Mining High-Level Imaging Genetic Associations via Clustering AD Candidate Variants with Similar Brain Association Patterns
Abstract
:1. Introduction
2. Material and Methods
2.1. Data Description
2.2. Data Preprocessing
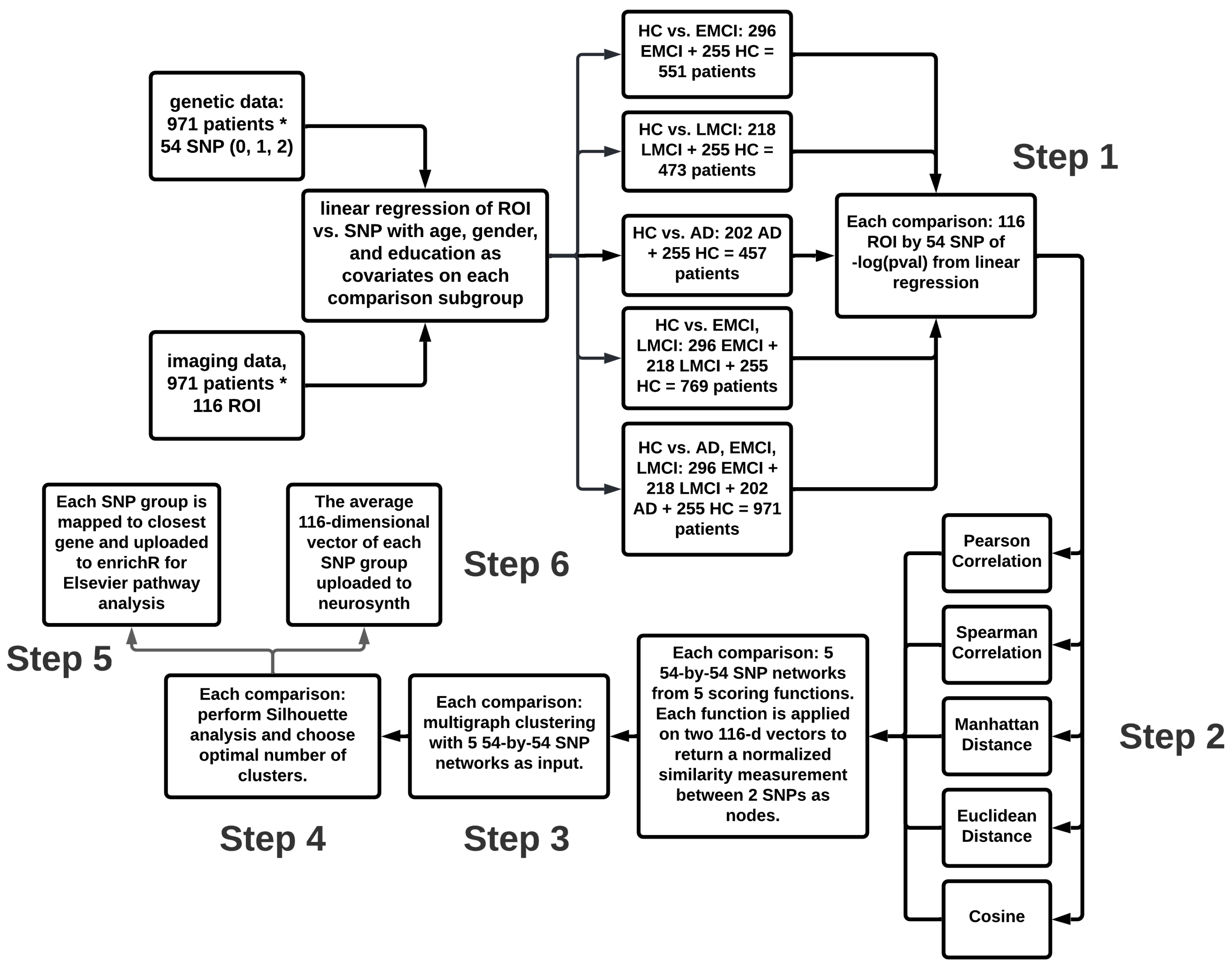
2.3. Method Overview
2.4. Step 1: Imaging Genetic Association Analysis
2.5. Step 2: SNP Networks with Different Similarity Measurements
2.6. Step 3: Multigraph Min-Max Graph Clustering
2.7. Step 4: Silhouette Scoring Analysis
2.8. Step 5: EnrichR Elsevier Pathway Analysis
2.9. Step 6: Neurovault Brain Region Analysis
3. Result
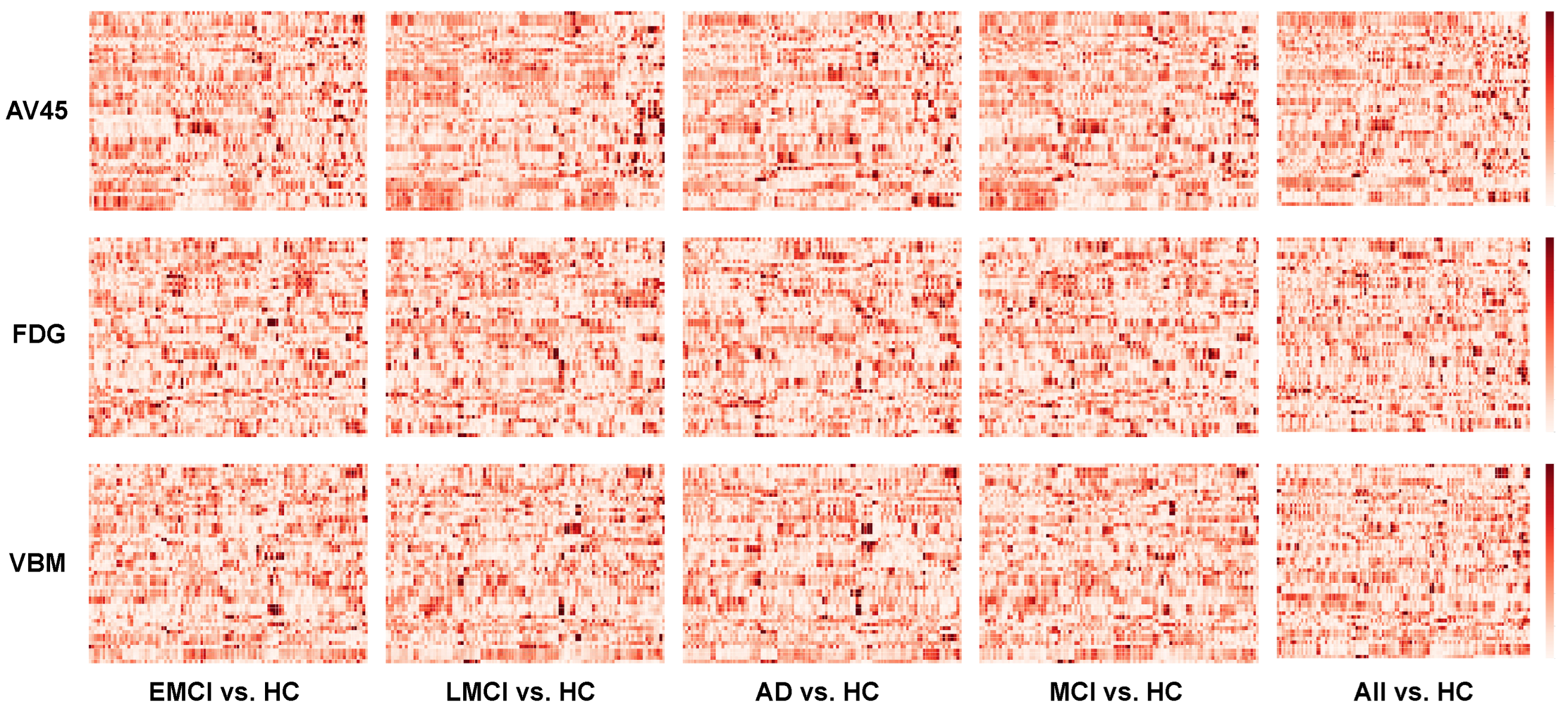
3.1. Imaging Genetic Association Maps
3.2. Multigraph vs. Single-Graph Silhouette Analysis
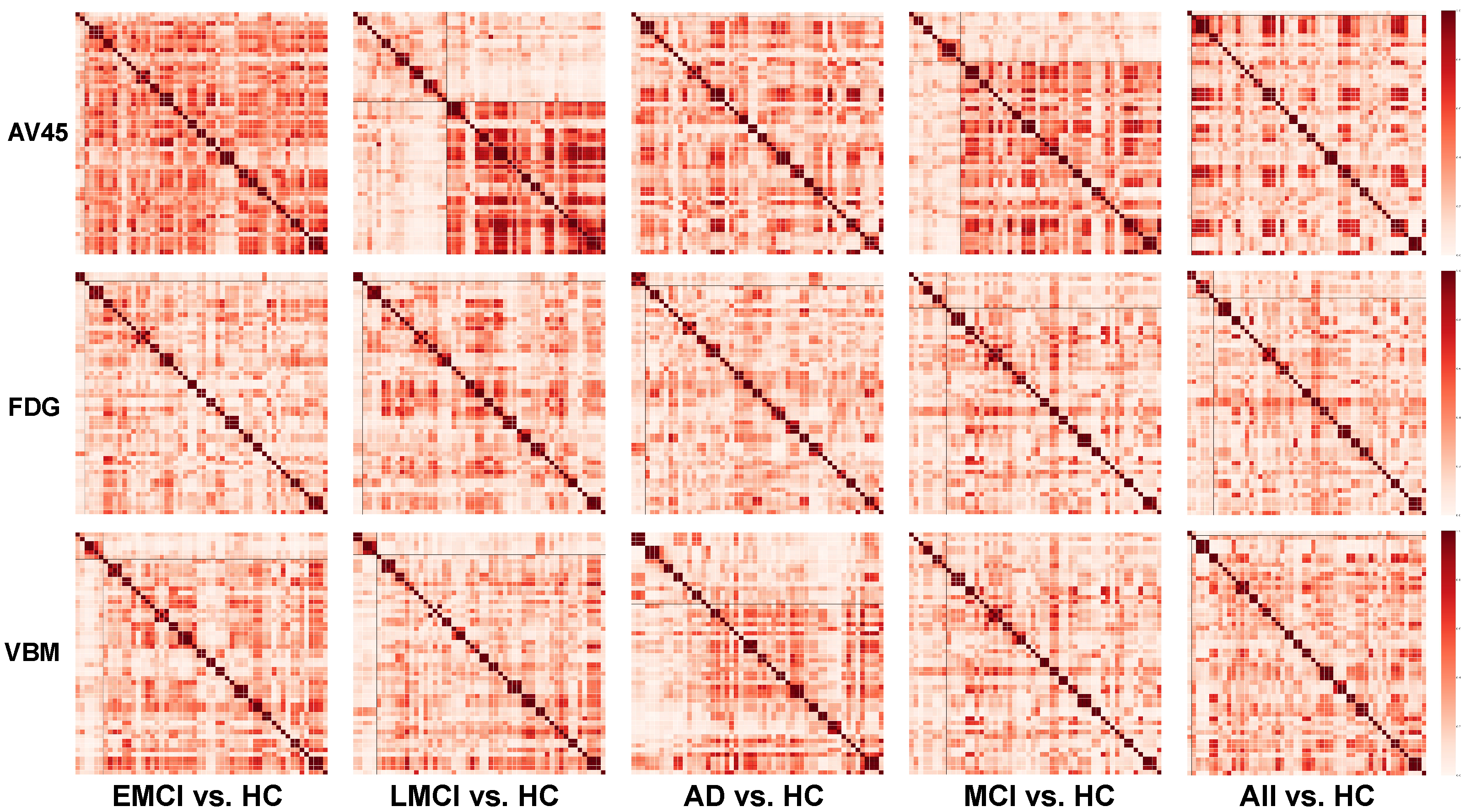
3.3. Clustering Results
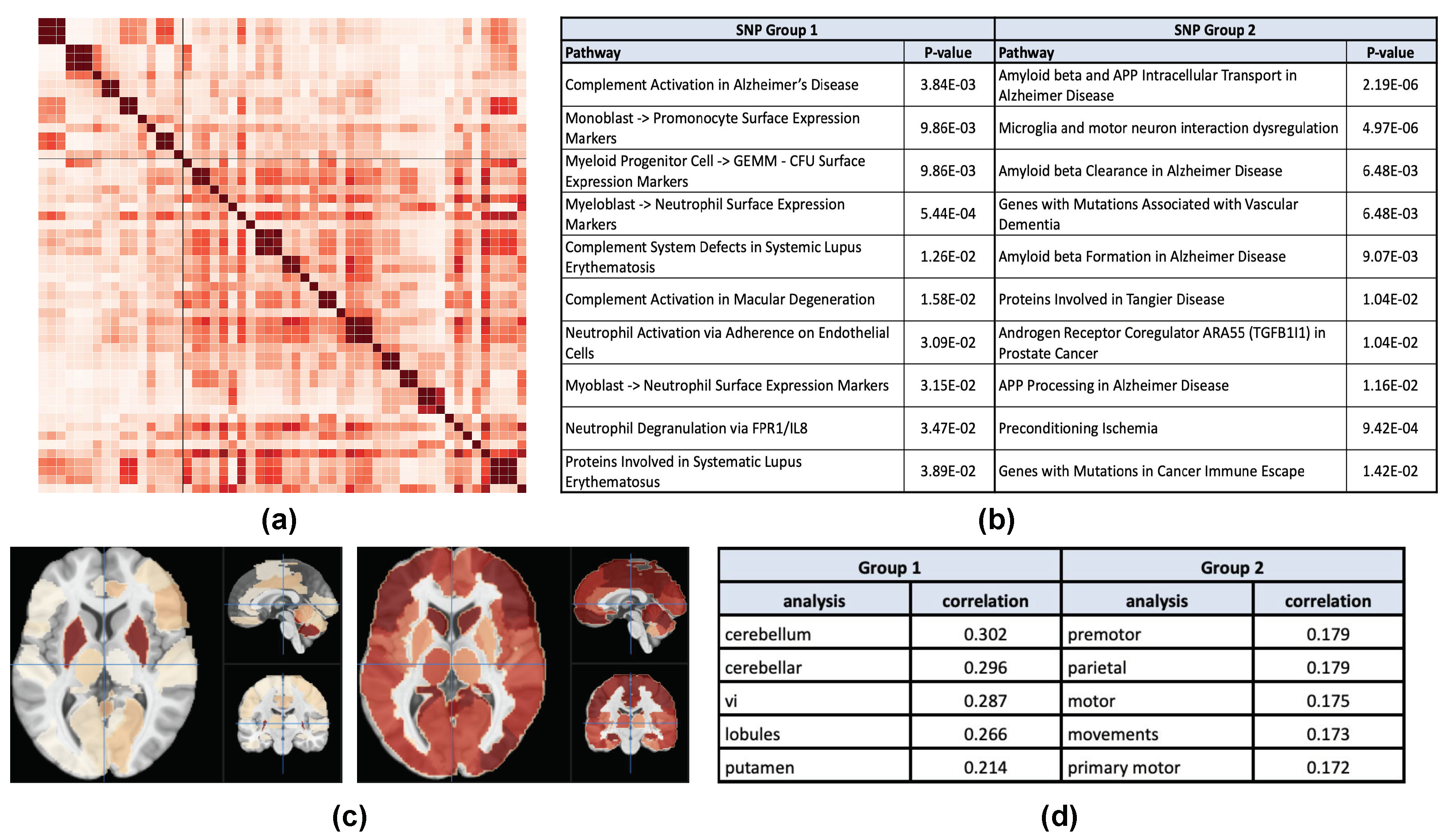
3.4. Case Study: Example AV45 Result
3.5. Case Study: Example VBM Result
4. Discussion
4.1. Comparison between Single-Graph and Multigraph Clusterings
4.2. AV45 Clustering Result
4.3. FDG Clustering Result
4.4. VBM Clustering Result
4.5. AV45 Case Study
4.6. VBM Case Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| GWAS | genome-wide association study |
| SNP | single nucleotide polymorphism |
| QT | quantitative traits |
| ROI | region of interest |
| MRI | magnetic resonance imaging |
| PET | positron emission tomography |
| HC | healthy control |
| EMCI | early mild cognitive impairment |
| LMCI | late mild cognitive impairment |
| AV45 | F-18 florbetapir |
| FDG | (18)F-fluorodeoxyglucose |
| VBM | voxel-based morphometry |
Appendix A
| HC | EMCI | LMCI | AD | Total | |
|---|---|---|---|---|---|
| Number of subject | 255 | 296 | 218 | 202 | 971 |
| Age | 76.35 ± 6.54 | 71.78 ± 7.28 | 74.71 ± 8.39 | 75.85 ± 7.67 | 74.48 ± 7.67 |
| Sex (Male/Female) | 132/123 | 167/129 | 129/89 | 123/79 | 551/420 |
| Education (Year) | 16.37 ± 2.64 | 12.12 ± 2.64 | 16.12 ± 2.94 | 15.83 ± 2.81 | 16.13 ± 2.75 |
| rs-ID | Chromosome | Position | Gene Symbol | rs-ID | Chromosome | Position | Gene Symbol |
|---|---|---|---|---|---|---|---|
| rs4575098 | chr1 | 161155392 | ADAMTS4 | rs7920721 | chr10 | 11720308 | ECHDC3 |
| rs6656401 | chr1 | 207692049 | CR1 | rs3740688 | chr11 | 47380340 | SPI1 |
| rs2093760 | chr1 | 207786828 | CR1 | rs10838725 | chr11 | 47557871 | CELF1 |
| rs4844610 | chr1 | 207802552 | CR1 | rs983392 | chr11 | 59923508 | MS4A6A |
