Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. IDH1 G105G SNP and Clinical/Pathological Characteristics
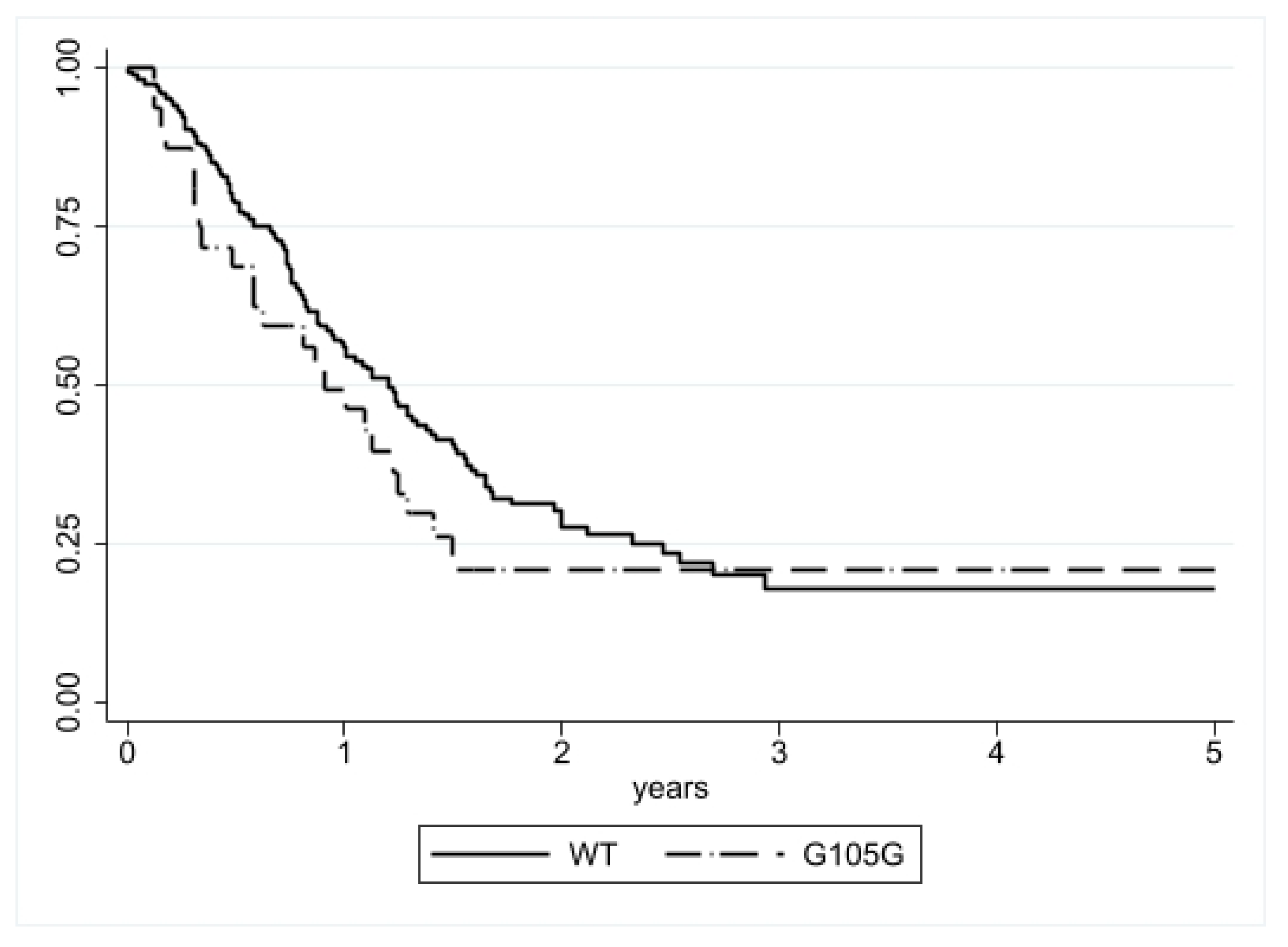
3.1.1. Progression-Free Survival (PFS) Analysis
3.1.2. Disease Specific Survival (DSS) Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Dang, L.; Yen, K.; Attar, E.C. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann. Oncol. 2016, 27, 599–608. [Google Scholar] [CrossRef]
- Issa, G.C.; DiNardo, C.D. Acute myeloid leukemia with IDH1 and IDH2 mutations: 2021 treatment algorithm. Blood Cancer J. 2021, 11, 107. [Google Scholar] [CrossRef] [PubMed]
- Acquaviva, G.; Visani, M.; De Biase, D.; Marucci, G.; Franceschi, E.; Tosoni, A.; Brandes, A.A.; Rhoden, K.J.; Pession, A.; Tallini, G. Prevalence of the single-nucleotide polymorphism rs11554137 (IDH1105GGT) in brain tumors of a cohort of Italian patients. Sci. Rep. 2018, 8, 4459. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.A.; Kopecky, K.J.; Alonzo, T.A.; Gerbing, R.B.; Miller, K.L.; Kuhn, J.; Zeng, R.; Ries, R.; Raimondi, S.C.; Hirsch, B.A.; et al. Prognostic implications of the IDH1 synonymous SNP rs11554137 in pediatric and adult AML: A report from the Children’s Oncology Group and SWOG. Blood 2011, 118, 4561–4566. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.; Damm, F.; Göhring, G.; Görlich, K.; Heuser, M.; Schäfer, I.; Ottmann, O.; Lübbert, M.; Heit, W.; Kanz, L.; et al. Impact of IDH1 R132 Mutations and an IDH1 Single Nucleotide Polymorphism in Cytogenetically Normal Acute Myeloid Leukemia: SNP rs11554137 Is an Adverse Prognostic Factor. J. Clin. Oncol. 2010, 28, 2356–2364. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Li, Y.; Lv, N.; Jing, Y.; Xu, Y.; Li, Y.; Li, W.; Yao, Z.; Chen, X.; Huang, S.; et al. Correlation Between Isocitrate Dehydrogenase Gene Aberrations and Prognosis of Patients with Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis. Clin. Cancer Res. 2017, 23, 4511–4522. [Google Scholar] [CrossRef]
- Wang, X.-W.; Boisselier, B.; Rossetto, M.; Marie, Y.; Idbaih, A.; Mokhtari, K.; Gousias, K.; Hoang-Xuan, K.; Delattre, J.-Y.; Simon, M.; et al. Prognostic impact of the isocitrate dehydrogenase 1 single-nucleotide polymorphism rs11554137 in malignant gliomas. Cancer 2013, 119, 806–813. [Google Scholar] [CrossRef]
- Mistry, A.M.; Vnencak-Jones, C.L.; Mobley, B.C. Clinical prognostic value of the isocitrate dehydrogenase 1 single-nucleotide polymorphism rs11554137 in glioblastoma. J. Neuro-Oncol. 2018, 138, 307–313. [Google Scholar] [CrossRef]
