Simple Sequence Repeat Markers Reveal Genetic Diversity and Population Structure of Bolivian Wild and Cultivated Tomatoes (Solanum lycopersicum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Planting, Sampling and DNA Extraction
2.2. Identification of SSRs in the Tomato Genome and Primer Design
2.3. PCR Amplification and Electrophoresis
2.4. Data Analysis
3. Results
3.1. SSR Markers
| Locus | TNA | Na | Ne | PIC | Ho | He | uHe | FIS | FIT | FST | GST | P(GST) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SSR22 a | 3 | 1.21 | 1.18 | 0.27 | 0.117 | 0.080 | 0.087 | −0.45 | 0.62 | 0.74 | 0.72 | 0.001 |
| SLR20 b | 3 | 1.18 | 1.09 | 0.19 | 0.020 | 0.056 | 0.063 | 0.65 | 0.90 | 0.72 | 0.67 | 0.001 |
| SLM6-11 c | 5 | 1.57 | 1.31 | 0.65 | 0.000 | 0.178 | 0.198 | 1.00 | 1.00 | 0.75 | 0.69 | 0.001 |
| LE20592 d | 4 | 1.43 | 1.28 | 0.51 | 0.198 | 0.150 | 0.165 | −0.32 | 0.65 | 0.73 | 0.71 | 0.001 |
| TomSatX2-2 | 2 | 1.04 | 1.02 | 0.05 | 0.000 | 0.011 | 0.013 | 1.00 | 1.00 | 0.79 | 0.75 | 0.001 |
| TomSatX7-1 | 3 | 1.18 | 1.12 | 0.30 | 0.080 | 0.069 | 0.077 | −0.16 | 0.76 | 0.79 | 0.77 | 0.001 |
| TomSatX7-2 | 2 | 1.04 | 1.04 | 0.10 | 0.012 | 0.018 | 0.021 | 0.33 | 0.88 | 0.82 | 0.80 | 0.001 |
| TomSatX8-1 | 2 | 1.29 | 1.14 | 0.33 | 0.042 | 0.090 | 0.102 | 0.53 | 0.90 | 0.79 | 0.75 | 0.001 |
| TomSatX9-2a | 2 | 1.07 | 1.02 | 0.13 | 0.007 | 0.015 | 0.017 | 0.53 | 0.95 | 0.89 | 0.87 | 0.001 |
| TomSatX9-2b | 2 | 1.07 | 1.02 | 0.13 | 0.007 | 0.015 | 0.017 | 0.53 | 0.95 | 0.89 | 0.87 | 0.001 |
| TomSatX11-1 | 5 | 1.18 | 1.10 | 0.49 | 0.013 | 0.049 | 0.055 | 0.75 | 0.98 | 0.91 | 0.89 | 0.001 |
| Mean | 1.21 | 1.12 | 0.29 | 0.045 | 0.067 | 0.074 | 0.40 | 0.87 | 0.80 | 0.77 | 0.001 | |
| SE | 0.025 | 0.017 | 0.19 | 0.010 | 0.009 | 0.010 | 0.15 | 0.04 | 0.02 | 0.03 |
3.2. Genetic Diversity of the Accessions
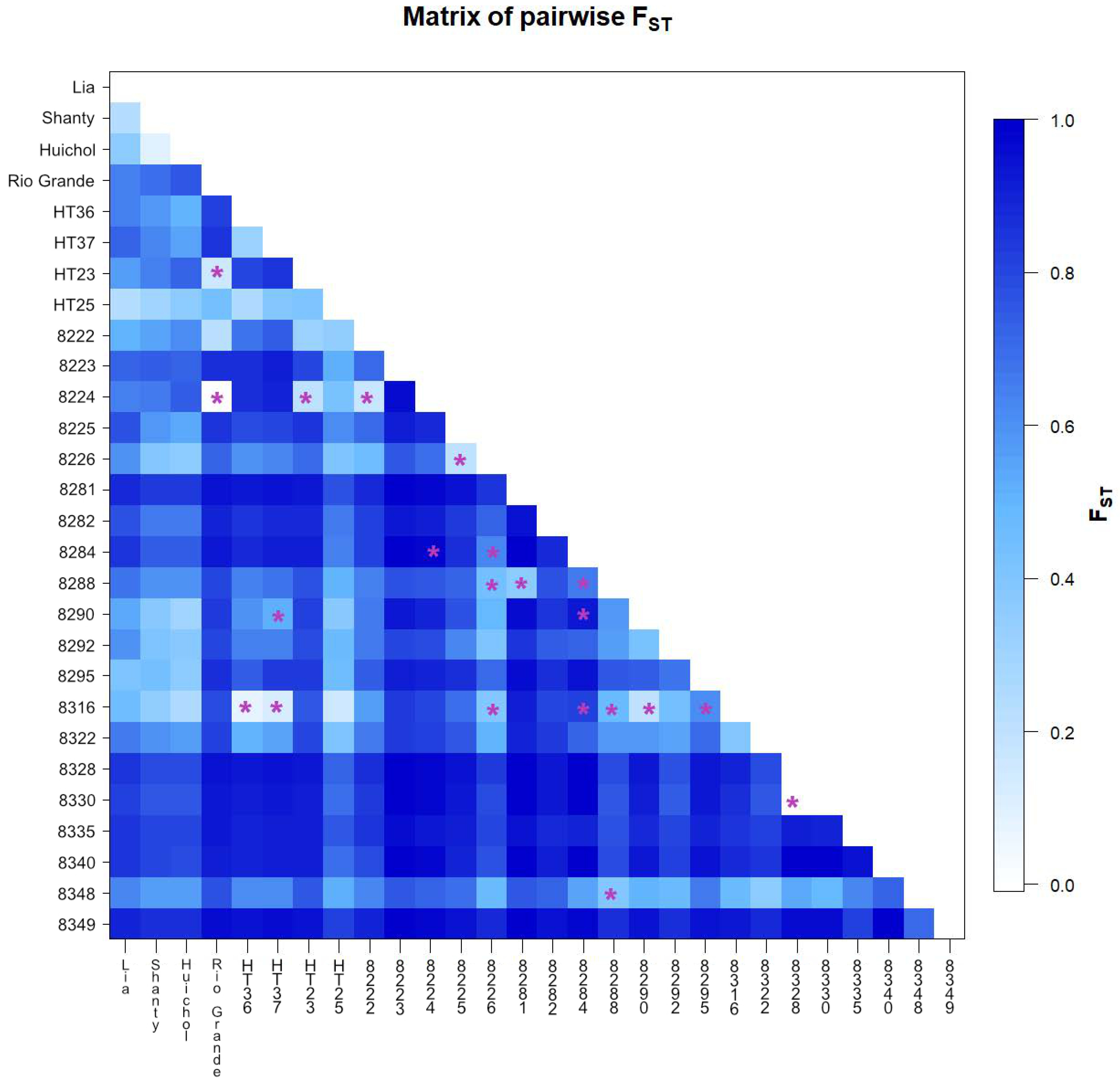
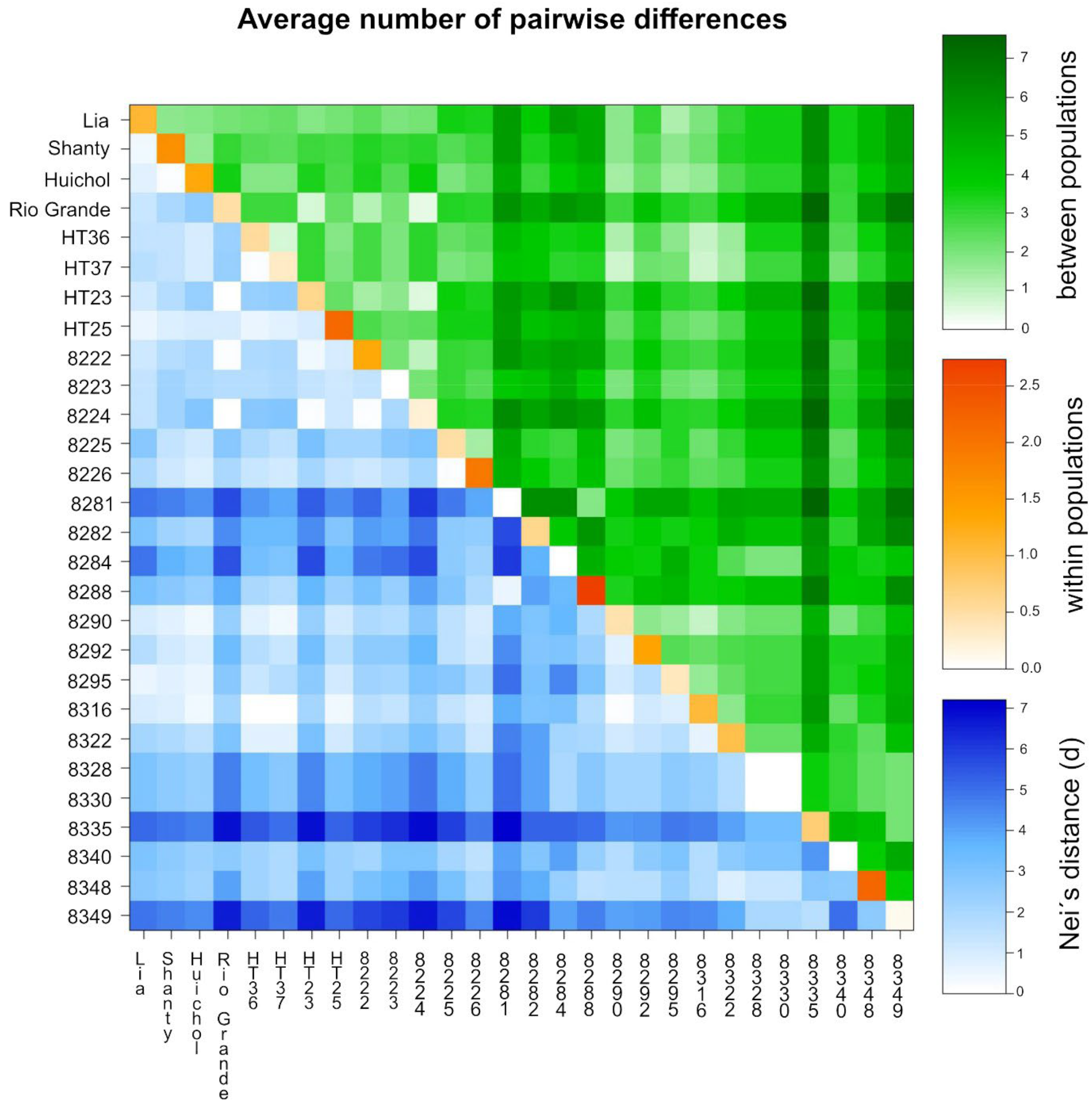