| rs4663105 | chr2 | 127891427 | BIN1 | rs7933202 | chr11 | 59936926 | MS4A2 |
| rs6733839 | chr2 | 127892810 | BIN1 | rs2081545 | chr11 | 59958380 | MS4A6A |
| rs10933431 | chr2 | 233981912 | INPP5D | rs867611 | chr11 | 85776544 | PICALM |
| rs35349669 | chr2 | 234068476 | INPP5D | rs10792832 | chr11 | 85867875 | PICALM |
| rs6448453 | chr4 | 11026028 | CLNK | rs3851179 | chr11 | 85868640 | PICALM |
| rs190982 | chr5 | 88223420 | MEF2C-AS1 | rs17125924 | chr14 | 53391680 | FERMT2 |
| rs9271058 | chr6 | 32575406 | HLA-DRB1 | rs17125944 | chr14 | 53400629 | FERMT2 |
| rs9473117 | chr6 | 47431284 | CD2AP | rs10498633 | chr14 | 92926952 | SLC24A4 |
| rs9381563 | chr6 | 47432637 | CD2AP | rs12881735 | chr14 | 92932828 | SLC24A4 |
| rs10948363 | chr6 | 47487762 | CD2AP | rs12590654 | chr14 | 92938855 | SLC24A4 |
| rs2718058 | chr7 | 37841534 | GPR141 | rs442495 | chr15 | 59022615 | ADAM10 |
| rs4723711 | chr7 | 37844263 | GPR141 | rs59735493 | chr16 | 31133100 | KAT8 |
| rs1859788 | chr7 | 99971834 | PILRA | rs113260531 | chr17 | 5138980 | SCIMP |
| rs1476679 | chr7 | 100004446 | ZCWPW1 | rs28394864 | chr17 | 47450775 | ABI3 |
| rs12539172 | chr7 | 100091795 | NYAP1 | rs111278892 | chr19 | 1039323 | ABCA7 |
| rs10808026 | chr7 | 143099133 | EPHA1 | rs3752246 | chr19 | 1056492 | ABCA7 |
| rs7810606 | chr7 | 143108158 | EPHA1-AS1 | rs4147929 | chr19 | 1063443 | ABCA7 |
| rs11771145 | chr7 | 143110762 | EPHA1-AS1 | rs41289512 | chr19 | 45351516 | PVRL2 |
| rs28834970 | chr8 | 27195121 | PTK2B | rs3865444 | chr19 | 51727962 | CD33 |
| rs73223431 | chr8 | 27219987 | PTK2B | rs6024870 | chr20 | 54997568 | CASS4 |
| rs4236673 | chr8 | 27464929 | CLU | rs6014724 | chr20 | 54998544 | CASS4 |
| rs9331896 | chr8 | 27467686 | CLU | rs7274581 | chr20 | 55018260 | CASS4 |
| rs11257238 | chr10 | 11717397 | ECHDC3 | rs429358 | chr19 | 45411941 | APOE |
| Index | Name | Index | Name | Index | Name | Index | Name |
|---|---|---|---|---|---|---|---|
| 1 | Precentral_L | 30 | Insula_R | 59 | Parietal_Sup_L | 88 | Temporal_Pole_Mid_R |
| 2 | Precentral_R | 31 | Cingulum_Ant_L | 60 | Parietal_Sup_R | 89 | Temporal_Inf_L |
| 3 | Frontal_Sup_L | 32 | Cingulum_Ant_R | 61 | Parietal_Inf_L | 90 | Temporal_Inf_R |
| 4 | Frontal_Sup_R | 33 | Cingulum_Mid_L | 62 | Parietal_Inf_R | 91 | Cerebelum_Crus1_L |
| 5 | Frontal_Sup_Orb_L | 34 | Cingulum_Mid_R | 63 | SupraMarginal_L | 92 | Cerebelum_Crus1_R |
