Effects of Exogenous Potassium (K+) Application on the Antioxidant Enzymes Activities in Leaves of Tamarix ramosissima under NaCl Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. NaCl Treatment and NaCl + KCl Treatment
2.3. Analysis of Leaf Salt Secretion under Different Treatments of T. ramosissima
2.4. Determination and Analysis of Antioxidant Enzyme Activity
2.5. Transcriptome Sequencing and Differentially Expressed Gene Screening
2.6. Validation of Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
3. Results
3.1. Analysis of Salt Secretion of T. ramosissima Leaves under Different Treatments
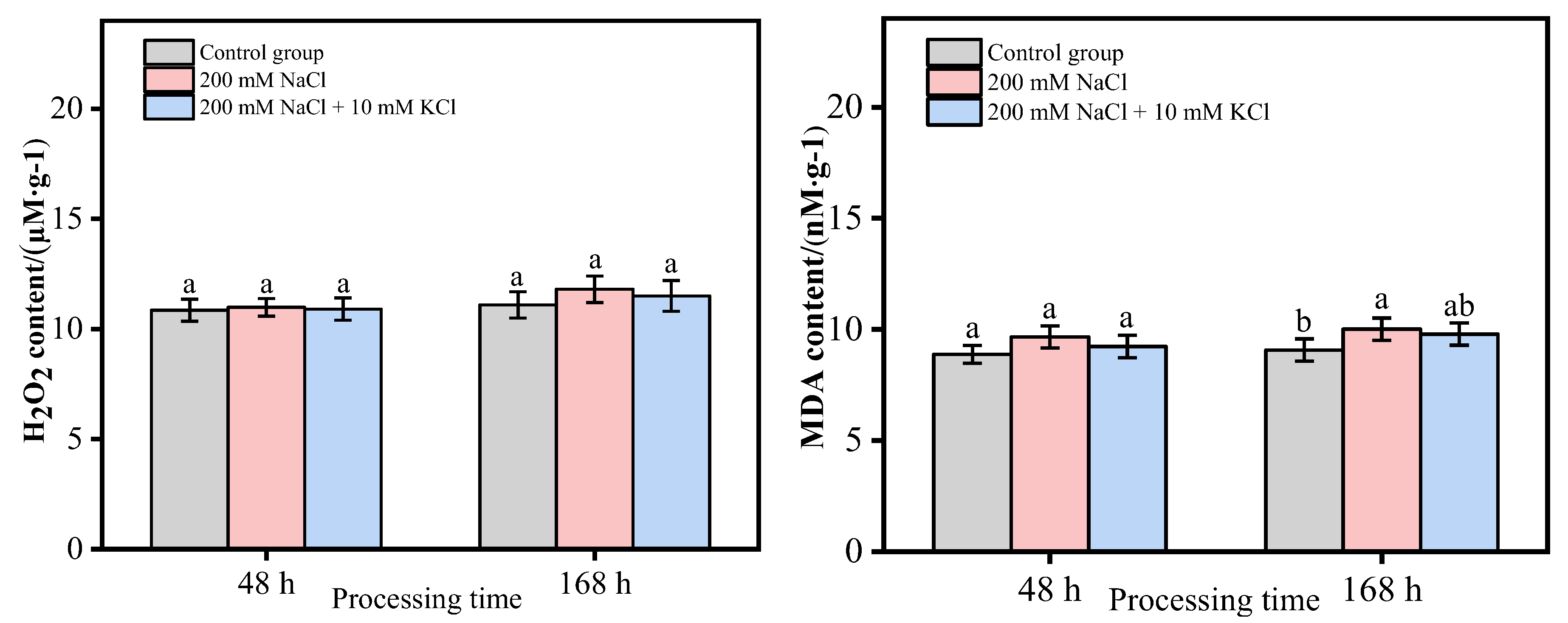
3.2. Analysis of H2O2 and MDA Content in T. ramosissima Leaves under Different Treatments
3.3. Antioxidant Enzyme Activity Analysis of T. ramosissima Leaves under Different Treatments
3.4. KEGG Pathway Analysis of DEGs
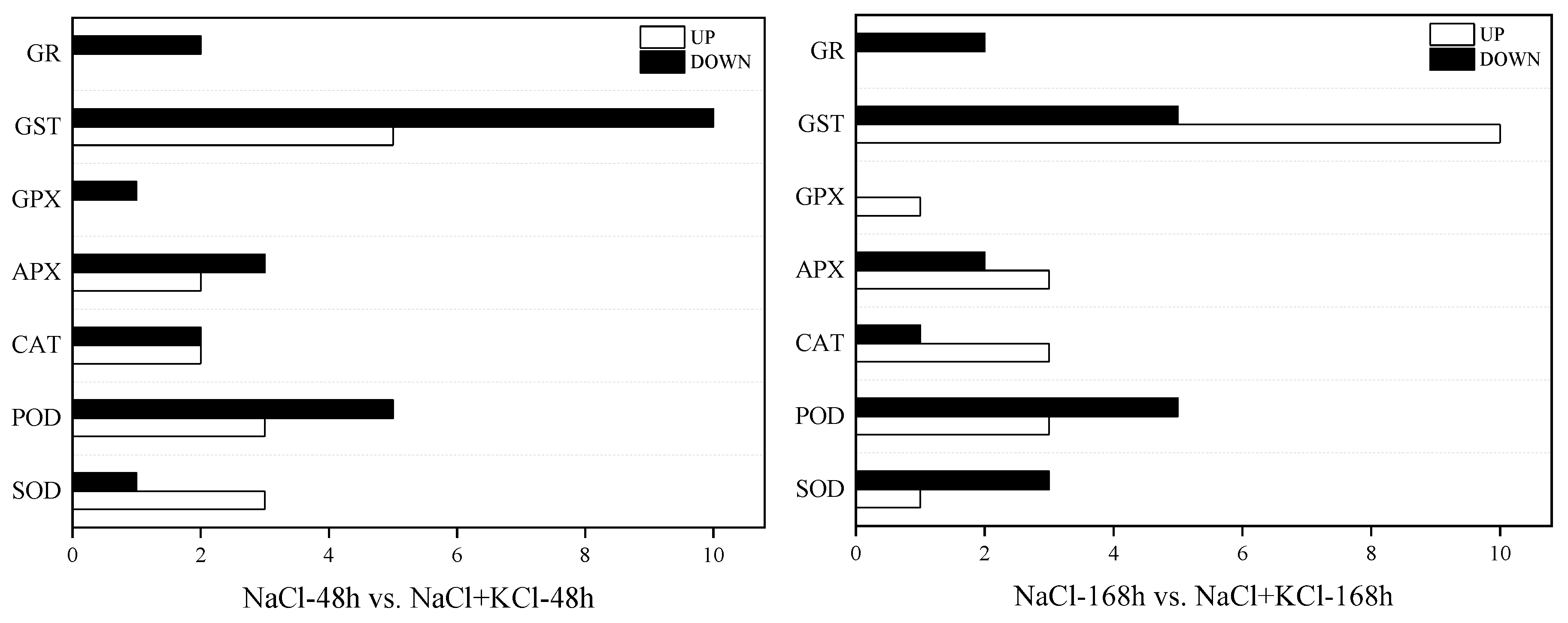
3.5. Analysis of DEGs in Leaves of T. ramosissima by Exogenous Potassium (K+) Application under NaCl Stress
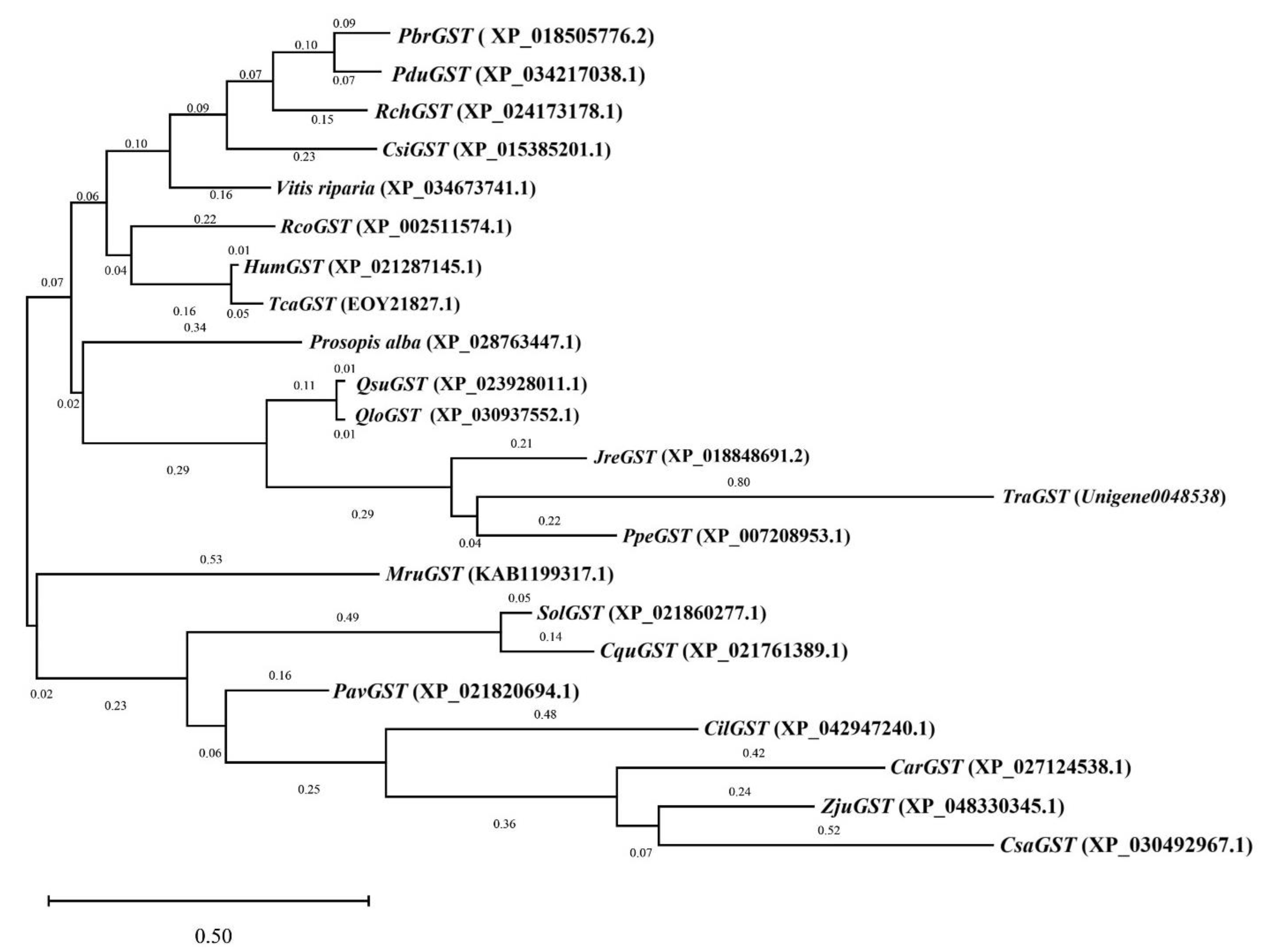
3.6. Phylogenetic Tree Analysis of Key Candidate Genes in Antioxidative Enzyme Activity
3.7. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Munns, R. Physiological processes limiting plant growth in saline soils: Some dogmas and hypotheses. Plant. Cell. Environ. 1993, 16, 15–24. [Google Scholar] [CrossRef]
- Anschutz, U.; Becker, D.; Shabala, S. Going beyond nutrition: Regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 2014, 171, 670–687. [Google Scholar] [CrossRef]
- Leigh, R.A.; Jones, R.G.W. A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol. 1984, 97, 1–13. [Google Scholar] [CrossRef]
- Mahmood, K. Salinity tolerance in barley (hordeum vulgare L.): Effects of varying NaCL, K+/Na+ and NaHCO3 levels on cultivars differing in tolerance. Pak. J. Bot. 2011, 43, 1651–1654. [Google Scholar]
- Bot, P.J.; Abbasi, G.H.; Akhtar, J.; Anwar-Ul-Haq, M.; Malik, W. Exogenous potassium differentially mitigates salt stress in tolerant and sensitive maize hybrids. Pak. J. Bot. 2015, 46, 135–146. [Google Scholar]
- Shabala, S.; Cuin, T.A. Potassium transport and plant salt tolerance. Physiol. Plantarum. 2008, 133, 651–669. [Google Scholar] [CrossRef]
- Wang, X.; Mohamed, I.; Ali, M.; Abbas, M.H.H.; Shah, G.M.; Chen, F. Potassium distribution in root and non-root zones of two cotton genotypes and its accumulation in their organs as affected by drought and potassium stress conditions. J. Plant Nutr. Soil Sci. 2019, 182, 72–81. [Google Scholar] [CrossRef]