3.3. Analysis of Molecular Variance (AMOVA)
3.4. Cluster Analysis and Principal Coordinate Analysis
3.5. Population Structure Analysis
4. Discussion
4.1. The SSR Markers in Revealing Tomato Genetic Diversity
4.2. AMOVA
4.3. Cluster, Principal Coordinate and Population Structure Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acquaah, G. Principles of Plant Genetics and Breeding: Acquaah/Principles of Plant Genetics and Breeding; John Wiley & Sons, Ltd.: Chichester, UK, 2012; ISBN 978-1-118-31371-8. [Google Scholar]
- Blanca, J.; Cañizares, J.; Cordero, L.; Pascual, L.; Diez, M.J.; Nuez, F. Variation Revealed by SNP Genotyping and Morphology Provides Insight into the Origin of the Tomato. PLoS ONE 2012, 7, e48198. [Google Scholar] [CrossRef] [PubMed]
- Knapp, S.; Peralta, I.E. The Tomato (Solanum lycopersicum L., Solanaceae) and Its Botanical Relatives. In The Tomato Genome; Compendium of Plant Genomes; Causse, M., Giovannoni, J., Bouzayen, M., Zouine, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 7–21. ISBN 978-3-662-53387-1. [Google Scholar]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.J.; Francis, D.; Causse, M.; van der Knaap, E.; Cañizares, J. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. 2015, 16, 257. [Google Scholar] [CrossRef] [PubMed]
- The GRIN Global Project Banco Nacional de Germoplasma Bolivia. Available online: http://germoplasma.iniaf.gob.bo/ (accessed on 27 July 2022).
- Mercado Rios, H.F. Manejo de Recursos Genéticos y Producción de Semillas en el Centro Nacional de Produccion de Semillas de Hortalizas (Genetic Resources Management and Seed Production in the National Center for Vegetable Seed Production); SoftGenetics, LLC.: State College, PA, USA, 2014; Volume 1, p. 3. [Google Scholar]
- INIAF Web-INIAF. Available online: https://www.iniaf.gob.bo/webiniaf/index.php/component/content/article/33-noticias-ultimos/104-gobierno-lanza-nuevas-variedades-para-fortalecer-la-produccion-de-trigo-zanahoria-y-tomate?Itemid=101 (accessed on 27 July 2022).