| 6 | Frontal_Sup_Orb_R | 35 | Cingulum_Post_L | 64 | SupraMarginal_R | 93 | Cerebelum_Crus2_L |
| 7 | Frontal_Mid_L | 36 | Cingulum_Post_R | 65 | Angular_L | 94 | Cerebelum_Crus2_R |
| 8 | Frontal_Mid_R | 37 | Hippocampus_L | 66 | Angular_R | 95 | Cerebelum_3_L |
| 9 | Frontal_Mid_Orb_L | 38 | Hippocampus_R | 67 | Precuneus_L | 96 | Cerebelum_3_R |
| 10 | Frontal_Mid_Orb_R | 39 | ParaHippocampal_L | 68 | Precuneus_R | 97 | Cerebelum_4_5_L |
| 11 | Frontal_Inf_Oper_L | 40 | ParaHippocampal_R | 69 | Paracentral_Lobule_L | 98 | Cerebelum_4_5_R |
| 12 | Frontal_Inf_Oper_R | 41 | Amygdala_L | 70 | Paracentral_Lobule_R | 99 | Cerebelum_6_L |
| 13 | Frontal_Inf_Tri_L | 42 | Amygdala_R | 71 | Caudate_L | 100 | Cerebelum_6_R |
| 14 | Frontal_Inf_Tri_R | 43 | Calcarine_L | 72 | Caudate_R | 101 | Cerebelum_7b_L |
| 15 | Frontal_Inf_Orb_L | 44 | Calcarine_R | 73 | Putamen_L | 102 | Cerebelum_7b_R |
| 16 | Frontal_Inf_Orb_R | 45 | Cuneus_L | 74 | Putamen_R | 103 | Cerebelum_8_L |
| 17 | Rolandic_Oper_L | 46 | Cuneus_R | 75 | Pallidum_L | 104 | Cerebelum_8_R |
| 18 | Rolandic_Oper_R | 47 | Lingual_L | 76 | Pallidum_R | 105 | Cerebelum_9_L |
| 19 | Supp_Motor_Area_L | 48 | Lingual_R | 77 | Thalamus_L | 106 | Cerebelum_9_R |
| 20 | Supp_Motor_Area_R | 49 | Occipital_Sup_L | 78 | Thalamus_R | 107 | Cerebelum_10_L |
| 21 | Olfactory_L | 50 | Occipital_Sup_R | 79 | Heschl_L | 108 | Cerebelum_10_R |
| 22 | Olfactory_R | 51 | Occipital_Mid_L | 80 | Heschl_R | 109 | Vermis_1_2 |
| 23 | Frontal_Sup_Medial_L | 52 | Occipital_Mid_R | 81 | Temporal_Sup_L | 110 | Vermis_3 |
| 24 | Frontal_Sup_Medial_R | 53 | Occipital_Inf_L | 82 | Temporal_Sup_R | 111 | Vermis_4_5 |
| 25 | Frontal_Med_Orb_L | 54 | Occipital_Inf_R | 83 | Temporal_Pole_Sup_L | 112 | Vermis_6 |
| 26 | Frontal_Med_Orb_R | 55 | Fusiform_L | 84 | Temporal_Pole_Sup_R | 113 | Vermis_7 |
| 27 | Rectus_L | 56 | Fusiform_R | 85 | Temporal_Mid_L | 114 | Vermis_8 |
| 28 | Rectus_R | 57 | Postcentral_L | 86 | Temporal_Mid_R | 115 | Vermis_9 |
| 29 | Insula_L | 58 | Postcentral_R | 87 | Temporal_Pole_Mid_L | 116 | Vermis_10 |
| Group 1 | Group 2 | ||||
|---|---|---|---|---|---|
| Index | SNP | Gene | Index | SNP | Gene |
| 1 | rs4575098_A | ADAMTS4 | 1 | rs6656401_A | CR1 |
| 2 | rs4663105_C | RP11-138I18.2 | 2 | rs2093760_A | CR1 |
| 3 | rs6733839_T | RP11-138I18.2 | 3 | rs4844610_A | CR1 |