- Gorham, J.U.C.O.; Hardy, C.; Jones, R.G.W.; Joppa, L.R.; Law, C.N. Chromosomal location of a K/Na discrimination character in the D genome of wheat. Theor. Appl. Genet. 1987, 74, 584–588. [Google Scholar] [CrossRef]
- Misra, N.; Gupta, A.K. Effect of salinity and different nitrogen sources on the activity of antioxidant enzymes and indole alkaloid content in Catharanthus roseus seedlings. J. Plant Physiol. 2006, 163, 11–18. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends. Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Ramachandra, R.A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef]
- Demidchik, V.; Shabala, S.N.; Coutts, K.B.; Tester, M.A.; Davies, J.M. Free oxygen radicals regulate plasma membrane Ca2+ and K+-permeable channels in plant root cells. J. Cell. Sci. 2003, 116, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Cuin, T.A.; Shabala, S. Compatible solutes reduce ROS-induced potassium efflux in Arabidopsis roots. Plant Cell. Environment. 2007, 30, 875–885. [Google Scholar] [CrossRef]
- Cakmak, I. The role of potassium in alleviating detrimental effects of abiotic stresses in plants. J. Plant Nutr. Soil. Sci. 2005, 168, 521–530. [Google Scholar] [CrossRef]
- Bosabalidis, A.M.; Thomson, W.W. Light microscopical studies on salt gland development in Tamarix aphylla L. Ann. Bot. 1984, 54, 169–174. [Google Scholar] [CrossRef]
- Wang, Z.; He, Z.; Xu, X.; Shi, X.; Ji, X.; Wang, Y. Revealing the salt tolerance mechanism of Tamarix hispida by large-scale identification of genes conferring salt tolerance. Tree Physiol. 2021, 41, 2153–2170. [Google Scholar] [CrossRef]
- Somaru, R.; Naidoo, Y.; Naidoo, G. Morphology and ultrastructure of the leaf salt glands of Odyssea paucinervis (Stapf) (Poaceae). Flora—Morphol. Distrib. Funct. Ecol. Plants 2002, 197, 67–75. [Google Scholar] [CrossRef]
- Garthwaite, A.J.; von Bothmer, R.; Colmer, T.D. Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl- into the shoots. J. Exp. Bot. 2005, 56, 2365–2378. [Google Scholar] [CrossRef]
- Lu, Y.; Lei, J.Q.; Zeng, F.J.; Xu, L.S.; Peng, S.L.; Gao, H.; Liu, G.J. Effects of NaCl treatment on growth and Ec-ophysiology Characteristics of Tamarix ramossisma. J. Des. Res. 2014, 34, 1509–1515. (In Chinese) [Google Scholar] [CrossRef]
