Genome-Wide Identification and Expression Analyses of the Cotton AGO Genes and Their Potential Roles in Fiber Development and Stress Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Gossypium AGO Genes
2.2. Phylogenetic, Gene Structure, and Conserved Domain Analyses
2.3. Chromosomal Mapping and Gene Duplication Analyses
2.4. Cis-Acting Regulatory Element Analysis
2.5. Transcriptome Data Analysis
2.6. Plant Materials and Stress Treatments
2.7. Quantitative Real-time PCR Analysis
2.8. Statistical Analysis
3. Results
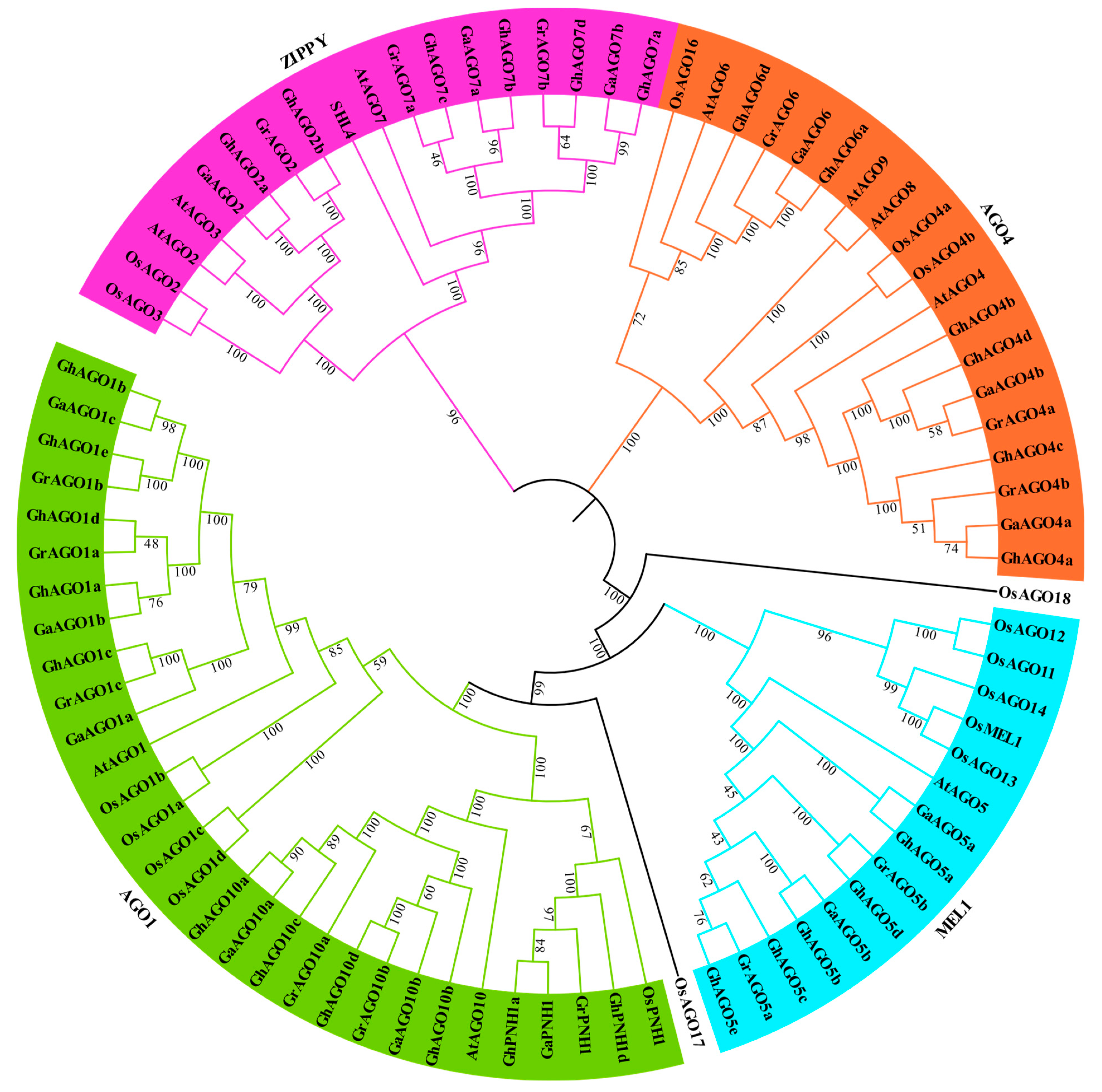
3.1. Identification and Phylogenetic Analysis of Gossypium AGO Genes
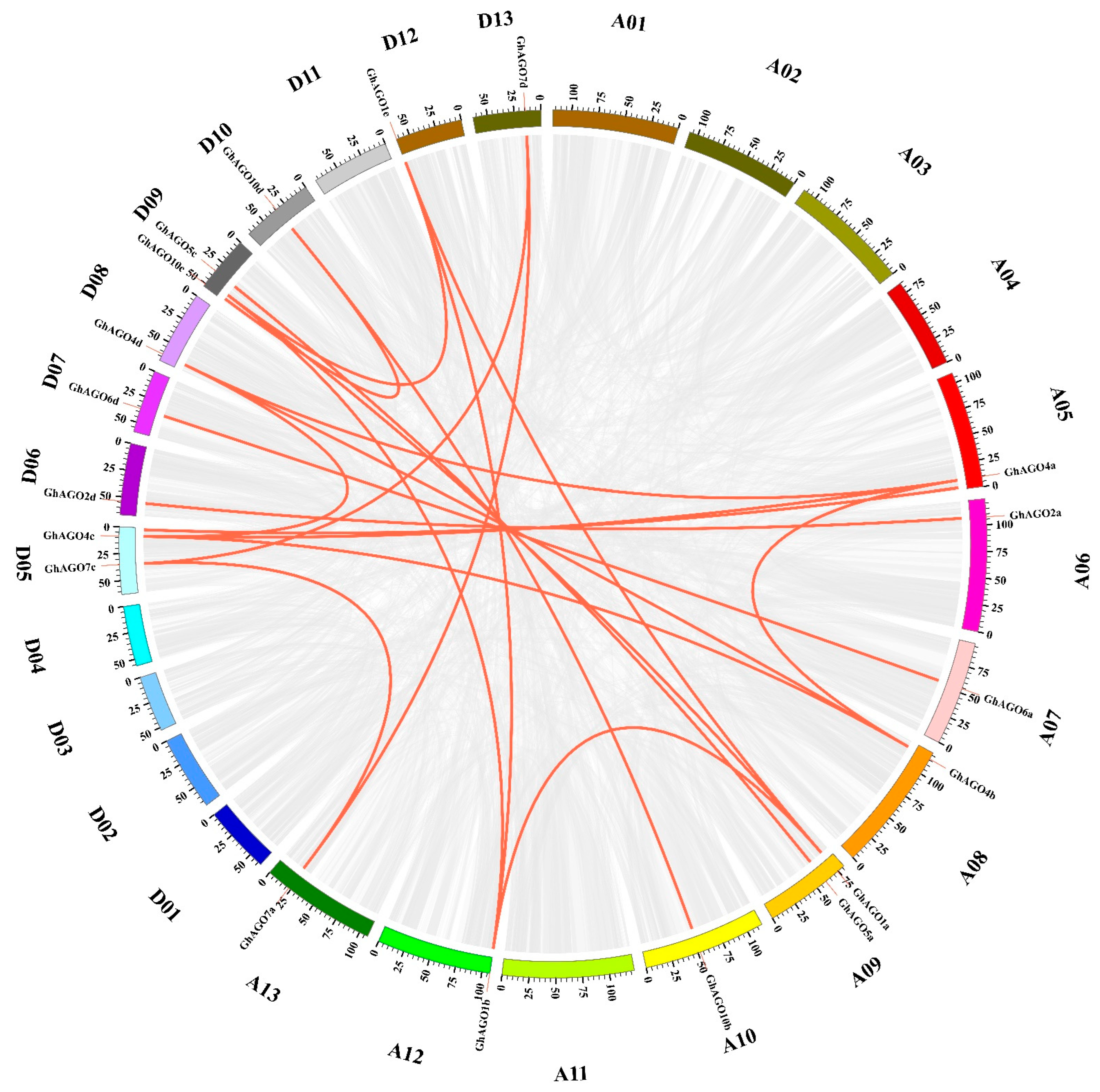
3.2. Genomic Localization and Gene Duplication Analysis of GhAGO Genes
3.3. Gene Structure and Conserved Domain Analysis of GhAGO Genes
3.4. Cis-Acting Regulatory Elements in Promoter Region of GhAGO Genes
3.5. GhAGO Gene Expression Patterns in Diverse Cotton Tissues
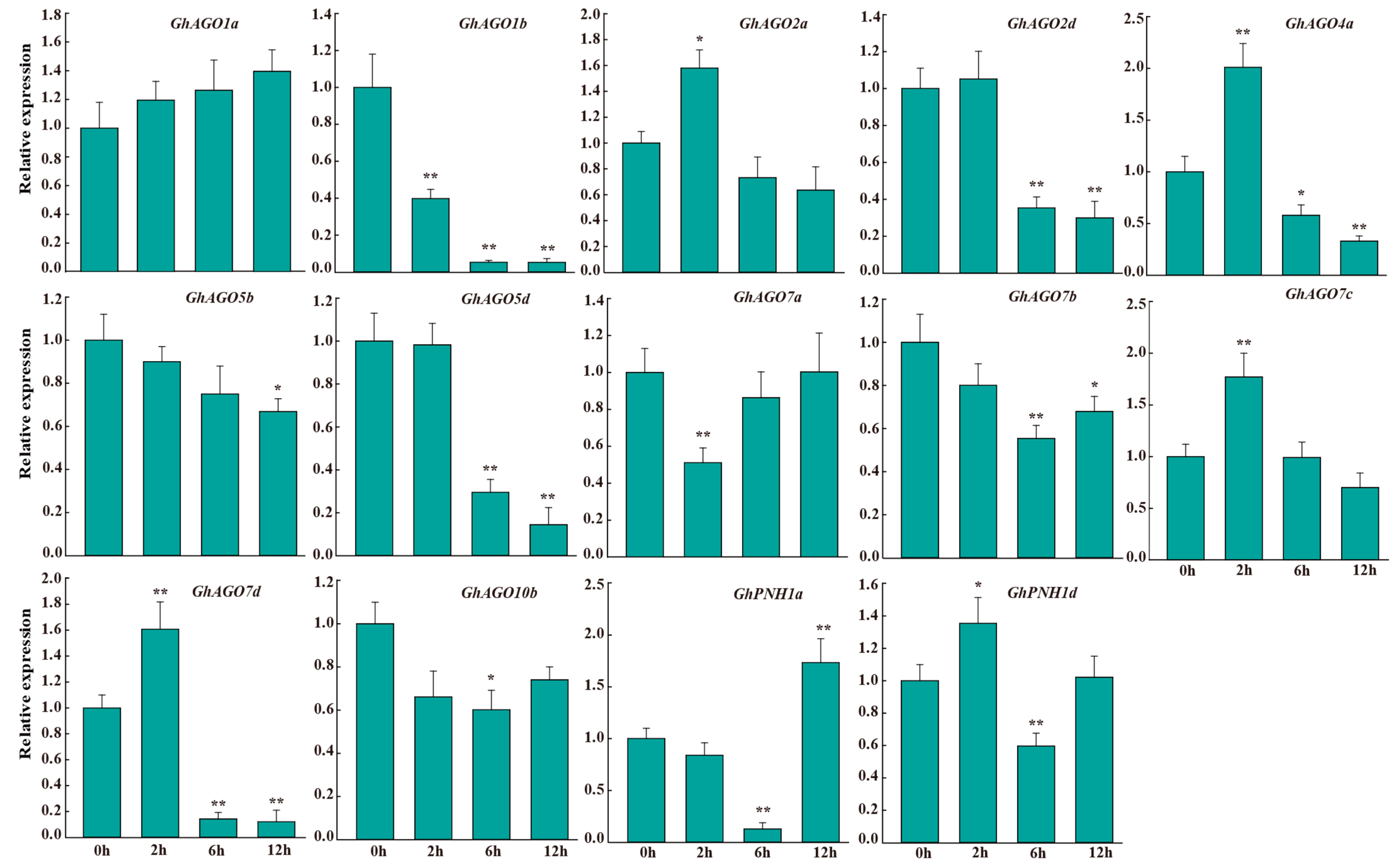
3.6. GhAGO Genes Were Influenced by Verticillium Wilt Infection
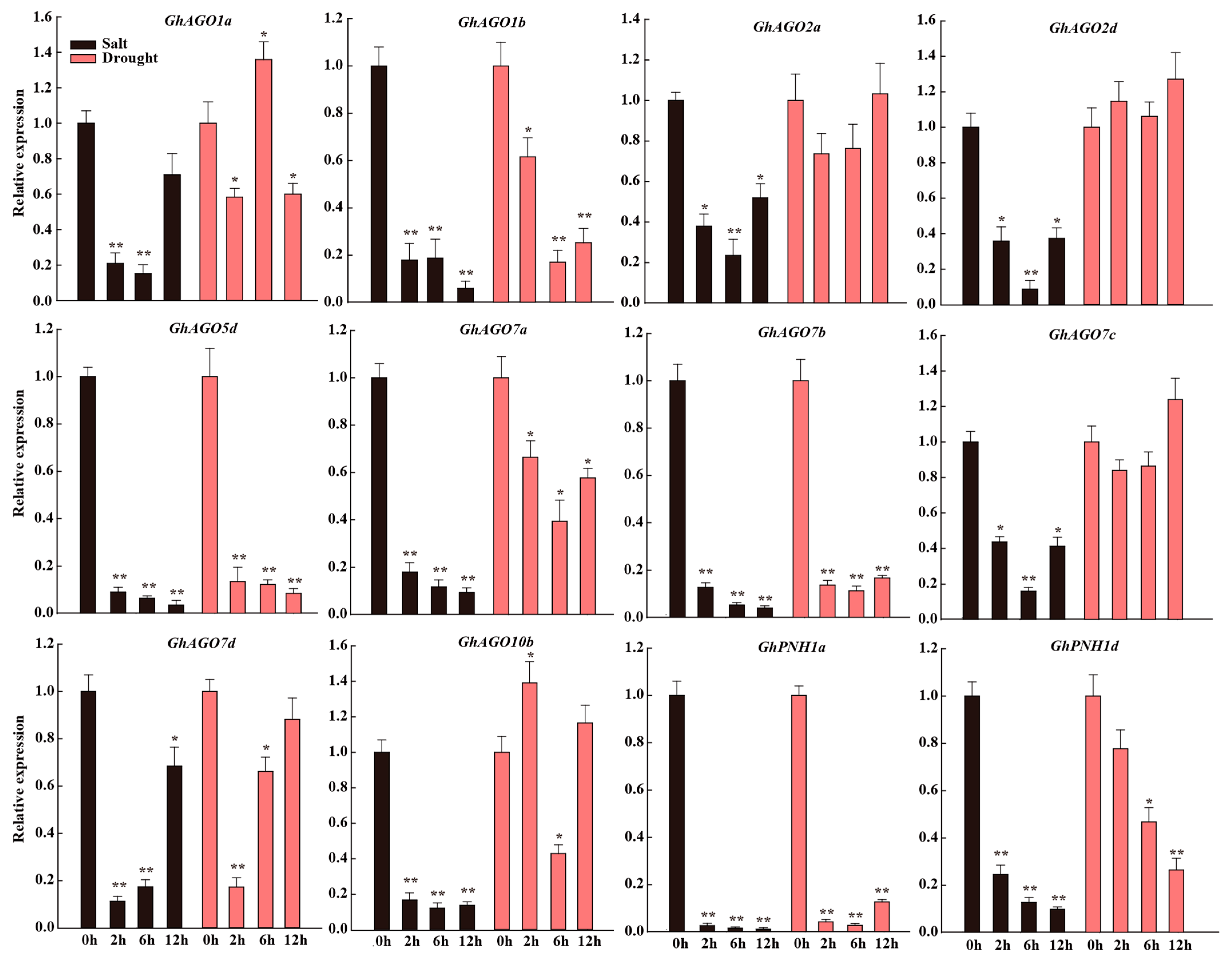
3.7. GhAGO Genes Were Modulated by Salt and Drought Stresses
4. Discussion
4.1. Characterization of Gossypium AGO Genes
4.2. Differential Expression of GhAGO Genes in Response to Multiple Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, C.; Xin, Y.; Xu, L.; Cai, Z.; Xue, Y.; Liu, Y.; Xie, D.; Liu, Y.; Qi, Y. Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev. Cell 2018, 44, 348–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute 10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnatreya, D.B.; Baruah, P.M.; Dowarah, B.; Chowrasia, S.; Mondal, T.K.; Agarwala, N. Genome-wide identification, evolutionary relationship and expression analysis of AGO, DCL and RDR family genes in tea. Sci. Rep. 2021, 11, 8679. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits mRNAs and siRNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 451. [Google Scholar] [CrossRef] [Green Version]
- Qian, Y.; Cheng, Y.; Cheng, X.; Jiang, H.; Zhu, S.; Cheng, B. Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep. 2011, 30, 1347–1363. [Google Scholar] [CrossRef]
- Bai, M.; Yang, G.S.; Chen, W.T.; Mao, Z.C.; Kang, H.X.; Chen, G.H.; Yang, Y.H.; Xie, B.Y. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef]
- Zhao, H.; Zhao, K.; Wang, J.; Chen, X.; Chen, Z.; Cai, R.; Xiang, Y. Comprehensive analysis of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in grapevine (Vitis vinifera). J. Plant Growth Regul. 2015, 34, 108–121. [Google Scholar] [CrossRef]
- Gan, D.; Liang, D.; Wu, J.; Zhan, M.; Yang, F.; Xu, W.; Zhu, S.; Shi, J. Genome-wide identification of the Dicer-like, Argonaute, and RNA-dependent RNA polymerase gene families in cucumber (Cucumis sativus L.). J. Plant Growth Regul. 2016, 35, 135–150. [Google Scholar] [CrossRef]
- Cao, J.Y.; Xu, Y.P.; Li, W.; Li, S.S.; Rahman, H.; Cai, X.Z. Genome-wide identification of Dicer-like, Argonaute, and RNA-dependent RNA polymerase gene families in Brassica Species and functional analyses of their Arabidopsis homologs in resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 1614. [Google Scholar] [CrossRef] [Green Version]