| 4 | rs6448453_A | AP001257.1 | 4 | rs10933431_G | SPI1 |
| 5 | rs9381563_C | RNU6-560P | 5 | rs35349669_T | CELF1 |
| 6 | rs2718058_G | FERMT2 | 6 | rs190982_G | MS4A6A |
| 7 | rs11257238_C | PVRL2 | 7 | rs9271058_A | MS4A6A |
| 8 | rs7920721_G | APOE | 8 | rs9473117_C | PICALM |
| 9 | rs10838725_C | BIN1 | 9 | rs10948363_G | RNU6-560P |
| 10 | rs983392_G | BIN1 | 10 | rs4723711_T | FERMT2 |
| 11 | rs7933202_C | INPP5D | 11 | rs1859788_A | SLC24A4 |
| 12 | rs2081545_A | INPP5D | 12 | rs1476679_C | SLC24A4 |
| 13 | rs867611_G | CASS4 | 13 | rs12539172_T | SLC24A4 |
| 14 | rs10792832_A | CASS4 | 14 | rs10808026_A | ADAM10 |
| 15 | rs3851179_T | CASS4 | 15 | rs7810606_T | KAT8 |
| 16 | rs10498633_T | HLA-DRB1 | 16 | rs11771145_A | RP11-333E1.1 |
| 17 | rs12881735_C | AL355353.1 | 17 | rs28834970_C | RP11-81K2.1 |
| 18 | rs12590654_A | AL355353.1 | 18 | rs73223431_T | CNN2 |
| 19 | rs113260531_A | EPDR1 | 19 | rs4236673_A | ABCA7 |
| 20 | rs28394864_A | GPR141 | 20 | rs9331896_C | ABCA7 |
| 21 | 21 | rs3740688_G | CD33 | ||
| 22 | 22 | rs17125924_G | RP11-61G19.1 | ||
| 23 | 23 | rs17125944_C | MEF2C-AS1 | ||
| 24 | 24 | rs442495_C | CD2AP | ||
| 25 | 25 | rs59735493_A | GPR141 | ||
| 26 | 26 | rs111278892_G | EPDR1 | ||
| 27 | 27 | rs3752246_G | PILRA | ||
| 28 | 28 | rs4147929_A | ZCWPW1 | ||
| 29 | 29 | rs41289512_G | NYAP1 | ||
| 30 | 30 | rs3865444_A | EPHA1 | ||
| 31 | 31 | rs6024870_A | EPHA1-AS1 | ||
| 32 | 32 | rs6014724_G | EPHA1-AS1 | ||
| 33 | 33 | rs7274581_C | PTK2B | ||
| 34 | 34 | rs429358_C | PTK2B |
| Group 1 | Group 2 | ||||
|---|---|---|---|---|---|
| Index | SNP | Gene | Index | SNP | Gene |
| 1 | rs6656401_A | CR1 | 1 | rs4575098_A | ADAMTS4 |
| 2 | rs2093760_A | CR1 | 2 | rs4663105_C | RP11-138I18.2 |
| 3 | rs4844610_A | CR1 | 3 | rs6733839_T | RP11-138I18.2 |
| 4 | rs1859788_A | SLC24A4 | 4 | rs10933431_G | SPI1 |
| 5 | rs1476679_C | SLC24A4 | 5 | rs35349669_T | CELF1 |
| 6 | rs12539172_T | SLC24A4 | 6 | rs6448453_A | AP001257.1 |
| 7 | rs11771145_A | RP11-333E1.1 | 7 | rs190982_G | MS4A6A |
| 8 | rs28834970_C | RP11-81K2.1 | 8 | rs9271058_A | MS4A6A |
| 9 | rs73223431_T | CNN2 | 9 | rs9473117_C | PICALM |
| 10 | rs4236673_A | ABCA7 | 10 | rs9381563_C | RNU6-560P |
| 11 | rs9331896_C | ABCA7 | 11 | rs10948363_G | RNU6-560P |
| 12 | rs3740688_G | CD33 | 12 | rs2718058_G | FERMT2 |
| 13 | rs113260531_A | EPDR1 | 13 | rs4723711_T | FERMT2 |