- Song, Z.; Su, Y. Distinctive potassium-accumulation capability of alligatorweed (Alternanthera philoxeroides) links to high-affinity potassium transport facilitated by K+-uptake systems. Weed Sci. 2012, 61, 77–84. [Google Scholar] [CrossRef]
- Song, Z.; Yang, S.; Zhu, H.; Jin, M.; Su, Y. Heterologous expression of an alligatorweed high-affinity potassium transporter gene enhances salinity tolerance in Arabidopsis thaliana. Am. J. Bot. 2014, 101, 840–850. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, G.; Zhang, H.; Zhang, N.; Jiang, J.; Song, Z. Transcriptome analysis of Tamarix ramosissima leaves in response to NaCl stress. PLoS ONE 2022, 17, e265653. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.; Catherine, A.B.; Judith, A.B.; David, B.; Heather, B.; Cherry, J.M.; Allan, P.D.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Wilson, H.; Mycock, D.; Weiersbye, I.M. The salt glands of Tamarix usneoides E. Mey. ex Bunge (South African Salt Cedar). Int. J. Phytoremediat. 2016, 19, 587–595. [Google Scholar] [CrossRef]
- Zhu, J. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 124, 941–948. [Google Scholar] [CrossRef]
- Ali, S.; Zeng, F.; Cai, S.; Qiu, B.; Zhang, G. The interaction of salinity and chromium in the influence of barley growth and oxidative stress. Plant Soil Environ. 2011, 57, 153–159. [Google Scholar] [CrossRef]
- Sun, J.; Li, T.; Xia, J.; Tian, J.; Lu, Z.; Wang, R. Influence of salt stress on ecophysiological parameters of Periploca sepium bunge. Plant Soil Environ. 2011, 57, 139–144. [Google Scholar] [CrossRef]
- Liang, Y.; Chen, Q.; Liu, Q.; Zhang, W.; Ding, R. Exogenous silicon (Si) increases antioxidant enzyme activity and reduces lipid peroxidation in roots of salt-stressed barley (Hordeum vulgare L.). J. Plant Physiol. 2003, 160, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.R.; Bewley, J.D. Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 1980, 65, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Reichheld, J.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Lazzarotto, F.; Teixeira, F.K.; Rosa, S.B.; Dunand, C.; Fernandes, C.L.; de Vasconcelos, F.A.; Silveira, J.; Verli, H.; Margis, R.; Margis-Pinheiro, M. Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol 2011, 191, 234–250. [Google Scholar] [CrossRef] [PubMed]
- Badawi, G.H.; Kawano, N.; Yamauchi, Y.; Shimada, E.; Sasaki, R.; Kubo, A.; Tanaka, K. Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol. Plant 2004, 121, 231–238. [Google Scholar] [CrossRef]