- Qin, L.; Mo, N.; Muhammad, T.; Liang, Y. Genome-wide analysis of DCL, AGO, and RDR gene families in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2018, 19, 1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabbione, A.; Daurelio, L.; Vegetti, A.; Talon, M.; Tadeo, F.; Dotto, M. Genome-wide analysis of AGO, DCL and RDR gene families reveals RNA-directed DNA methylation is involved in fruit abscission in Citrus sinensis. BMC Plant Biol. 2019, 19, 401. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Wang, L.M.; Zhao, L.Z.; Wang, W.; Zhang, H.X. Genome-wide identification and evolutionary analysis of Argonaute genes in hexaploidy bread wheat. Biomed. Res. Int. 2021, 2021, 9983858. [Google Scholar]
- Alvarez-Diaz, J.C.; Richard, M.M.S.; Thareau, V.; Teano, G.; Paysant-Le-Roux, C.; Rigaill, G.; Pflieger, S.; Gratias, A.; Geffroy, V. Genome-Wide Identification of Key Components of RNA Silencing in Two Phaseolus vulgaris Genotypes of Contrasting Origin and Their Expression Analyses in Response to Fungal Infection. Genes 2022, 13, 64. [Google Scholar] [CrossRef]
- Liao, Z.; Hoden, K.P.; Singh, R.K.; Dixelius, C. Genome-wide identification of Argonautes in Solanaceae with emphasis on potato. Sci. Rep. 2020, 10, 20577. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.F.; Hossen, M.I.; Sarkar, M.A.R.; Konak, J.N.; Zohra, F.T.; Shoyeb, M.; Mondal, S. Genome-wide identification of DCL, AGO, and RDR gene families and their associated functional regulatory elements analyses in banana (Musa acuminata). PLoS ONE 2021, 16, e0256873. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Abdelraheem, A.; Esmaeili, N.; O’Connell, M.; Zhang, J. Progress and perspective on drought and salt stress tolerance in cotton. Ind. Crop. Prod. 2019, 130, 118–129. [Google Scholar] [CrossRef]
- Song, J.; Pei, W.; Wang, N.; Ma, J.; Xin, Y.; Yang, S.; Wang, W.; Chen, Q.; Zhang, J.; Yu, J.; et al. Transcriptome analysis and identification of genes associated with oil accumulation in upland cotton. Physiol. Plant. 2022, 174, e13701. [Google Scholar] [CrossRef]