| 14 | rs3752246_G | PILRA | 14 | rs10808026_A | ADAM10 |
| 15 | rs4147929_A | ZCWPW1 | 15 | rs7810606_T | KAT8 |
| 16 | rs3865444_A | EPHA1 | 16 | rs11257238_C | PVRL2 |
| 17 | 17 | rs7920721_G | APOE | ||
| 18 | 18 | rs10838725_C | BIN1 | ||
| 19 | 19 | rs983392_G | BIN1 | ||
| 20 | 20 | rs7933202_C | INPP5D | ||
| 21 | 21 | rs2081545_A | INPP5D | ||
| 22 | 22 | rs867611_G | CASS4 | ||
| 23 | 23 | rs10792832_A | CASS4 | ||
| 24 | 24 | rs3851179_T | CASS4 | ||
| 25 | 25 | rs17125924_G | RP11-61G19.1 | ||
| 26 | 26 | rs17125944_C | MEF2C-AS1 | ||
| 27 | 27 | rs10498633_T | HLA-DRB1 | ||
| 28 | 28 | rs12881735_C | AL355353.1 | ||
| 29 | 29 | rs12590654_A | AL355353.1 | ||
| 30 | 30 | rs442495_C | CD2AP | ||
| 31 | 31 | rs59735493_A | GPR141 | ||
| 32 | 32 | rs28394864_A | GPR141 | ||
| 33 | 33 | rs111278892_G | EPDR1 | ||
| 34 | 34 | rs41289512_G | NYAP1 | ||
| 35 | 35 | rs6024870_A | EPHA1-AS1 | ||
| 36 | 36 | rs6014724_G | EPHA1-AS1 | ||
| 37 | 37 | rs7274581_C | PTK2B | ||
| 38 | 38 | rs429358_C | PTK2B |
References
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Feldman, H.H.; Frisoni, G.B.; Hampel, H.; Jagust, W.J.; Johnson, K.A.; Knopman, D.S.; et al. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 2016, 87, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Jansen, I.E.; Savage, J.E.; Watanabe, K.; Bryois, J.; Williams, D.M.; Steinberg, S.; Sealock, J.; Karlsson, I.K.; Hägg, S.; Athanasiu, L.; et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 2019, 51, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2020, 16, 391–460. [Google Scholar] [CrossRef] [PubMed]
- Gatz, M.; Reynolds, C.A.; Fratiglioni, L.; Johansson, B.; Mortimer, J.A.; Berg, S.; Fiske, A.; Pedersen, N.L. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 2006, 63, 168–174. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; Van Der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; Jun, G.; DeStefano, A.L.; Bis, J.C.; Beecham, G.W.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Ramanan, V.; Saykin, A. Pathways to neurodegeneration: Mechanistic insights from GWAS in Alzheimer’s disease, Parkinson’s disease, and related disorders. Am. J. Neurodegener. Dis. 2013, 2, 145. [Google Scholar]
- Gandhi, S.; Wood, N.W. Genome-wide association studies: The key to unlocking neurodegeneration? Nat. Neurosci. 2010, 13, 789–794. [Google Scholar] [CrossRef]