- Soleimanzadeh, H.; Habibi, D.; Ardakani, M.R.; Paknejad, F.; Rejali, F. Effect of potassium levels on antioxidant enzymes and malondialdehyde content under drought stress in sunflower (Helianthus annuus L.). Am. J. Agric. Biol. Sci. 2010, 5, 56–61. [Google Scholar] [CrossRef]
- Guo, P.; Yang, X.; Mao, H.; Wang, X.; Shen, Z.; Liu, D.; Yang, A.; Wang, Y. Overexpression and knockout of NtPHGPx altered salt tolerance of tobacco seedlings. Chin. Tob. Sci. 2021, 42, 8–14. (In Chinese) [Google Scholar] [CrossRef]
- Chen, S.; Vaghchhipawala, Z.; Li, W.; Asard, H.; Dickman, M.B. Tomato phospholipid hydroperoxide glutathione peroxidase inhibits cell death induced by Bax and oxidative stresses in yeast and plants. Plant Physiol. 2004, 135, 1630–1641. [Google Scholar] [CrossRef]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell. Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef]
- Le Martret, B.; Poage, M.; Shiel, K.; Nugent, G.D.; Dix, P.J. Tobacco chloroplast transformants expressing genes encoding dehydroascorbate reductase, glutathione reductase, and glutathione-S-transferase, exhibit altered anti-oxidant metabolism and improved abiotic stress tolerance. Plant Biotechnol. J. 2011, 9, 661–673. [Google Scholar] [CrossRef]
- Hao, Y.; Xu, S.; Lyu, Z.; Wang, H.; Kong, L.; Sun, S. Comparative analysis of the Glutathione S-Transferase gene family of four triticeae species and transcriptome analysis of GST genes in common wheat responding to salt stress. Int. J. Genom. 2021, 2021, 6289174. [Google Scholar] [CrossRef] [PubMed]
- Jha, B.; Sharma, A.; Mishra, A. Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol. Biol. Rep. 2011, 38, 4823–4832. [Google Scholar] [CrossRef] [PubMed]
- Hussain, T.; Li, J.; Feng, X.; Asrar, H.; Gul, B.; Liu, X. Salinity induced alterations in photosynthetic and oxidative regulation are ameliorated as a function of salt secretion. J. Plant Res. 2021, 134, 779–796. [Google Scholar] [CrossRef]
- Yue, C.; Cao, H.; Zhou, Y.; Wang, L.; Hao, X.; Wang, X.; Yang, Y. Cloning and expression analysis of glutathione reductase genes (CsGRs) in tea plant (Camellia sinensis). Sci. Agric. Sin. 2014, 47, 3279–3291. (In Chinese) [Google Scholar] [CrossRef]


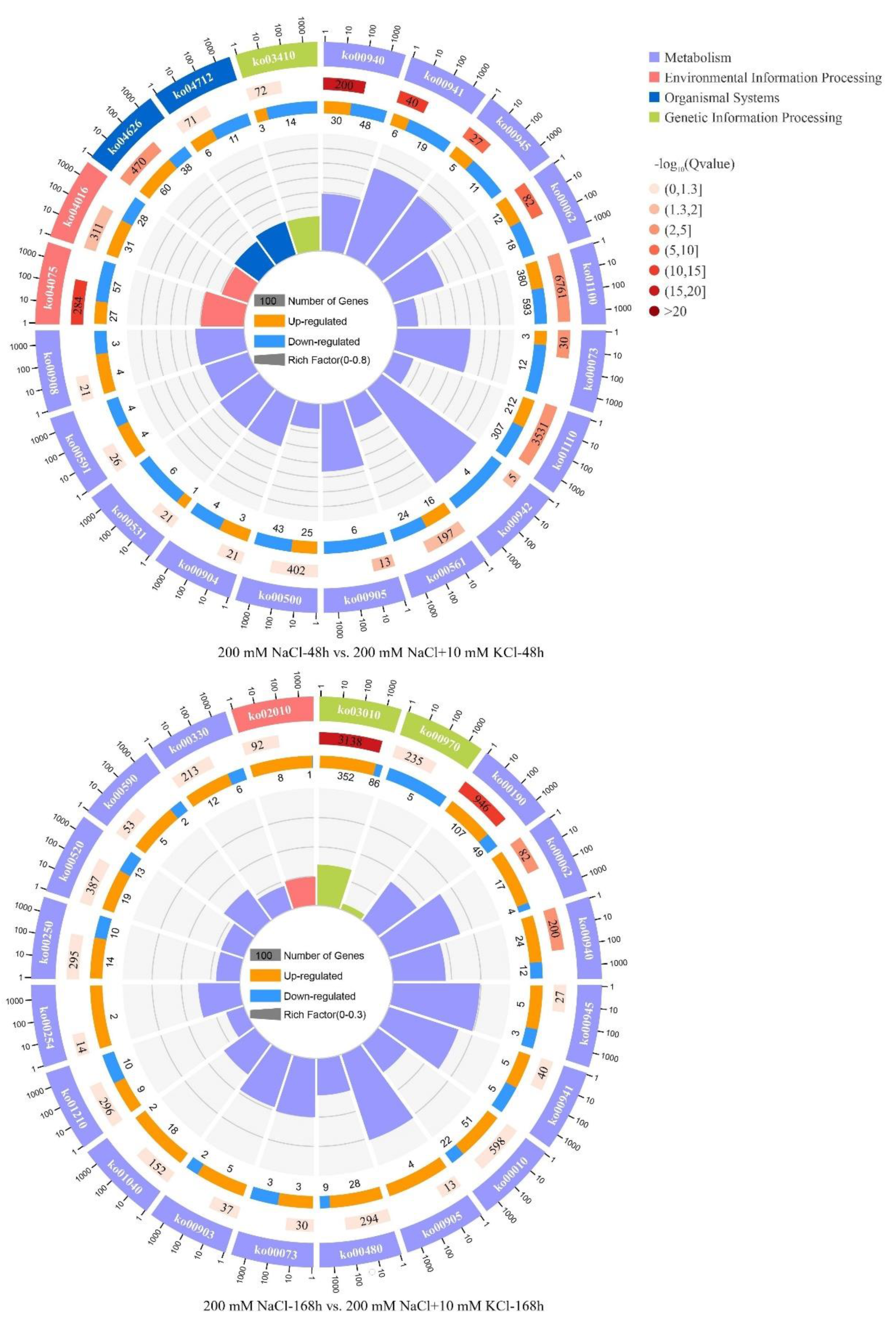
| ID | Gene Numbers | Class | KEGG Pathway | p-Value | Up | Down |
|---|---|---|---|---|---|---|
| 200 mM NaCl-48 h vs. 200 mM NaCl + 10 mM KCl-48 h | ||||||
| Phenylpropanoid biosynthesis | 78 | Metabolism | ko00940 | 0.000000 | 30 | 48 |