- Choque, E. Caracterización Y Evaluación Química de Fruto en Germoplasma Nativo E Introducido de Tomate (Solanum Lycopersicum L.) Del Banco de Germoplasma Del Iniaf (Instituto Nacional de Innovación Agropecuaria Y Forestal) (Characterization and Chemical Evaluation of Fruit in Native and Introduced Germplasm of Tomato (Solanum Lycopersicum L.) From the Iniaf (National Institute of Agricultural and Forestry Innovation) Germplasm Bank). Master’s Thesis, Universidad Mayor de San Simon, Cochabamba, Bolivia, 2014. [Google Scholar]
- Torrico, A.; Crespo, M.; Rojas-Beltran, J. Estudio morfológico y molecular de la diversidad genética del tomate silvestre (Solanum spp.) boliviano (Morphological and molecular study of the genetic diversity of the Bolivian wild tomato (Solanum spp.)). Rev. Agric. 2015, 55, 20–28. [Google Scholar]
- Rick, C.M.; Holle, M. Andean Lycopersicon esculentum var. cerasiforme: Genetic Variation and Its Evolutionary Significance. Econ. Bot. 1990, 44, 69–78. [Google Scholar] [CrossRef]
- Sunilkumar, M.K.; Bommesh, J.C.; Vijeth, S.; Gokavi, N.; Shankargouda, S.; Ravi, Y. Molecular characterisation in tomato (solanum lycopersicum L.)—A review. Int. J. Agricul. Environ. Biotech. 2016, 9, 339. [Google Scholar] [CrossRef]
- The Tomato Genome Consortium; Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar]
- The 100 Tomato Genome Sequencing Consortium; Aflitos, S.; Schijlen, E.; de Jong, H.; de Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. 2014, 80, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.J. Molecular Markers in Plants; John Wiley & Sons: Hoboken, NJ, USA, 2012; ISBN 978-0-470-95951-0. [Google Scholar]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- FAO Organizations. Available online: http://www.fao.org/wiews/data/organizations/en/?no_cache=1&instcode=BOL316 (accessed on 27 July 2022).