- Pihlstrom, L.; Wiethoff, S.; Houlden, H. Chapter 22—Genetics of neurodegenerative diseases: An overview. In Handbook of Clinical Neurology; Kovacs, G.G., Alafuzoff, I., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 145, pp. 309–323. [Google Scholar] [CrossRef]
- Tsuji, S. Genetics of neurodegenerative diseases: Insights from high-throughput resequencing. Hum. Mol. Genet. 2010, 19, R65–R70. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.; Wang, X.; Maruyama, T.; Ma, Y.; Zhang, X.; Mez, J.; Sherva, R.; Takeyama, H.; The Alzheimer’s Disease Neuroimaging Initiative; Lunetta, K.L.; et al. Genome-wide association study of Alzheimer’s disease endophenotypes at prediagnosis stages. Alzheimer’s Dement. 2018, 14, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Waring, S.C.; Rosenberg, R.N. Genome-Wide Association Studies in Alzheimer Disease. Arch. Neurol. 2008, 65, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Harold, D.; Abraham, R.; Hollingworth, P.; Sims, R.; Gerrish, A.; Hamshere, M.L.; Pahwa, J.S.; Moskvina, V.; Dowzell, K.; Williams, A.; et al. Genome-Wide Association Study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 2009, 41, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Thompson, P.M. Brain Imaging Genomics: Integrated Analysis and Machine Learning. Proc. IEEE 2020, 108, 125–162. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Thompson, P.M.; Potkin, S.G.; Bertram, L.; Farrer, L.A.; Foroud, T.M.; Green, R.C.; Hu, X.; Huentelman, M.J.; Kim, S.; et al. Genetic analysis of quantitative phenotypes in AD and MCI: Imaging, cognition and biomarkers. Brain Imaging Behav. 2014, 8, 183–207. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Kim, S.; Risacher, S.L.; Nho, K.; Swaminathan, S.; West, J.D.; Foroud, T.; Pankratz, N.; Moore, J.H.; Sloan, C.D.; et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage 2010, 53, 1051–1063. [Google Scholar] [CrossRef]
- Ferreira, D.; Nordberg, A.; Westman, E. Biological subtypes of Alzheimer disease. Neurology 2020, 94, 436–448. [Google Scholar] [CrossRef]
- Jellinger, K.A. Pathobiological Subtypes of Alzheimer Disease. Dement. Geriatr. Cogn. Disord. 2021, 49, 321–333. [Google Scholar] [CrossRef]
- Yao, X.; Yan, J.; Kim, S.; Nho, K.; Risacher, S.L.; Inlow, M.; Moore, J.H.; Saykin, A.J.; Shen, L. Two-dimensional enrichment analysis for mining high-level imaging genetic associations. Brain Inform. 2017, 4, 27–37. [Google Scholar] [CrossRef]