| Plant hormone signal transduction | 84 | Environmental Information Processing | ko04075 | 0.000000 0.000000 | 27 | 57 |
| Flavonoid biosynthesis | 25 | Metabolism | ko00941 | 0.000000 | 6 | 19 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 16 | Metabolism | ko00945 | 0.000000 | 5 | 11 |
| Fatty acid elongation | 30 | Metabolism | ko00062 | 0.000000 | 12 | 18 |
| Plant–pathogen interaction | 98 | Organismal Systems | ko04626 | 0.000000 | 60 | 38 |
| Metabolic pathways | 973 | Metabolism | ko01100 | 0.000000 | 380 | 593 |
| Cutin, suberine and wax biosynthesis | 15 | Metabolism | ko00073 | 0.000000 | 3 | 12 |
| Biosynthesis of secondary metabolites | 519 | Metabolism | ko01110 | 0.000193 | 212 | 307 |
| Anthocyanin biosynthesis | 4 | Metabolism | ko00942 | 0.001236 | 0 | 4 |
| MAPK signaling pathway—plant | 59 | Environmental Information Processing | ko04016 | 0.001374 | 31 | 28 |
| Glycerolipid metabolism | 40 | Metabolism | ko00561 | 0.002193 | 16 | 24 |
| Brassinosteroid biosynthesis | 6 | Metabolism | ko00905 | 0.003482 | 0 | 6 |
| Circadian rhythm-plant | 17 | Organismal Systems | ko04712 | 0.007701 | 6 | 11 |
| Base excision repair | 17 | Genetic Information Processing | ko03410 | 0.008901 | 3 | 14 |
| Starch and sucrose metabolism | 68 | Metabolism | ko00500 | 0.010989 | 25 | 43 |
| Glycosaminoglycan degradation | 7 | Metabolism | ko00531 | 0.013165 | 1 | 6 |
| Diterpenoid biosynthesis | 7 | Metabolism | ko00904 | 0.013165 | 3 | 4 |
| Zeatin biosynthesis | 7 | Metabolism | ko00908 | 0.013165 | 4 | 3 |
| Linoleic acid metabolism | 8 | Metabolism | ko00591 | 0.013753 | 4 | 4 |
| 200 mM NaCl-168 h vs. 200 mM NaCl + 10 mM KCl-168 h | ||||||
| Ribosome | 438 | Genetic Information Processing | ko03010 | 0.000000 | 352 | 86 |
| Oxidative phosphorylation | 156 | Metabolism | ko00190 | 0.000000 | 107 | 49 |
| Fatty acid elongation | 21 | Metabolism | ko00062 | 0.000000 | 17 | 4 |
| Phenylpropanoid biosynthesis | 36 | Metabolism | ko00940 | 0.000000 | 24 | 12 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 8 | Metabolism | ko00945 | 0.002351 | 5 | 3 |
| Flavonoid biosynthesis | 10 | Metabolism | ko00941 | 0.002846 | 5 | 5 |
| Glycolysis/Gluconeogenesis | 73 | Metabolism | ko00010 | 0.008388 | 51 | 22 |
| Brassinosteroid biosynthesis | 4 | Metabolism | ko00905 | 0.026406 | 4 | 0 |
| Glutathione metabolism | 37 | Metabolism | ko00480 | 0.033437 | 28 | 9 |