- Martins, W.S.; Lucas, D.C.S.; Neves, K.F.S.; Bertioli, D.J. WebSat—A Web Software for MicroSatellite marker development. Bioinformation 2009, 3, 282–283. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Smulders, M.J.M.; Bredemeijer, G.; Rus-Kortekaas, W.; Arens, P.; Vosman, B. Use of short microsatellites from database sequences to generate polymorphisms among Lycopersicon esculentum cultivars and accessions of other Lycopersicon species. Theor. Appl. Genet. 1997, 94, 264–272. [Google Scholar] [CrossRef]
- Frary, A.; Xu, Y.; Liu, J.; Mitchell, S.; Tedeschi, E.; Tanksley, S. Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theor. Appl. Genet. 2005, 111, 291–312. [Google Scholar] [CrossRef] [PubMed]
- Geethanjali, S.; Chen, K.-Y.; Pastrana, D.V.; Wang, J.-F. Development and characterization of tomato SSR markers from genomic sequences of anchored BAC clones on chromosome 6. Euphytica 2010, 173, 85–97. [Google Scholar] [CrossRef]
- Korir, N.K.; Diao, W.; Tao, R.; Li, X.; Kayesh, E.; Li, A.; Zhen, W.; Wang, S. Genetic diversity and relationships among different tomato varieties revealed by EST-SSR markers. Genet. Mol. Res. 2014, 13, 43–53. [Google Scholar] [CrossRef]
- Ballard, L.; Adams, P.; Bao, Y.; Bartley, D.; Bintzler, D.; Kasch, L.; Petukhova, L.; Rosato, C. Strategies for genotyping: Effectiveness of tailing primers to increase accuracy in short tandem repeat determinations. J. Biomol. Tech. JBT 2002, 13, 20. [Google Scholar]
- Geleta, M.; Herrera, I.; Monzón, A.; Bryngelsson, T. Genetic Diversity of Arabica Coffee (Coffea arabica L.) in Nicaragua as Estimated by Simple Sequence Repeat Markers. Sci. World J. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef]
- Softgenetics GeneMarker 2016. Available online: https://softgenetics.com/products/genemarker/ (accessed on 27 July 2022).
- Yeh, F.C.; Boyle, T.B.J. Population Genetic Analysis of Codominant and Dominant Markers and Quantitative Traits. Belg. J. Bot. 1997, 129, 157–163. [Google Scholar]
- Peakall, R.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Sneath, P.H.A.; Sokal, R.R. Numerical Taxonomy: The Principles and Practice of Numerical Classification; WH Freeman & Co: San Francisco, CA, USA, 1973; p. 573. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Li, Y.-L.; Liu, J.-X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Panguluri, S.K.; Kumar, A.A. Phenotyping for Plant Breeding Applications of Phenotyping Methods for Crop Improvement, 1st ed.; Springer: New York, NY, USA, 2013; ISBN 1-4614-8320-4. [Google Scholar]
- Bauchet, G.; Causse, M. Genetic Diversity in Tomato (Solanum lycopersicum) and Its Wild Relatives. In Genetic Diversity in Plants; Caliskan, M., Ed.; InTech: Singapore, 2012; pp. 133–162. ISBN 978-953-51-0185-7. [Google Scholar]
- Byrne, P.F.; Volk, G.M.; Gardner, C.; Gore, M.A.; Simon, P.W.; Smith, S. Sustaining the Future of Plant Breeding: The Critical Role of the USDA-ARS National Plant Germplasm System. Crop Sci. 2018, 58, 451–468. [Google Scholar] [CrossRef]
- Aguirre, N.C.; López, W.; Orozco-Cárdenas, M.; Coronado, Y.M.; Vallejo-Cabrera, F. Use of microsatellites for evaluation of genetic diversity in cherry tomato. Bragantia 2017, 76, 220–228. [Google Scholar] [CrossRef]
- Gonias, E.D.; Ganopoulos, I.; Mellidou, I.; Bibi, A.C.; Kalivas, A.; Mylona, P.V.; Osanthanunkul, M.; Tsaftaris, A.; Madesis, P.; Doulis, A.G. Exploring genetic diversity of tomato (Solanum lycopersicum L.) germplasm of genebank collection employing SSR and SCAR markers. Genet. Resour. Crop Evol. 2019, 66, 1295–1309. [Google Scholar] [CrossRef]
- Benor, S.; Zhang, M.; Wang, Z.; Zhang, H. Assessment of genetic variation in tomato (Solanum lycopersicum L.) inbred lines using SSR molecular markers. J. Genet. Genom. 2008, 35, 373–379. [Google Scholar] [CrossRef]