- Weiner, M.W.; Veitch, D.P.; Aisen, P.S.; Beckett, L.A.; Cairns, N.J.; Green, R.C.; Harvey, D.; Jack, C.R.; Jagust, W.; Liu, E.; et al. The Alzheimer’s Disease Neuroimaging Initiative: A review of papers published since its inception. Alzheimer’s Dement. 2013, 9, e111–e194. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Wu, R.; Yao, X.; Saykin, A.J.; Moore, J.H.; Shen, L.; Alzheimer’s Disease Neuroimaging Initiative. Identifying genetic markers enriched by brain imaging endophenotypes in Alzheimer’s disease. BMC Med. Genom. 2022, 15, 168. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; He, X.; Zha, H.; Gu, M.; Simon, H. A min-max cut algorithm for graph partitioning and data clustering. In Proceedings of the Proceedings 2001 IEEE International Conference on Data Mining, San Jose, CA, USA, 29 November–2 December 2001; pp. 107–114. [Google Scholar] [CrossRef]
- De, W.; Wang, Y.; Nie, F.; Yan, J.; Cai, W.; Saykin, A.J.; Shen, L.; Huang, H. Human connectome module pattern detection using a new multi-graph MinMax cut model. Med. Image Comput. Comput. Assist. Interv. 2014, 17, 313–320. [Google Scholar] [PubMed]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Gorgolewski, K.J.; Varoquaux, G.; Rivera, G.; Schwarz, Y.; Ghosh, S.S.; Maumet, C.; Sochat, V.V.; Nichols, T.E.; Poldrack, R.A.; Poline, J.B.; et al. NeuroVault.org: A web-based repository for collecting and sharing unthresholded statistical maps of the human brain. Front. Neuroinform. 2015, 9, 8. [Google Scholar] [CrossRef]
- Jun, G.; Ibrahim-Verbaas, C.A.; Vronskaya, M. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol. Psychiatry 2016, 21, 108–117. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Raja V, V.; Huang, Z.; Amico, E.; Nho, K.; Fang, S.; Sporns, O.; Wu, Y.C.; Saykin, A.; Goni, J.; et al. Brain-wide structural connectivity alterations under the control of Alzheimer risk genes. Int. J. Comput. Biol. Drug Des. 2020, 13, 58–70. [Google Scholar] [CrossRef]