- Franceschi, E.; De Biase, D.; Di Nunno, V.; Pession, A.; Tosoni, A.; Gatto, L.; Lodi, R.; Tallini, G.; Visani, M.; Bartolini, S.; et al. IDH1105GGT single nucleotide polymorphism improves progression free survival in patients with IDH mutated grade II and III gliomas. Pathol. Res. Pract. 2021, 221, 153445. [Google Scholar] [CrossRef] [PubMed]
- Fairbrother, W.G.; Yeh, R.-F.; Sharp, P.A.; Burge, C.B. Predictive Identification of Exonic Splicing Enhancers in Human Genes. Science 2002, 297, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.-W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “Silent” Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science 2007, 315, 525–528. [Google Scholar] [CrossRef] [PubMed]
- Rudà, R.; Bruno, F.; Ius, T.; Silvani, A.; Minniti, G.; Pace, A.; Lombardi, G.; Bertero, L.; Pizzolitto, S.; Pollo, B.; et al. IDH wild-type grade 2 diffuse astrocytomas: Prognostic factors and impact of treatments within molecular subgroups. Neuro-Oncology 2022, 24, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.; Baborie, A.; Alam, F.; Joyce, K.; Moxham, M.; Sibson, R.; Crooks, D.; Husband, D.; Shenoy, A.; Brodbelt, A.; et al. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br. J. Cancer 2009, 101, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Macdonald, D.R.; Reardon, D.A.; Cloughesy, T.F.; Sorensen, A.G.; DeGroot, E.G.; Wick, W.; Gilbert, M.R.; Lassman, A.B.; Tsien, C.; et al. Updated Response Assessment Criteria for High-Grade Gliomas: Response Assessment in Neuro-Oncology Working Group. J. Clin. Oncol. 2010, 28, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Gessler, F.; Bernstock, J.D.; Braczynski, A.; Lescher, S.; Baumgarten, P.; Harter, P.N.; Mittelbronn, M.; Wu, T.; Seifert, V.; Senft, C. Surgery for Glioblastoma in Light of Molecular Markers: Impact of Resection and MGMT Promoter Methylation in Newly Diagnosed IDH-1 Wild-Type Glioblastomas. Neurosurgery 2019, 84, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Pala, A.; Schmitz, A.L.; Knoll, A.; Schneider, M.; Hlavac, M.; König, R.; Wirtz, C.R.; Coburger, J. Is MGMT promoter methylation to be considered in the decision making for recurrent surgery in glioblastoma patients? Clin. Neurol. Neurosurg. 2018, 167, 6–10. [Google Scholar] [CrossRef]

| Total (n = 211) | G105G IDH | ||||
|---|---|---|---|---|---|
| NO | YES | p | |||
| Age | Median | 66 | 66 | 64 | 0.868 |
| Interval | 23–84 | 23–84 | 32–80 | ||
| <55 | 47 (22.3%) | 40 (85.1%) | 7 (14.9%) | 0.953 | |
| >55 | 164 (77.7%) | 139 (84.7%) | 25 (15.3%) | ||
| Sex | F | 80 (37.9%) | 65 (81.2%) | 15 (18.8%) | 0.257 |
| M | 131 (62.1%) | 114 (87.0%) | 17 (13.0%) | ||
| Tumor site (missing: 2) | Hemispheric | 200 (95.7%) | 170 (85.0%) | 30 (15.0%) | 0.679 |
| Midline | 6 (2.9%) | 5 (83.3%) | 1 (16.7%) | ||