- Meng, F.; Xu, X.; Huang, F.; Li, J. Analysis of Genetic Diversity in Cultivated and Wild Tomato Varieties in Chinese Market by RAPD and SSR. Agric. Sci. China 2010, 9, 1430–1437. [Google Scholar] [CrossRef]
- Caramante, M.; Rouphael, Y.; Corrado, G. The Genetic Diversity and Structure of Tomato Landraces from the Campania Region (Southern Italy) Uncovers a Distinct Population Identity. Agronomy 2021, 11, 564. [Google Scholar] [CrossRef]
- Athinodorou, F.; Foukas, P.; Tsaniklidis, G.; Kotsiras, A.; Chrysargyris, A.; Delis, C.; Kyratzis, A.C.; Tzortzakis, N.; Nikoloudakis, N. Morphological Diversity, Genetic Characterization, and Phytochemical Assessment of the Cypriot Tomato Germplasm. Plants 2021, 10, 1698. [Google Scholar] [CrossRef]
- Causse, M.; Zhao, J.; Diouf, I.; Wang, J.; Lefebvre, V.; Caromel, B.; Génard, M.; Bertin, N. Genomic Designing for Climate-Smart Tomato. In Genomic Designing of Climate-Smart Vegetable Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 47–159. ISBN 978-3-319-97414-9. [Google Scholar]
- Nakazato, T.; Franklin, R.A.; Kirk, B.C.; Housworth, E.A. Population structure, demographic history, and evolutionary patterns of a green-fruited tomato, Solanum peruvianum (Solanaceae), revealed by spatial genetics analyses. Am. J. Bot. 2012, 99, 1207–1216. [Google Scholar] [CrossRef]
- Politov, D.V.; Belokon, M.M.; Belokon, Y.S. Dynamics of allozyme heterozygosity in Siberian dwarf pine Pinus pumila (Pall.) Regel populations of the Russian Far East: Comparison of embryos and maternal plants. Russ. J. Genet. 2006, 42, 1127–1136. [Google Scholar] [CrossRef]
- Ranc, N.; Muños, S.; Santoni, S.; Causse, M. A clarified position for Solanum lycopersicum var. cerasiformein the evolutionary history of tomatoes (solanaceae). BMC Plant Biol. 2008, 8, 130. [Google Scholar] [CrossRef]
- Rao, E.S.; Kadirvel, P.; Symonds, R.C.; Geethanjali, S.; Ebert, A.W. Using SSR markers to map genetic diversity and population structure of Solanum pimpinellifolium for development of a core collection. Plant Genet. Res. 2012, 10, 38–48. [Google Scholar] [CrossRef]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef]
- Fox, C.W. Problems in Measuring among-Family Variation in Inbreeding Depression. Am. J. Bot. 2005, 92, 1929–1932. [Google Scholar] [CrossRef] [PubMed]
- Pailles, Y.; Ho, S.; Pires, I.S.; Tester, M.; Negrão, S.; Schmöckel, S.M. Genetic Diversity and Population Structure of Two Tomato Species from the Galapagos Islands. Front. Plant Sci. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Lindhout, P. Domestication and Breeding of Tomatoes: What have We Gained and What Can We Gain in the Future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.; Sharma, S.; Panthee, D.R. Diversity among Modern Tomato Genotypes at Different Levels in Fresh-Market Breeding. Int. J. Agron. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Tan, J.; Zhou, Z.; Niu, Y.; Sun, X.; Deng, Z. Identification and Functional Characterization of Tomato CircRNAs Derived from Genes Involved in Fruit Pigment Accumulation. Sci. Rep. 2017, 7, 8594. [Google Scholar] [CrossRef]
- Carbonell, P.; Alonso, A.; Grau, A.; Salinas, J.; García-Martínez, S.; Ruiz, J. Twenty Years of Tomato Breeding at EPSO-UMH: Transfer Resistance from Wild Types to Local Landraces—From the First Molecular Markers to Genotyping by Sequencing (GBS). Diversity 2018, 10, 12. [Google Scholar] [CrossRef]
- Geleta, M.; Ortiz, R. Molecular and Genomic Tools Provide Insights on Crop Domestication and Evolution. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2016; Volume 135, pp. 181–223. ISBN 978-0-12-804693-7. [Google Scholar]
- Rannala, B.; Mountain, J.L. Detecting Immigration by Using Multilocus Genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 9197–9201. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Pasam, R.K.; Sharma, R.; Walther, A.; Özkan, H.; Graner, A.; Kilian, B. Genetic diversity and population structure in a legacy collection of spring barley landraces adapted to a wide range of climates. PLoS ONE 2014, 9, e116164. [Google Scholar] [CrossRef]