| Limonene and pinene degradation | 7 | Metabolism | ko00903 | 0.050223 | 5 | 2 |
| Cutin, suberine and wax biosynthesis | 6 | Metabolism | ko00073 | 0.053552 | 3 | 3 |
| Biosynthesis of unsaturated fatty acids | 20 | Metabolism | ko01040 | 0.068133 | 18 | 2 |
| Diterpenoid biosynthesis | 4 | Metabolism | ko00904 | 0.123174 | 1 | 3 |
| Citrate cycle (TCA cycle) | 52 | Metabolism | ko00020 | 0.149175 | 33 | 19 |
| Ascorbate and aldarate metabolism | 18 | Metabolism | ko00053 | 0.172390 | 11 | 7 |
| Fatty acid metabolism | 40 | Metabolism | ko01212 | 0.177221 | 35 | 5 |
| Sesquiterpenoid and triterpenoid biosynthesis | 5 | Metabolism | ko00909 | 0.185229 | 2 | 3 |
| Plant hormone signal transduction | 31 | Environmental Information Processing | ko04075 | 0.189067 | 21 | 10 |
| Arachidonic acid metabolism | 7 | Metabolism | ko00590 | 0.215454 | 5 | 2 |
| Flavone and flavonol biosynthesis | Metabolism | ko00944 | 0.252624 | 0 | 1 | |
| Pathway | Gene ID | Description | Log2 Fold-Change | |
|---|---|---|---|---|
| 200 mM NaCl-48 h vs. 200 mM NaCl + 10 mM KCl-48 h | 200 mM NaCl-168 h vs. 200 mM NaCl + 10 mM KCl-168 h | |||
| SOD | ||||
| ko04146 | Unigene0033269 | SOD4 protein, partial | 0.69 | −0.06 |
| Unigene0049419 | Superoxide dismutase [Mn] | −7.82 | −9.54 | |
| Unigene0050462 | Superoxide dismutase | 0.73 | 0.71 | |
| Unigene0082550 | Superoxide dismutase | 0.25 | −0.25 | |
| POD | ||||
| ko01100; ko01110; ko00940 | Unigene0009260 | Peroxidase 20 | −1.66 | −0.10 |
| Unigene0013825 | Peroxidase | −1.87 | 0.99 | |
| Unigene0013827 | Peroxidase | −0.65 | −0.09 | |
| Unigene0014843 | Peroxidase | 3.20 | 1.25 | |
| Unigene0029752 | Peroxidase 17 | −2.04 | 0.03 | |
| Unigene0049353 | Peroxidase 5 | 4.50 | −1.35 | |
| Unigene0086491 | Peroxidase 52 | −1.70 | −0.59 | |
| Unigene0094375 | Peroxidase 31 | 1.10 | −0.42 | |
| CAT | ||||
| ko01100; ko01110; ko01200; ko00630; ko04146; ko04016; ko00380 | Unigene0046159 | Catalase isozyme 1 | 0.77 | 0.25 |
| Unigene0046160 | Catalase, partial | 0.93 | 0.40 | |
| Unigene0087092 | Leaf catalase | −0.75 | 0.38 | |
| Unigene0103080 | Catalase isozyme 1 | −5.74 | −12.41 | |
| APX | ||||
| ko01100; ko00480 | Unigene0008032 | L-ascorbate peroxidase 3 | 0.01 | 0.21 |
| Unigene0008033 | L-ascorbate peroxidase 3 | −0.34 | −0.43 | |
| Unigene0008513 | Peroxidase domain-containing | −1.50 | 0.47 | |
| Unigene0048033 | Cytosolic ascorbate peroxidase | 0.62 | 0.13 | |
| Unigene0105664 | Thylakoid ascorbate Peroxidase precursor, partial | −0.35 | −0.21 | |
