A Comprehensive Characterization of Small RNA Profiles by Massively Parallel Sequencing in Six Forensic Body Fluids/Tissue
Abstract
:1. Introduction
2. Materials and Methods
2.1. Body Fluids and Skin Tissue Sampling
2.2. RNA Extraction and Quantification
2.3. Small RNA Library Preparation and Sequencing
2.4. Bioinformatics Analysis
2.5. Body Fluids/Tissue-Specific RNA Analysis
3. Results and Discussion
3.1. Small RNA Sequencing Results
3.2. Small RNA Expression Profiles
3.3. Body Fluids/Tissue-Specific RNAs
3.4. Reference Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virkler, K.; Lednev, I.K. Analysis of Body Fluids for Forensic Purposes: From Laboratory Testing to Non-Destructive Rapid Confirmatory Identification at a Crime Scene. Forensic Sci. Int. 2009, 188, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Kakizaki, E.; Takahama, K. A Sandwich Enzyme Immunoassay for Brain S-100 Protein and Its Forensic Application. Forensic Sci. Int. 1997, 87, 145–154. [Google Scholar] [CrossRef]
- Takahama, K. Forensic Application of Organ-Specific Antigens. Forensic Sci. Int. 1996, 80, 63–69. [Google Scholar] [CrossRef]
- Lee, H.Y.; Jung, S.E.; Lee, E.H.; Yang, W.I.; Shin, K.J. DNA Methylation Profiling for a Confirmatory Test for Blood, Saliva, Semen, Vaginal Fluid and Menstrual Blood. Forensic Sci. Int. Genet. 2016, 24, 75–82. [Google Scholar] [CrossRef]
- Silva, D.S.B.S.; Antunes, J.; Balamurugan, K.; Duncan, G.; Alho, C.S.; McCord, B. Developmental Validation Studies of Epigenetic DNA Methylation Markers for the Detection of Blood, Semen and Saliva Samples. Forensic Sci. Int. Genet. 2016, 23, 55–63. [Google Scholar] [CrossRef]
- Díez López, C.; Montiel González, D.; Haas, C.; Vidaki, A.; Kayser, M. Microbiome-Based Body Site of Origin Classification of Forensically Relevant Blood Traces. Forensic Sci. Int. Genet. 2020, 47, 102280. [Google Scholar] [CrossRef]
- Blackman, S.; Stafford-Allen, B.; Hanson, E.K.; Panasiuk, M.; Brooker, A.L.; Rendell, P.; Ballantyne, J.; Wells, S. Developmental Validation of the ParaDNA® Body Fluid ID System—A Rapid Multiplex MRNA-Profiling System for the Forensic Identification of Body Fluids. Forensic Sci. Int. Genet. 2018, 37, 151–161. [Google Scholar] [CrossRef]
- Ingold, S.; Dørum, G.; Hanson, E.; Berti, A.; Branicki, W.; Brito, P.; Elsmore, P.; Gettings, K.B.; Giangasparo, F.; Gross, T.E.; et al. Body Fluid Identification Using a Targeted MRNA Massively Parallel Sequencing Approach–Results of a EUROFORGEN/EDNAP Collaborative Exercise. Forensic Sci. Int. Genet. 2018, 34, 105–115. [Google Scholar] [CrossRef]
- Seashols-Williams, S.; Lewis, C.; Calloway, C.; Peace, N.; Harrison, A.; Hayes-Nash, C.; Fleming, S.; Wu, Q.; Zehner, Z.E. High-Throughput MiRNA Sequencing and Identification of Biomarkers for Forensically Relevant Biological Fluids. Electrophoresis 2016, 37, 2780–2788. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Tao, R.; Wang, M.; Liu, J.; He, G.; Yang, Y.; Xie, M.; Zou, X.; Hou, Y. Expression Profile Analysis of Piwi-Interacting RNA in Forensically Relevant Biological Fluids. Forensic Sci. Int. Genet. 2019, 42, 171–180. [Google Scholar] [CrossRef]
- An, J.H.; Shin, K.J.; Yang, W.I.; Lee, H.Y. Body Fluid Identification in Forensics. BMB Rep. 2012, 45, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Rocchi, A.; Chiti, E.; Maiese, A.; Turillazzi, E.; Spinetti, I. MicroRNAs: An Update of Applications in Forensic Science. Diagnostics 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhao, M.; Zhu, Y.; Zhang, L.; Zheng, Z.; Wang, Q.; Li, Y.; Zhang, P.; Zhu, S.; Ding, S.; et al. The Persistence and Stability of MiRNA in Bloodstained Samples under Different Environmental Conditions. Forensic Sci. Int. 2021, 318, 110594. [Google Scholar] [CrossRef]
- Kakimoto, Y.; Tanaka, M.; Kamiguchi, H.; Ochiai, E.; Osawa, M. MicroRNA Stability in FFPE Tissue Samples: Dependence on Gc Content. PLoS ONE 2016, 11, e0163125. [Google Scholar] [CrossRef]
