine, A.-M.; Eggermont, H.; Cocquyt, C.; Francus, P.; Cazet, J.-P.; Fagot, M.; Rumes, B.; Russell, J.M.; et al. Climate-driven ecosystem succession in the Sahara: The past 6000 years. Science 2008, 320, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.B. Origins and Spread of Pastoralism in Africa. Annu. Rev. Anthropol. 1992, 21, 125–141. [Google Scholar] [CrossRef]
- Pereira, F.; Amorim, A.; Pereira, F.; Amorim, A. Origin and Spread of Goat Pastoralism. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2010; ISBN 9780470015902. [Google Scholar]
- Ben Jemaa, S.; Kdidi, S.; Boussaha, M.; Rebours, E.; YahyAoui, M.H. Genomic inbreeding and population structure in rams of Tunisian D’man sheep. J. New Sci. 2019, 54, 531–537. [Google Scholar] [CrossRef]
- Pereira, L.; Cerný, V.; Cerezo, M.; Silva, N.M.; Hájek, M.; Vasíková, A.; Kujanová, M.; Brdicka, R.; Salas, A. Linking the sub-Saharan and West Eurasian gene pools: Maternal and paternal heritage of the Tuareg nomads from the African Sahel. Eur. J. Hum. Genet. 2010, 18, 915–923. [Google Scholar] [CrossRef]
- Spangler, G.L.; Rosen, B.D.; Ilori, M.B.; Hanotte, O.; Kim, E.S.; Sonstegard, T.S.; Burke, J.M.; Morgan, J.L.M.; Notter, D.R.; Van Tassell, C.P. Whole genome structural analysis of Caribbean hair sheep reveals quantitative link to West African ancestry. PLoS ONE 2017, 12, e0179021. [Google Scholar] [CrossRef]
- Olivieri, C.; Ermini, L.; Rizzi, E.; Corti, G.; Luciani, S.; Marota, I.; de Bellis, G.; Rollo, F. Phylogenetic position of a copper age sheep (Ovis aries) mitochondrial DNA. PLoS ONE 2012, 7, e33792. [Google Scholar] [CrossRef]
- Grömer, K.; Saliari, K.; Erich Pucher, F. Dressing Central European prehistory-the sheep’s contribution An interdisciplinary study about archaeological textile finds and archaeozoology. Ann. Naturhist. Mus. Wien Ser. A 2018, 120, 127–156. [Google Scholar]
- Vila, E.; Helmer, D. The Expansion of Sheep Herding and the Development of Wool Production in the Ancient Near East: In Wool Economy in the Ancient Near East; Breniquet, C., Cécile, M., Eds.; Oxbow Books: Oxford, UK, 2019; pp. 22–40. [Google Scholar]
- Gleba, M. Sheep to Textiles: Approaches to Investigating Ancient Wool Trade. In Textile Trading and Distribution in Antiquity; Harrassowitz: Wiesbaden, Germany, 2014; Volume 73, pp. 123–133. [Google Scholar]
- Lawrence, D.; Philip, G.; de Gruchy, M.W. Climate change and early urbanism in Southwest Asia: A review. Wiley Interdiscip. Rev. Clim. Chang. 2022, 13, e741. [Google Scholar] [CrossRef]