| Cerebellar | 3 (1.4%) | 2 (66.7%) | 1 (33.3%) | ||
| Main involved lobe(missing: 11) | Frontal | 48 (24.0%) | 42 (87.5%) | 6 (12.5%) | 0.792 |
| Temporal | 49 (24.5%) | 42 (85.7%) | 7 (14.3%) | ||
| Parietal | 13 (6.5%) | 12 (92.3%) | 1 (7.7%) | ||
| Occipital | 81 (40.5%) | 66 (81.5%) | 15 (18.5%) | ||
| No predominant lobe | 9 (4.5%) | 8 (88.9%) | 1 (11.1%) | ||
| Number ofinvolved lobes (missing: 12) | 1 | 119 (59.8%) | 104 (87.4%) | 15 (12.6%) | 0.486 |
| 2 | 60 (30.2%) | 49 (81.7%) | 11 (18.3%) | ||
| 3 | 20 (10.1%) | 16 (80.0%) | 4 (20.0%) | ||
| Multifocaltumor | No | 190 (90.1%) | 163 (85.8%) | 27 (14.2%) | 0.245 |
| Yes | 21 (10.0%) | 16 (76.2%) | 5 (23.8%) | ||
| Type of surgery | Biopsy | 24 (11.4%) | 19 (79.2%) | 5 (20.8%) | 0.448 |
| Partial | 74 (35.0%) | 61 (82.4%) | 13 (17.6%) | ||
| Gross | 113 (53.6%) | 99 (87.6%) | 14 (12.4%) | ||
| Progression (missing: 39) | No | 33 (19.2%) | 28 (84.8%) | 5 (15.2%) | 0.995 |
| Yes | 139 (80.8%) | 118 (84.9%) | 21 (15.1%) | ||
| Type ofprogression (missing: 73) | Local | 116 (84.0%) | 98 (84.5%) | 18 (15.5%) | 0.284 |
| Distant | 2 (1.4%) | 1 (0.5%) | 1 (0.5%) | ||
| Local + Distant | 17 (12.3%) | 16 (94.1%) | 1 (5.9%) | ||
| Leptomeningeal dissemination | 3 (2.2%) | 2 (66.7%) | 1 (33.3%) | ||
| Outcome at last follow-up | Alive | 71 (33.6%) | 64 (90.1%) | 7 (9.9%) | 0.126 |
| Deceased | 140 (66.4%) | 115 (82.1%) | 25 (17.9%) | ||
| Total (n = 211) | G105G IDH | ||||
|---|---|---|---|---|---|
| NO | YES | p | |||
| MGMT(missing: 3) | Median | 8 | 8 | 6 | 0.594 |
| Interval | 1–81 | 1–81 | 1–61 | ||
| MGMTpromoter methylation status(missing: 3) | <9% | 108 (51.9%) | 90 (83.3%) | 18 (16.7%) | 0.324 |
| 9–29% | 33 (15.9%) | 26 (78.8%) | 7 (21.2%) | ||
| ≥30% | 67 (32.2%) | 60 (89.6%) | 7 (10.4%) | ||
| Mitotic count | Median | 11 | 11 | 14 | 0.672 |
| Interval | 2–72 | 2–72 | 4–51 | ||
| Ki-67 | Median | 30 | 30 | 27 | 0.120 |
| Interval | 5–90 | 5–90 | 15–75 | ||
| HR 1 | CI | p | ||
|---|---|---|---|---|
| IDH1 G105G SNP | Present vs. Absent | 1.01 | 0.62–1.62 | 0.977 |
| Sex | M vs. F | 1.13 | 0.81–1.61 | 0.460 |
| Age | Linear | 1.01 | 1.00–1.03 | 0.008 |
| >55 vs. <55 | 1.41 | 0.92–2.12 | 0.106 | |
| MGMT promotermethylation status | <9% | 1 | ||
| 9–29% | 0.44 | 0.26–0.76 | 0.003 | |
| ≥30% | 0.54 | 0.37–0.79 | 0.001 | |
| Mitotic count | Linear | 1.01 | 0.46–1.85 | 0.813 |
| Tumor site | Hemispheric | 1 | ||
| Midline | 0.60 | 0.22–1.64 | 0.324 | |
| Cerebellar | 41.5 | 4.64–371 | 0.001 | |
| Main involved lobe | Frontal | 1 | ||
| Temporal | 0.58 | 0.36–0.93 | 0.025 | |
| Parietal | 0.51 | 0.23–1.16 | 0.111 | |