| GPX | ||||
| ko01100; ko00480; ko00590 | Unigene0035407 | Glutathione peroxidase | −1.32 | 0.34 |
| GST | ||||
| ko01100; ko00480 | Unigene0001041 | Glutathione S-transferase | −1.61 | 5.20 |
| Unigene0004890 | Glutathione S-transferase T1-like | 1.03 | 0.58 | |
| Unigene0007072 | Glutathione S-transferase U17-like | −1.13 | −1.16 | |
| Unigene0012650 | Glutathione S-transferase Mu 1-like | −4.15 | 0.13 | |
| Unigene0015109 | Glutathione S-transferase U8-like | 1.48 | 0.44 | |
| Unigene0020552 | Glutathione S-transferase | 0.48 | 0.12 | |
| Unigene0041633 | Microsomal glutathione S-transferase 3-like | −0.24 | −0.04 | |
| Unigene0048538 | Glutathione S-transferase U10-like | 3.56 | 3.19 | |
| Unigene0056773 | Glutathione S-transferase | −0.04 | −0.44 | |
| Unigene0064942 | Glutathione S-transferase L3 | −0.47 | 0.33 | |
| Unigene0069058 | Glutathione-S-transferase | 0.72 | −0.06 | |
| Unigene0069060 | Glutathione S-transferase L3-like | −1.55 | 0.48 | |
| Unigene0081745 | Glutathione S-transferase U10-like | −1.08 | −0.63 | |
| Unigene0082147 | Glutathione S-transferase F11-like | −3.80 | 0.49 | |
| Unigene0098941 | Glutathione S-transferase U9 | −0.83 | 5.91 | |
| GR | ||||
| ko01100; ko00480 | Unigene0075696 | Glutathione reductase | −0.50 | −0.10 |
| Unigene0098587 | Glutathione reductase-like | −8.98 | −10.52 | |
| Family | Species | Description | Gene | Protein ID | CDS (bp) | ORF Length (aa) |
|---|---|---|---|---|---|---|
| Amaranthaceae | Spinacia oleracea | Glutathione S-transferase U10-like | SolGST | XP_021860277.1 | 693 | 230 |
| Euphorbiaceae | Ricinus communis | Glutathione S-transferase U9 | RcoGST | XP_002511574.1 | 696 | 231 |
| Rhamnaceae | Ziziphus jujuba var. spinosa | Glutathione S-transferase U9-like | ZjuGST | XP_048330345.1 | 693 | 230 |
| Malvaceae | Herrania umbratica | Glutathione S-transferase U9-like | HumGST | XP_021287145.1 | 693 | 230 |
| Juglandaceae | Carya illinoinensis | Glutathione S-transferase U9 | CilGST | XP_042947240.1 | 705 | 234 |
| Amaranthaceae | Chenopodium quinoa | Glutathione S-transferase U10-like | CquGST | XP_021761389.1 | 693 | 230 |
| Malvaceae | Theobroma cacao | Glutathione S-transferase tau 9 | TcaGST | EOY21827.1 | 693 | 230 |
| Rosaceae | Pyrus× bretschneideri | Glutathione S-transferase U9 | PbrGST | XP_018505776.2 | 693 | 230 |
| Juglandaceae | Juglans regia | Glutathione S-transferase U9-like | JreGST | XP_018848691.2 | 690 | 229 |