- Jones, M.D.; Abu-Jaber, N.; AlShdaifat, A.; Baird, D.; Cook, B.I.; Cuthbert, M.O.; Dean, J.R.; Djamali, M.; Eastwood, W.; Fleitmann, D.; et al. 20,000 years of societal vulnerability and adaptation to climate change in southwest Asia. Wiley Interdiscip. Rev. Water 2019, 6, e1330. [Google Scholar] [CrossRef]
- Demars, J.; Cano, M.; Drouilhet, L.; Plisson-Petit, F.; Bardou, P.; Fabre, S.; Servin, B.; Sarry, J.; Woloszyn, F.; Mulsant, P.; et al. Genome-Wide Identification of the Mutation Underlying Fleece Variation and Discriminating Ancestral Hairy Species from Modern Woolly Sheep. Mol. Biol. Evol. 2017, 34, 1722–1729. [Google Scholar] [CrossRef]
- Becker, C.; Benecke, N.; Grabundžija, A.; Küchelmann, H.-C.; Pollock, S.; Schier, W.; Schoch, C.; Schrakamp, I.; Schütt, B.; Schumacher, M. The textile revolution. Research into the origin and spread of wool production between the Near East and Central Europe. eTopoi J. Anc. Stud. 2016, 6, 102–151. [Google Scholar]
- Sabatini, S.; Bergerbrant, S.; Brandt, L.; Margaryan, A.; Allentoft, M.E. Approaching sheep herds origins and the emergence of the wool economy in continental Europe during the Bronze Age. Archaeol. Anthropol. Sci. 2019, 11, 4909–4925. [Google Scholar] [CrossRef]
- Trixl, S. The biometry of prehistoric Alpine sheep: Exploring four millennia of human-sheep interaction by means of osteometry. Anthropozoologica 2022, 57, 117–139. [Google Scholar] [CrossRef]
- Wallis Budge, E.A. From Fetish to God in Ancient Egypt; Oxford University Press: Oxford, UK; London, UK, 1934. [Google Scholar]
- Mason, T.L. The Sheep Breeds of the Mediterranean; FAO CAB: Edinburgh, UK, 1967; p. 215. [Google Scholar]
- Babelon, E. Carthage et l’Archéologie Punique en Tunisie. Am. J. Archaeol. Hist. Fine Arts 1885, 1, 173–181. Available online: https://www.jstor.org/stable/pdf/496350.pdf (accessed on 10 June 2022). [CrossRef]
- McIntosh, M.A. The History of Ancient Canaan (Palestine). Time Maps World Hist. Encycl. 2018. Available online: https://brewminate.com/the-history-of-ancient-canaan-palestine (accessed on 10 June 2022).
- Delile, H.; Pleuger, E.; Blichert-Toft, J.; Goiran, J.P.; Fagel, N.; Gadhoum, A.; Abichou, A.; Ben Jerbania, I.; Fentress, E.; Wilson, A.I. Economic resilience of Carthage during the Punic Wars: Insights from sediments of the Medjerda delta around Utica (Tunisia). Proc. Natl. Acad. Sci. USA 2019, 116, 9764–9769. [Google Scholar] [CrossRef]
- Stephens, L.; Fuller, D.; Boivin, N.; Rick, T.; Gauthier, N.; Kay, A.; Marwick, B.; Armstrong, C.G.D.; Barton, C.M.; Denham, T.; et al. Archaeological assessment reveals Earth’s early transformation through land use. Science 2019, 365, 897–902. [Google Scholar] [CrossRef]
- Deng, J.; Xie, X.L.; Wang, D.F.; Zhao, C.; Lv, F.H.; Li, X.; Yang, J.; Yu, J.L.; Shen, M.; Gao, L.; et al. Paternal Origins and Migratory Episodes of Domestic Sheep. Curr. Biol. 2020, 30, 4085–4095.e6. [Google Scholar] [CrossRef]