| Occipital | 0.84 | 0.54–1.30 | 0.431 | |
| No predominant lobe | 1.60 | 0.70–3.62 | 0.261 | |
| Number ofinvolved lobes | 1 | 1 | ||
| 2 | 0.95 | 0.64–1.41 | 0.789 | |
| 3 | 2.37 | 1.31–4.28 | 0.004 | |
| Multifocal tumor | Yes vs. No | 2.37 | 1.43–3.90 | 0.001 |
| Surgery type | Biopsy | 1 | ||
| Partial | 1.10 | 0.64–1.87 | 0.729 | |
| Gross | 0.61 | 0.37–1.01 | 0.058 |
| HR 1 | CI | p | ||
|---|---|---|---|---|
| IDH1 G105G SNP | Present vs. Absent | 1.34 | 0.87–2.08 | 0.185 |
| Sex | M vs. F | 1.07 | 0.76–1.51 | 0.704 |
| Age | Linear | 1.01 | 0.99–1.03 | 0.056 |
| >55 vs. <55 | 1.31 | 0.87–1.97 | 0.189 | |
| MGMT promotermethylation status | <9% | 1 | ||
| 9–29% | 0.53 | 0.32–0.89 | 0.016 | |
| ≥30% | 0.43 | 0.28–0.64 | <0.001 | |
| Mitotic count | Linear | 1.00 | 0.98–1.02 | 0.858 |
| Tumor site | Hemispheric | 1 | ||
| Median line | 0.76 | 0.28–2.08 | 0.604 | |
| Cerebellar | 5.55 | 1.35–22.9 | 0.018 | |
| Main involved lobe | Frontal | 1 | ||
| Temporal | 0.57 | 0.34–0.95 | 0.030 | |
| Parietal | 0.74 | 0.36–1.51 | 0.411 | |
| Occipital | 0.97 | 0.63–1.49 | 0.876 | |
| No predominant lobe | 1.03 | 0.40–2.64 | 0.953 | |
| Number ofinvolved lobes | 1 | 1 | ||
| 2 | 1.09 | 0.74–1.61 | 0.645 | |
| 3 | 2.28 | 1.31–4.00 | 0.004 | |
| Multifocal tumor | Yes vs. No | 2.25 | 1.38–3.66 | 0.001 |
| Surgery type | Biopsy | 1 | ||
| Partial | 0.88 | 0.53–1.46 | 0.623 | |
| Gross | 0.53 | 0.33–0.86 | 0.010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saaid, A.; Monticelli, M.; Ricci, A.A.; Orlando, G.; Botta, C.; Zeppa, P.; Bianconi, A.; Osella-Abate, S.; Bruno, F.; Pellerino, A.; et al. Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma. Genes 2022, 13, 1439. https://doi.org/10.3390/genes13081439
Saaid A, Monticelli M, Ricci AA, Orlando G, Botta C, Zeppa P, Bianconi A, Osella-Abate S, Bruno F, Pellerino A, et al. Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma. Genes. 2022; 13(8):1439. https://doi.org/10.3390/genes13081439
Chicago/Turabian StyleSaaid, Ayoub, Matteo Monticelli, Alessia Andrea Ricci, Giulia Orlando, Cristina Botta, Pietro Zeppa, Andrea Bianconi, Simona Osella-Abate, Francesco Bruno, Alessia Pellerino, and et al. 2022. "Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma" Genes 13, no. 8: 1439. https://doi.org/10.3390/genes13081439
APA StyleSaaid, A., Monticelli, M., Ricci, A. A., Orlando, G., Botta, C., Zeppa, P., Bianconi, A., Osella-Abate, S., Bruno, F., Pellerino, A., Rudà, R., Cassoni, P., Garbossa, D., Cofano, F., & Bertero, L. (2022). Prognostic Analysis of the IDH1 G105G (rs11554137) SNP in IDH-Wildtype Glioblastoma. Genes, 13(8), 1439. https://doi.org/10.3390/genes13081439