| Rosaceae | Prunus avium | Glutathione S-transferase U10-like | PavGST | XP_021820694.1 | 696 | 231 |
| Fagaceae | Quercus suber | Glutathione S-transferase U9-like | QsuGST | XP_023928011.1 | 699 | 232 |
| Rosaceae | Rosa chinensis | Glutathione S-transferase U10 | RchGST | XP_024173178.1 | 702 | 233 |
| Fabaceae | Prosopis alba | Glutathione S-transferase U9-like | PalGST | XP_028763447.1 | 699 | 232 |
| Rosaceae | Prunus dulcis | Glutathione S-transferase U9-like | PduGST | XP_034217038.1 | 693 | 230 |
| Fagaceae | Quercus lobata | Glutathione S-transferase U9 | QloGST | XP_030937552.1 | 699 | 232 |
| Rutaceae | Citrus sinensis | Glutathione S-transferase U9-like | CsiGST | XP_015385201.1 | 693 | 230 |
| Rosaceae | Prunus persica | Glutathione S-transferase U10 | PpeGST | XP_007208953.1 | 699 | 232 |
| Rubiaceae | Coffea arabica | Glutathione S-transferase U9-like | CarGST | XP_027124538.1 | 705 | 234 |
| Myricaceae | Morella rubra | Glutathione S-transferase U9 | MruGST | KAB1199317.1 | 696 | 231 |
| Cannabaceae | Cannabis sativa | Glutathione S-transferase U9 | CsaGST | XP_030492967.1 | 693 | 230 |
| Vitaceae | Vitis riparia | glutathione S-transferase U9-like | VriGST | XP_034673741.1 | 696 | 231 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Zhang, S.; Du, S.; Zhang, X.; Wang, G.; Huang, J.; Jiang, J. Effects of Exogenous Potassium (K+) Application on the Antioxidant Enzymes Activities in Leaves of Tamarix ramosissima under NaCl Stress. Genes 2022, 13, 1507. https://doi.org/10.3390/genes13091507
Chen Y, Zhang S, Du S, Zhang X, Wang G, Huang J, Jiang J. Effects of Exogenous Potassium (K+) Application on the Antioxidant Enzymes Activities in Leaves of Tamarix ramosissima under NaCl Stress. Genes. 2022; 13(9):1507. https://doi.org/10.3390/genes13091507
Chicago/Turabian StyleChen, Yahui, Shiyang Zhang, Shanfeng Du, Xiaomian Zhang, Guangyu Wang, Jiefan Huang, and Jiang Jiang. 2022. "Effects of Exogenous Potassium (K+) Application on the Antioxidant Enzymes Activities in Leaves of Tamarix ramosissima under NaCl Stress" Genes 13, no. 9: 1507. https://doi.org/10.3390/genes13091507
APA StyleChen, Y., Zhang, S., Du, S., Zhang, X., Wang, G., Huang, J., & Jiang, J. (2022). Effects of Exogenous Potassium (K+) Application on the Antioxidant Enzymes Activities in Leaves of Tamarix ramosissima under NaCl Stress. Genes, 13(9), 1507. https://doi.org/10.3390/genes13091507