- Sarson, M. Les ovins dans l’antiquité d’après les vestiges phéniciens et romains en Tunisie et en Algérie. Doc. Tech. INRAT 1973, 65, 30. [Google Scholar]
- Gaouar, S.B.S.; Lafri, M.; Djaout, A.; El-Bouyahiaoui, R.; Bouri, A.; Bouchatal, A.; Maftah, A.; Ciani, E.; Da Silva, A.B. Genome-wide analysis highlights genetic dilution in Algerian sheep. Heredity 2016, 118, 293–301. [Google Scholar] [CrossRef]



| Region | Breed | Tail | Horn | Fleece/Coat Color | Photos * |
|---|---|---|---|---|---|
| Maghrebian | BAR | Fat tail | Presence in (  ) rare in ( ) rare in (  ) ) | Woolly (white or brown fleece)/black or red head and legs or mixed (white and black or red legs, muzzle and eye area) |  |
| QFO | Thin tail | Frequent (  ) low frequency ( ) low frequency (  ) ) | Woolly (white fleece)/white coat |  | |
| CRO | Intermediate tail | - | BAR-like fleece and coat |  | |
| NTH | Thin tail | Hornless in both sex | Woolly (dark fleece)/dark coat |  | |
| SS | Thin tail | Presence in both sex | Woolly (white fleece)/white coat |  | |
| DM | Long and thin tail | Hornless in both sex | Mixed hairy and woolly with natural molting and presence of hairy man in rams/colored and spotted coat |  | |
| Italian | ALP | Long and thin tail | Hornless in both sex | Woolly (white fleece)/brown-spotted coat |  |
| BRO | Long and thin tail | Rarely present in (  ) ) | Woolly (white fleece)/red-spotted coat |  | |
| FOZ | Thin tail | Hornless in both sex | Woolly (white fleece)/black-spotted coat |  | |
| LAM | Long and thin tail | Hornless in both sex | Woolly (white fleece)/dark brown Spotted coat |  | |
| APP | Long and thin tail | Hornless in both sex | Woolly (white fleece)/white coat |  |
| Locus | Size | Chr | TNA | AR | PIC |
|---|---|---|---|---|---|
| Inra023 | 195–221 | 1 | 17 | 10.25 | 0.87 |
| Inra063 | 168–208 | 14 | 28 | 10.40 | 0.82 |
| OarCP49 | 71–137 | 17 | 31 | 13.34 | 0.85 |
| OarFCB304 | 145–201 | 19 | 24 | 8.96 | 0.76 |
| OarFCB20 | 85–121 | 2 | 19 | 10.26 | 0.84 |
| MAF65 | 113–139 | 15 | 21 | 7.75 | 0.75 |
| ILST087 | 134–184 | 6 | 26 | 13.17 | 0.88 |
| OarAE119 | 145–185 | 19 | 20 | 9.51 | 0.82 |
| MCM527 | 164–190 | 5 | 17 | 7.89 | 0.78 |
| MAF214 | 182–262 | 16 | 38 | 6.96 | 0.61 |
| OarAE129 | 135–165 | 5 | 16 | 5.26 | 0.63 |
| OarCP34 | 93–117 | 3 | 18 | 6.62 | 0.77 |
| OarAE54 | 120–152 | 25 | 19 | 10.44 | 0.82 |
| TGLA | 125–163 | 12 | 16 | 9.67 | 0.84 |
| URB | 159–211 | 13 | 24 | 9.93 | 0.86 |
| CSRD | 208–262 | 14 | 28 | 9.74 | 0.84 |
| HSC | 260–296 | 20 | 21 | 9.33 | 0.85 |
| Average | - | - | 22.53 | 9.38 | 0.80 |