- Corrado, G.; Piffanelli, P.; Caramante, M.; Coppola, M.; Rao, R. SNP genotyping reveals genetic diversity between cultivated landraces and contemporary varieties of tomato. BMC Genom. 2013, 14, 835. [Google Scholar] [CrossRef] [PubMed]





| Accession/Commercial Name | Germ-Plasm Provider | Country of Origin | Region of Sampling Site in Bolivia | Domestication/Breeding Status | Geographical Position of Sampling Site | Altitude of Sampling Site (Masl) | Fruit Shape | Fruit Size | Fruit Colour | Plant Flowering Type |
|---|---|---|---|---|---|---|---|---|---|---|
| Lia | Hazera | Israel | -- | Cultivated | Not applicable | Not applicable | Cylindrical | Intermediate | Red | Determinate |
| Shanty | Hazera | Israel | -- | Cultivated | Not applicable | Not applicable | Cylindrical | Intermediate | Red | Determinate |
| Huichol | Seminis | Thailand | -- | Cultivated | Not applicable | Not applicable | Cylindrical | Intermediate | Red | Determinate |
| Rio Grande | CNPSH | Bolivia | Cochabamba | Cultivated | 17°26′24″ S; 66°20′47″ W | 2548 | Cylindrical | Intermediate | Red | Determinate |
| HT-36 | CNPSH | Bolivia | Cochabamba | Advanced line | 17°26′24″ S; 66°20′47″ W | 2548 | Rounded | Very large | Red | Determinate |
| HT-37 | CNPSH | Bolivia | Cochabamba | Advanced line | 17°26′24″ S; 66°20′47″ W | 2548 | Rounded | Large | Red | Determinate |
| HT-23 | CNPSH | Bolivia | Cochabamba | Advanced line | 17°26′24″ S; 66°20′7″ W | 2548 | Cylindrical | Intermediate | Red | Determinate |
| HT-25 | CNPSH | Bolivia | Cochabamba | Advanced line | 17°26′24″ S; 66°20′47″ W | 2548 | Cylindrical | Intermediate | Red | Determinate |
| BOL-8222-HT | BGH-BNG | Bolivia | Cochabamba | Cultivated | 17°23′03″ S; 66°08′05″ W | 2858 | High rounded | Intermediate | Red | Determinate |
| BOL-8223-HT | BGH-BNG | Bolivia | Sucre | Cultivated | 19°17′43″ S; 64°22′33″ W | 2201 | High rounded | Very small | Yellow | Semi-determinate |
| BOL-8224-HT | BGH-BNG | Bolivia | Santa Cruz | Cultivated | 17°24′00″ S; 63°53′00″ W | 300 | High rounded | Small, intermediate | Red | Determinate |
| BOL-8225-HT | BGH-BNG | Bolivia | Sucre | Cultivated | 19°44′26″ S; 63°52′24″ W | 1165 | Slightly flattened | Very small | Pink | Semi-determinate |
| BOL-8226-HT | BGH-BNG | Bolivia | Sucre | Cultivated | 19°48′26″ S; 64°00′29″ W | 1143 | Rounded | Very small | Yellow | Semi-determinate |
| BOL-8281-HT | BGH-BNG | Bolivia | Beni | Wild | 14°52′10.7″ S; 61°04′42.3″ W | 227 | Rounded | Very small | Red | Indeterminate |
| BOL-8282-HT | BGH-BNG | Bolivia | Beni | Cultivated | Not reported | 227 | Slightly flattened | Small | Red | Indeterminate |
| BOL-8284-HT | BGH-BNG | Bolivia | Beni | Cultivated | 15°08′47.4″ S; 61°02′15″ W | 259 | Slightly flattened | Very small | Red | Indeterminate |
| BOL-8288-HT | BGH-BNG | Bolivia | La Paz | Wild | 15°47′32″ S; 60°58′41″ W | 498 | Rounded | Very small | Red | Indeterminate |
| BOL-8290-HT | BGH-BNG | Bolivia | La Paz | Wild | 15°48′28″ S; 61°37′27″ W | 594 | Slightly flattened | Very small | Yellow | Semi-determinate |
| BOL-8292-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°15′47″ S; 61°41′44″ W | 1676 | Slightly flattened | Very small | Yellow | Semi-determinate |
| BOL-8295-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°11′21.6″ S; 67°43′29″ W | 599 | Slightly flattened | Small, Intermediate | Red | Determinate |
| BOL-8316-HT | BGH-BNG | Bolivia | La Paz | Cultivated | 15°58′12″ S; 67°27′44″ W | 961–1030 | Slightly flattened | Intermediate | Red | Semi-determinate |
| BOL-8322-HT | BGH-BNG | Bolivia | La Paz | Cultivated | 16°11′53″ S; 67°42′16″ W | 1725–1690 | Slightly flattened | Very small | Red | Semi-determinate |
| BOL-8328-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°15′31″ S; 67°41′32″ W | 1853–1870 | Flattened | Very small | Red | Semi-determinate |