- Reiser, L.; Subramaniam, S.; Li, D.; Huala, E. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. Bioinf. 2017, 60, 1.11.1–1.11.45. [Google Scholar] [CrossRef]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Thibaud-Nissen, F.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR rice genome resource: Improvements and new features. Nucl. Acids Res. 2007, 35, D883–D887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucl. Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [Green Version]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucl. Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Fu, M.; Li, H.; Chen, Y.; Wang, L.; Liu, R. Systematic analysis of NAC transcription factors in Gossypium barbadense uncovers their roles in response to Verticillium wilt. PeerJ 2019, 7, e7995. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinf. 2006, 4, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, J.Y.; Huang, J.Q.; Li, N.Y.; Ma, X.F.; Wang, J.L.; Liu, C.; Liu, Y.F.; Liang, Y.; Bao, Y.M.; Dai, X.F. Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to verticillium wilt. BMC Plant Biol. 2015, 15, 148. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhang, D.; Zhu, G.; Mi, X.; Guo, W. Combining genome-wide and transcriptome-wide analyses reveal the evolutionary conservation and functional diversity of aquaporins in cotton. BMC Genom. 2019, 20, 538. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Qi, Y. RNAi in plants: An Argonaute-centered view. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Li, W.; Guo, M.; Liu, S.; Liu, L.; Yu, Y.; Mo, B.; Chen, X.; Gao, L. Origin, evolution and diversification of plant ARGONAUTE proteins. Plant J. 2022, 109, 1086–1097. [Google Scholar] [CrossRef] [PubMed]