| SD | - | - | 6.07 | 2.09 | 0.08 |
| FST | LAM | FOZ | ALP | BRO | DM | CRO | NTH | QFO | SS | BAR | APP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LAM | - | 0.042 | 0.038 | 0.039 | 0.102 | 0.118 | 0.114 | 0.105 | 0.118 | 0.118 | 0.126 |
| FOZ | - | 0.053 | 0.041 | 0.098 | 0.110 | 0.098 | 0.096 | 0.111 | 0.110 | 0.116 | |
| ALP | - | 0.040 | 0.114 | 0.130 | 0.126 | 0.124 | 0.132 | 0.137 | 0.147 | ||
| BRO | - | 0.105 | 0.114 | 0.113 | 0.106 | 0.122 | 0.116 | 0.128 | |||
| DM | - | 0.029 | 0.034 | 0.021 | 0.043 | 0.032 | 0.073 | ||||
| CRO | - | 0.018 | 0.006 * | 0.023 | 0.008 * | 0.070 | |||||
| NTH | - | 0.014 | 0.020 | 0.024 | 0.069 | ||||||
| QFO | - | 0.018 | 0.005 * | 0.052 | |||||||
| SS | - | 0.028 | 0.057 | ||||||||
| BAR | - | 0.067 | |||||||||
| APP | - |
| fij/Nm | DM | CRO | NTH | QFO | SS | BAR | APP | LAM | FOZ | ALP | BRO |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DM | - | 8.63 | 7.12 | 11.23 | 5.60 | 7.33 | 3.20 | 2.03 | 2.27 | 1.80 | 1.74 |
| CRO | 0.17 | - | 13.89 | 50.65 | 10.87 | 30.54 | 3.33 | 1.80 | 2.08 | 1.49 | 1.66 |
| NTH | 0.16 | 0.18 | - | 18.13 | 11.80 | 10.24 | 3.39 | 1.78 | 2.23 | 1.52 | 1.64 |
| QFO | 0.16 | 0.18 | 0.17 | - | 13.59 | 43.91 | 4.55 | 2.02 | 2.36 | 1.60 | 1.87 |
| SS | 0.15 | 0.17 | 0.17 | 0.16 | - | 8.64 | 4.06 | 1.76 | 1.97 | 1.45 | 1.53 |
| BAR | 0.16 | 0.19 | 0.17 | 0.18 | 0.17 | - | 3.44 | 1.70 | 2.02 | 1.43 | 1.63 |
| APP | 0.15 | 0.17 | 0.16 | 0.17 | 0.17 | 0.17 | - | 1.60 | 1.85 | 1.25 | 1.37 |
| LAM | 0.10 | 0.10 | 0.09 | 0.10 | 0.09 | 0.09 | 0.12 | - | 5.27 | 3.98 | 3.24 |
| FOZ | 0.10 | 0.10 | 0.10 | 0.10 | 0.09 | 0.09 | 0.11 | 0.17 | - | 3.73 | 4.12 |
| ALP | 0.12 | 0.11 | 0.10 | 0.10 | 0.09 | 0.09 | 0.11 | 0.20 | 0.18 | - | 3.06 |
| BRO | 0.10 | 0.10 | 0.10 | 0.10 | 0.09 | 0.10 | 0.11 | 0.18 | 0.17 | 0.21 | - |
| (δµ)2/DS | LAM | FOZ | ALP | BRO | DM | CRO | NTH | QFO | SS | BAR | APP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LAM | - | 0.186 | 0.142 | 0.162 | 0.486 | 0.541 | 0.532 | 0.509 | 0.564 | 0.555 | 0.534 |
| FOZ | 1.652 | - | 0.205 | 0.169 | 0.475 | 0.510 | 0.461 | 0.474 | 0.536 | 0.521 | 0.492 |
| ALP | 1.757 | 0.932 | - | 0.147 | 0.482 | 0.538 | 0.528 | 0.538 | 0.563 | 0.585 | 0.571 |
| BRO | 1.753 | 1.116 | 1.679 | - | 0.478 | 0.499 | 0.501 | 0.492 | 0.558 | 0.521 | 0.523 |
| DM | 5.949 | 6.402 | 5.635 | 6.442 | - | 0.132 | 0.156 | 0.108 | 0.201 | 0.146 | 0.298 |
| CRO | 2.752 | 2.841 | 3.102 | 3.391 | 2.470 | - | 0.079 | 0.030 | 0.104 | 0.037 | 0.275 |
| NTH | 3.069 | 3.141 | 3.801 | 4.471 | 3.578 | 0.458 | - | 0.065 | 0.090 | 0.105 | 0.273 |
| QFO | 2.503 | 2.829 | 3.146 | 3.633 | 3.508 | 0.692 | 0.540 | - | 0.089 | 0.025 | 0.213 |