| BOL-8330-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°11′19″ S; 67°43′29″ W | 1716–1720 | Slightly flattened | Very small | Red | Semi-determinate |
| BOL-8335-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°20′21″ S; 67°26′38″ W | 1124–1190 | Slightly flattened | Very small | Red | Semi-determinate |
| BOL-8340-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°26′09″ S; 67°28′26″ W | 1492–1550 | Slightly flattened | Very small | Red | Semi-determinate |
| BOL-8348-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°28′35″ S; 67°26′59″ W | 2021–2012 | Rounded | Very small | Yellow | Semi-determinate |
| BOL-8349-HT | BGH-BNG | Bolivia | La Paz | Wild | 16°28′35″ S; 67°26′59″ W | 2021–2010 | Rounded | Very small | Red | Semi-determinate |
| Genotype | Na | Ne | NPA | NLCA ≤ 0.25 | NLCA ≤ 0.50 | PPL | I | Ho | He | uHe | F |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ‘Lia’ | 1.18 | 1.18 | 0.00 | 0.09 | 0.27 | 0.18 | 0.13 | 0.18 | 0.09 | 0.10 | −1.00 |
| ‘Shanty’ | 1.27 | 1.27 | 0.00 | 0.18 | 0.27 | 0.27 | 0.19 | 0.27 | 0.14 | 0.15 | −1.00 |
| ‘Huichol’ | 1.27 | 1.21 | 0.00 | 0.09 | 0.27 | 0.27 | 0.16 | 0.18 | 0.11 | 0.12 | −0.33 |
| ‘Rio Grande’ | 1.36 | 1.05 | 0.00 | 0.27 | 0.46 | 0.36 | 0.08 | 0.03 | 0.04 | 0.04 | 0.21 |
| ‘HT-36′ | 1.09 | 1.09 | 0.00 | 0.00 | 0.18 | 0.09 | 0.06 | 0.09 | 0.05 | 0.05 | −1.00 |
| ‘HT-37’ | 1.18 | 1.03 | 0.00 | 0.00 | 0.18 | 0.18 | 0.05 | 0.01 | 0.03 | 0.03 | 0.47 |
| ‘HT-23’ | 1.18 | 1.12 | 0.00 | 0.27 | 0.36 | 0.09 | 0.09 | 0.00 | 0.05 | 0.06 | 1.00 |
| ‘HT-25’ | 1.64 | 1.34 | 0.00 | 0.27 | 0.64 | 0.55 | 0.29 | 0.24 | 0.18 | 0.20 | −0.17 |
| BOL-8222-HT | 1.36 | 1.24 | 0.09 | 0.18 | 0.36 | 0.27 | 0.18 | 0.05 | 0.11 | 0.12 | 0.52 |
| BOL-8223-HT | 1.00 | 1.00 | 0.00 | 0.09 | 0.18 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | na |
| BOL-8224-HT | 1.09 | 1.03 | 0.00 | 0.18 | 0.36 | 0.09 | 0.03 | 0.02 | 0.02 | 0.02 | −0.14 |
| BOL-8225-HT | 1.18 | 1.07 | 0.09 | 0.18 | 0.27 | 0.09 | 0.07 | 0.00 | 0.04 | 0.04 | 1.00 |
| BOL-8226-HT | 1.36 | 1.25 | 0.00 | 0.27 | 0.46 | 0.36 | 0.22 | 0.03 | 0.15 | 0.18 | 0.70 |
| BOL-8281-HT | 1.00 | 1.00 | 0.00 | 0.36 | 0.36 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | na |
| BOL-8282-HT | 1.18 | 1.07 | 0.09 | 0.27 | 0.36 | 0.18 | 0.08 | 0.00 | 0.05 | 0.06 | 1.00 |
| BOL-8284-HT | 1.00 | 1.00 | 0.00 | 0.18 | 0.36 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | na |
| BOL-8288-HT | 1.46 | 1.38 | 0.00 | 0.46 | 0.64 | 0.45 | 0.29 | 0.03 | 0.21 | 0.25 | 0.87 |
| BOL-8290-HT | 1.09 | 1.06 | 0.00 | 0.00 | 0.09 | 0.09 | 0.05 | 0.00 | 0.03 | 0.04 | 1.00 |
| BOL-8292-HT | 1.36 | 1.18 | 0.00 | 0.18 | 0.36 | 0.36 | 0.18 | 0.03 | 0.12 | 0.13 | 0.81 |
| BOL-8295-HT | 1.09 | 1.05 | 0.00 | 0.00 | 0.18 | 0.09 | 0.05 | 0.01 | 0.03 | 0.03 | 0.58 |
| BOL-8316-HT | 1.18 | 1.15 | 0.00 | 0.00 | 0.18 | 0.18 | 0.12 | 0.00 | 0.08 | 0.10 | 1.00 |
| BOL-8322-HT | 1.27 | 1.12 | 0.00 | 0.00 | 0.27 | 0.27 | 0.13 | 0.00 | 0.08 | 0.09 | 1.00 |
| BOL-8328-HT | 1.00 | 1.00 | 0.00 | 0.09 | 0.18 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | na |
| BOL-8330-HT | 1.00 | 1.00 | 0.00 | 0.09 | 0.18 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | na |
| BOL-8335-HT | 1.27 | 1.08 | 0.09 | 0.27 | 0.36 | 0.27 | 0.11 | 0.04 | 0.06 | 0.07 | 0.26 |
| BOL-8340-HT | 1.00 | 1.00 | 0.00 | 0.18 | 0.18 | 0 | 0.00 | 0.00 | 0.00 | 0.00 | na |
| BOL-8348-HT | 1.55 | 1.37 | 0.09 | 0.36 | 0.55 | 0.45 | 0.29 | 0.04 | 0.19 | 0.20 | 0.81 |
| BOL-8349-HT | 1.09 | 1.01 | 0.00 | 0.36 | 0.46 | 0.09 | 0.02 | 0.01 | 0.01 | 0.01 | −0.07 |
| 1.21 | 1.12 | 0.02 | 0.18 | 0.32 | 0.19 | 0.10 | 0.05 | 0.07 | 0.07 | 0.36 | |
| 0.03 | 0.02 | 0.02 | 0.10 | 0.15 | 0.03 | 0.01 | 0.01 | 0.01 | 0.01 | 0.04 |
| Grouping Factor | Source of Variation | Degrees of Freedom | Sum of Squares | Variance Components | Percentage of Variation | Fixation Indices | Probability (P) Value |