- Murphy, M.P.; LeVine, H. Alzheimer’s disease and the amyloid-beta peptide. J. Alzheimer’s Dis. 2010, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.O.; Mett, J.; Stahlmann, C.P.; Grösgen, S.; Haupenthal, V.J.; Blümel, T.; Hundsdörfer, B.; Zimmer, V.C.; Mylonas, N.T.; Tanila, H.; et al. APP intracellular domain derived from amyloidogenic β- and γ-secretase cleavage regulates neprilysin expression. Front. Aging Neurosci. 2015, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Papuć, E.; Rejdak, K. The role of myelin damage in Alzheimer’s disease pathology. Arch. Med. Sci. 2020, 16, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Kolev, M.V.; Ruseva, M.M.; Harris, C.L.; Morgan, B.P.; Donev, R.M. Implication of complement system and its regulators in Alzheimer’s disease. Curr. Neuropharmacol. 2009, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Koldamova, R.; Fitz, N.F.; Lefterov, I. The role of ATP-binding cassette transporter A1 in Alzheimer’s disease and neurodegeneration. Biochim. Biophys. Acta 2010, 1801, 824–830. [Google Scholar] [CrossRef] [Green Version]


| Measurement | Scoring Function | Normalized Similarity |
|---|---|---|
| Pearson correlation | ||
| Spearman correlation | ||
| Manhattan distance | ||
| Euclidean distance | ||
| Cosine |
| Graph Cut Algorithm for Cluster Analysis | Objective Function |
|---|---|
| Single-graph min-max cut | |
| Multigraph min-max cut |
| Measure | Calculation |
|---|---|
| mean distance | |
| mean dissimilarity | |
| Silhouette value |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, R.; Bao, J.; Kim, M.; Saykin, A.J.; Moore, J.H.; Shen, L.; on behalf of ADNI. Mining High-Level Imaging Genetic Associations via Clustering AD Candidate Variants with Similar Brain Association Patterns. Genes 2022, 13, 1520. https://doi.org/10.3390/genes13091520
Wu R, Bao J, Kim M, Saykin AJ, Moore JH, Shen L, on behalf of ADNI. Mining High-Level Imaging Genetic Associations via Clustering AD Candidate Variants with Similar Brain Association Patterns. Genes. 2022; 13(9):1520. https://doi.org/10.3390/genes13091520
Chicago/Turabian StyleWu, Ruiming, Jingxuan Bao, Mansu Kim, Andrew J. Saykin, Jason H. Moore, Li Shen, and on behalf of ADNI. 2022. "Mining High-Level Imaging Genetic Associations via Clustering AD Candidate Variants with Similar Brain Association Patterns" Genes 13, no. 9: 1520. https://doi.org/10.3390/genes13091520
APA StyleWu, R., Bao, J., Kim, M., Saykin, A. J., Moore, J. H., Shen, L., & on behalf of ADNI. (2022). Mining High-Level Imaging Genetic Associations via Clustering AD Candidate Variants with Similar Brain Association Patterns. Genes, 13(9), 1520. https://doi.org/10.3390/genes13091520