| SS | 2.118 | 2.266 | 3.074 | 3.208 | 4.055 | 0.925 | 0.717 | 0.998 | - | 0.125 | 0.230 |
| BAR | 3.165 | 3.023 | 3.325 | 3.258 | 3.378 | 1.150 | 1.390 | 0.864 | 2.149 | - | 0.267 |
| APP | 3.220 | 3.852 | 5.869 | 2.954 | 6.609 | 3.340 | 3.756 | 3.899 | 2.366 | 4.166 | - |
| Y. Ago | LAM | FOZ | ALP | BRO | DM | CRO | NTH | QFO | SS | BAR | APP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LAM | |||||||||||
| FOZ | 2753.3 | ||||||||||
| ALP | 2928.3 | 1553.3 | |||||||||
| BRO | 2921.7 | 1860.0 | 2798.8 | ||||||||
| DM | 9915.0 | 10,670.0 | 9392.0 | 10,736.3 | |||||||
| CRO | 4586.7 | 4735.0 | 5169.2 | 5652.4 | 4117.4 | ||||||
| NTH | 5115.0 | 5235.0 | 6334.2 | 7451.7 | 5963.3 | 763.8 | |||||
| QFO | 4171.7 | 4715.0 | 5243.7 | 6055.0 | 5847.4 | 1154.0 | 900.8 | ||||
| SS | 3530.0 | 3776.7 | 5123.7 | 5346.4 | 6758.7 | 1541.2 | 1195.6 | 1663.6 | |||
| BAR | 5275.0 | 5038.3 | 5542.5 | 5430.6 | 5629.6 | 1916.6 | 2316.3 | 1440.8 | 3582.3 | ||
| APP | 5366.7 | 6420.0 | 9781.8 | 4922.8 | 11,015.0 | 5566.3 | 6260.8 | 6498.1 | 3943.4 | 6943.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Sassi-Zaidy, Y.; Mohamed-Brahmi, A.; Chaouch, M.; Maretto, F.; Cendron, F.; Charfi-Cheikhrouha, F.; Ben Abderrazak, S.; Djemali, M.; Cassandro, M. Historical Westward Migration Phases of Ovis aries Inferred from the Population Structure and the Phylogeography of Occidental Mediterranean Native Sheep Breeds. Genes 2022, 13, 1421. https://doi.org/10.3390/genes13081421
Ben Sassi-Zaidy Y, Mohamed-Brahmi A, Chaouch M, Maretto F, Cendron F, Charfi-Cheikhrouha F, Ben Abderrazak S, Djemali M, Cassandro M. Historical Westward Migration Phases of Ovis aries Inferred from the Population Structure and the Phylogeography of Occidental Mediterranean Native Sheep Breeds. Genes. 2022; 13(8):1421. https://doi.org/10.3390/genes13081421
Chicago/Turabian StyleBen Sassi-Zaidy, Yousra, Aziza Mohamed-Brahmi, Melek Chaouch, Fabio Maretto, Filippo Cendron, Faouzia Charfi-Cheikhrouha, Souha Ben Abderrazak, Mnaour Djemali, and Martino Cassandro. 2022. "Historical Westward Migration Phases of Ovis aries Inferred from the Population Structure and the Phylogeography of Occidental Mediterranean Native Sheep Breeds" Genes 13, no. 8: 1421. https://doi.org/10.3390/genes13081421
APA StyleBen Sassi-Zaidy, Y., Mohamed-Brahmi, A., Chaouch, M., Maretto, F., Cendron, F., Charfi-Cheikhrouha, F., Ben Abderrazak, S., Djemali, M., & Cassandro, M. (2022). Historical Westward Migration Phases of Ovis aries Inferred from the Population Structure and the Phylogeography of Occidental Mediterranean Native Sheep Breeds. Genes, 13(8), 1421. https://doi.org/10.3390/genes13081421