- Hock, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 210, 210. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wu, H.; Qanmber, G.; Ali, F.; Wang, L.; Liu, Z.; Yu, D.; Wang, Q.; Xu, A.; Yang, Z. Genome-wide study of the GATL gene family in Gossypium hirsutum L. reveals that GhGATL genes act on pectin synthesis to regulate plant growth and fiber elongation. Genes 2020, 11, 64. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Leal, D.; Castillo-Cobian, A.; Rodriguez-Arevalo, I.; Vielle-Calzada, J.P. A primary sequence analysis of the ARGONAUTE protein family in plants. Front. Plant Sci. 2016, 7, 1347. [Google Scholar] [CrossRef] [Green Version]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.C.; Katiyar-Agarwal, S.; Huang, H.D.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 regulates innate immunity via miRNA393*-mediated silencing of a Golgi-localized SNARE gene MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Agorio, A.; Vera, P. ARGONAUTE4 is required for resistance to Pseudomonas syringe in Arabidopsis. Plant Cell 2007, 19, 3778–3790. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, M.; Pandey, P.; Baldwin, I.T.; Pandey, S.P. Argonaute4 modulates resistance to Fusarium brachygibbosum infection by regulating jasmonic acid signaling. Plant Physiol. 2020, 184, 1128–1152. [Google Scholar] [CrossRef]