|---|---|---|---|---|---|---|---|
| Among accessions | 27 | 464.92 | 1.349 Va | 77.29 | FST = 0.77 | Va & FST = 0.000 | |
| AIWA * | 146 | 75.86 | 0.123 Vb | 7.07 | FIS = 0.31 | Vb & FIS = 0.000 | |
| Within individuals | 174 | 47.50 | 0.273 Vc | 15.64 | FIT = 0.84 | Vc & FIT = 0.000 | |
| Total | 347 | 588.28 | 1.745 | ||||
| Geographical | a Among groups | 1 | 83.48 | 0.438 Va | 21.71 | FCT = 0.22 | Va & FCT = 0.000 |
| region of origin | AAWGr ** | 23 | 351.54 | 1.232 Vb | 61.07 | FSC = 0.78 | Vb & FSC = 0.000 |
| Within accessions | 281 | 97.61 | 0.347 Vc | 17.22 | FST = 0.82 | Vc & FST = 0.000 | |
| Total | 305 | 532.63 | 2.018 | ||||
| Altitude | b Among groups | 3 | 54.43 | −0.027 Va | −1.43 | FCT = −0.01 | Va & FCT = 0.566 |
| groups | AAWGr | 15 | 260.65 | 1.602 Vb | 85.08 | FSC = 0.84 | Vb & FSC = 0.000 |
| Within accessions | 189 | 58.19 | 0.308 Vc | 16.35 | FST = 0.84 | Vc & FST = 0.000 | |
| Total | 207 | 373.27 | 1.883 | ||||
| Cultivation | c Among groups | 1 | 61.93 | 0.361 Va | 17.92 | FCT = 0.18 | Va & FCT = 0.005 |
| status | AAWGr | 21 | 332.80 | 1.276 Vb | 63.36 | FSC = 077 | Vb & FSC = 0.000 |
| Within accessions | 259 | 97.61 | 0.377 Vc | 18.72 | FST = 0.81 | Vc & FST = 0.000 | |
| Total | 281 | 492.34 | 2.013 | ||||
| Fruit shape | d Among groups | 2 | 71.59 | 0.282 Va | 16.75 | FCT = 0.16 | Va & FCT = 0.000 |
| AAWGr | 16 | 195.54 | 0.948 Vb | 56.25 | FSC = 0.67 | Vb & FSC = 0.000 | |
| Within accessions | 221 | 100.56 | 0.455 Vc | 26.99 | FST = 0.73 | Vc & FST = 0.000 | |
| Total | 239 | 367.69 | 1.686 | ||||
| Fruit colour | e Among groups | 2 | 12.73 | −0.034 Va | −2.05 | FCT = −0.02 | Va & FCT = 0.533 |
| AAWGr | 24 | 378.92 | 1.263 Vb | 77.11 | FSC = 0.76 | Vb & FSC = 0.000 | |
| Within accessions | 292 | 119.28 | 0.408 Vc | 24.94 | FST = 0.75 | Vc & FST = 0.000 | |
| Total | 317 | 510.93 | 1.634 | ||||
| Fruit size | f Among groups | 1 | 70.74 | 0.440 Va | 22.53 | FCT = 0.23 | Va & FCT = 0.000 |
| AAWGr | 19 | 252.18 | 1.076 Vb | 55.08 | FSC = 0.71 | Vb & FSC = 0.000 | |
| Within accessions | 233 | 101.97 | 0.438 Vc | 22.39 | FST = 0.77 | Vc & FST = 0.000 | |
| Total | 253 | 424.89 | 1.954 | ||||
| Growth type | g Among groups | 2 | 111.77 | 0.420 Va | 21.98 | FCT = 0.22 | Va & FCT = 0.000 |
| AAWGr | 25 | 353.13 | 1.106 Vb | 57.85 | FSC = 0.74 | Vb & FSC = 0.000 | |
| Within accessions | 320 | 123.36 | 0.385 Vc | 20.16 | FST = 0.79 | Vc & FST = 0.000 | |
| Total | 347 | 588.28 | 1.912 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villanueva-Gutierrez, E.E.; Johansson, E.; Prieto-Linde, M.L.; Centellas Quezada, A.; Olsson, M.E.; Geleta, M. Simple Sequence Repeat Markers Reveal Genetic Diversity and Population Structure of Bolivian Wild and Cultivated Tomatoes (Solanum lycopersicum L.). Genes 2022, 13, 1505. https://doi.org/10.3390/genes13091505
Villanueva-Gutierrez EE, Johansson E, Prieto-Linde ML, Centellas Quezada A, Olsson ME, Geleta M. Simple Sequence Repeat Markers Reveal Genetic Diversity and Population Structure of Bolivian Wild and Cultivated Tomatoes (Solanum lycopersicum L.). Genes. 2022; 13(9):1505. https://doi.org/10.3390/genes13091505
Chicago/Turabian StyleVillanueva-Gutierrez, Evelyn E., Eva Johansson, Maria Luisa Prieto-Linde, Alberto Centellas Quezada, Marie E. Olsson, and Mulatu Geleta. 2022. "Simple Sequence Repeat Markers Reveal Genetic Diversity and Population Structure of Bolivian Wild and Cultivated Tomatoes (Solanum lycopersicum L.)" Genes 13, no. 9: 1505. https://doi.org/10.3390/genes13091505
APA StyleVillanueva-Gutierrez, E. E., Johansson, E., Prieto-Linde, M. L., Centellas Quezada, A., Olsson, M. E., & Geleta, M. (2022). Simple Sequence Repeat Markers Reveal Genetic Diversity and Population Structure of Bolivian Wild and Cultivated Tomatoes (Solanum lycopersicum L.). Genes, 13(9), 1505. https://doi.org/10.3390/genes13091505