- Zhai, L.; Teng, F.; Zheng, K.; Xiao, J.; Deng, W.; Sun, W. Expression analysis of Argonaute genes in maize (Zea mays L.) in response to abiotic stress. Hereditas 2019, 156, 27. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, C.; Ren, Y.; Wu, M.; Wu, Z.; Chen, Y.; He, L.; Tang, B.; Huang, X.; Shabala, S.; et al. An RNA-binding protein MUG13.4 interacts with AtAGO2 to modulate salinity tolerance in Arabidopsis. Plant Sci. 2019, 288, 110218. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, S.; Jia, G.; Schnable, J.C.; Su, H.; Tang, C.; Zhi, H.; Diao, X. The C-terminal motif of SiAGO1b is required for the regulation of growth, development and stress responses in foxtail millet (Setaria italica (L.) P. Beauv). J. Exp. Bot. 2016, 67, 3237–3249. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, M.; Chen, Y.; Li, H.; Wang, L.; Liu, R.; Liu, Z. Genome-Wide Identification and Expression Analyses of the Cotton AGO Genes and Their Potential Roles in Fiber Development and Stress Response. Genes 2022, 13, 1492. https://doi.org/10.3390/genes13081492
Fu M, Chen Y, Li H, Wang L, Liu R, Liu Z. Genome-Wide Identification and Expression Analyses of the Cotton AGO Genes and Their Potential Roles in Fiber Development and Stress Response. Genes. 2022; 13(8):1492. https://doi.org/10.3390/genes13081492
Chicago/Turabian StyleFu, Mingchuan, Yizhen Chen, Hao Li, Liguo Wang, Renzhong Liu, and Zhanji Liu. 2022. "Genome-Wide Identification and Expression Analyses of the Cotton AGO Genes and Their Potential Roles in Fiber Development and Stress Response" Genes 13, no. 8: 1492. https://doi.org/10.3390/genes13081492
APA StyleFu, M., Chen, Y., Li, H., Wang, L., Liu, R., & Liu, Z. (2022). Genome-Wide Identification and Expression Analyses of the Cotton AGO Genes and Their Potential Roles in Fiber Development and Stress Response. Genes, 13(8), 1492. https://doi.org/10.3390/genes13081492