- He, H.; Han, N.; Ji, C.; Zhao, Y.; Hu, S.; Kong, Q.; Ye, J.; Ji, A.; Sun, Q. Identification of Five Types of Forensic Body Fluids Based on Stepwise Discriminant Analysis. Forensic Sci. Int. Genet. 2020, 48, 102337. [Google Scholar] [CrossRef]
- He, H.; Ji, A.; Zhao, Y.; Han, N.; Hu, S.; Kong, Q.; Jiang, L.; Ye, J.; Liu, Y.; Sun, Q. A Stepwise Strategy to Distinguish Menstrual Blood from Peripheral Blood by Fisher’s Discriminant Function. Int. J. Legal Med. 2020, 134, 845–851. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Diao, L.; Han, L. Small Non-Coding RNAs in Human Cancer: Function, Clinical Utility, and Characterization. Oncogene 2021, 40, 1570–1577. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [Green Version]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-Interacting Small RNAs: The Vanguard of Genome Defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.H.; Chen, L.L. Processing and Roles of SnoRNA-Ended Long Noncoding RNAs. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 596–606. [Google Scholar] [CrossRef]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-Coding RNAs: Lessons from the Small Nuclear and Small Nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Sauer, E.; Reinke, A.K.; Courts, C. Differentiation of Five Body Fluids from Forensic Samples by Expression Analysis of Four MicroRNAs Using Quantitative PCR. Forensic Sci. Int. Genet. 2016, 22, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bai, P.; Peng, D.; Wang, H.; Guo, Y.; Jiang, Y.; He, W.; Tian, H.; Yang, Y.; Huang, Y.; et al. Screening and Confirmation of MicroRNA Markers for Distinguishing between Menstrual and Peripheral Blood. Forensic Sci. Int. Genet. 2017, 30, 24–33. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, D.; Cao, Y.; Hu, Z.; Zhang, S.; Bian, Y.; Hou, Y.; Li, C. Characterization of MicroRNA Expression Profiles in Blood and Saliva Using the Ion Personal Genome Machine® System (Ion PGMTM System). Forensic Sci. Int. Genet. 2016, 20, 140–146. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Xu, M.; You, Y.; Hunsicker, P.; Hori, T.; Small, C.; Griswold, M.D.; Hecht, N.B. Mice Deficient for a Small Cluster of Piwi-Lnteracting RNAs Implicate Piwi-Interacting RNAs in Transposon Control. Biol. Reprod. 2008, 79, 51–57. [Google Scholar] [CrossRef]
- Lestrade, L.; Weber, M.J. SnoRNA-LBME-Db, a Comprehensive Database of Human H/ACA and C/D Box SnoRNAs. Nucleic Acids Res. 2006, 34, D158–D162. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-Fold Faster RNA Homology Searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering MicroRNAs from Deep Sequencing Data Using MiRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Kivioja, T.; Vähärautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting Absolute Numbers of Molecules Using Unique Molecular Identifiers. Nat. Methods 2012, 9, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zeng, Z.; Xiong, Y.; Guo, W.; Du, H. ERgene: Python Library for Screening Endogenous Reference Genes. Sci. Rep. 2020, 10, 18557. [Google Scholar] [CrossRef]
- Salzmann, A.P.; Russo, G.; Aluri, S.; Haas, C. Transcription and Microbial Profiling of Body Fluids Using a Massively Parallel Sequencing Approach. Forensic Sci. Int. Genet. 2019, 43, 102149. [Google Scholar] [CrossRef]
- Haas, C.; Muheim, C.; Kratzer, A.; Bär, W.; Maake, C. MRNA Profiling for the Identification of Sperm and Seminal Plasma. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 534–535. [Google Scholar] [CrossRef]
- Sauer, E.; Extra, A.; Cachée, P.; Courts, C. Identification of Organ Tissue Types and Skin from Forensic Samples by MicroRNA Expression Analysis. Forensic Sci. Int. Genet. 2017, 28, 99–110. [Google Scholar] [CrossRef]
- Hall, J.S.; Taylor, J.; Valentine, H.R.; Irlam, J.J.; Eustace, A.; Hoskin, P.J.; Miller, C.J.; West, C.M.L. Enhanced Stability of MicroRNA Expression Facilitates Classification of FFPE Tumour Samples Exhibiting near Total MRNA Degradation. Br. J. Cancer 2012, 107, 684–694. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Schaefer, A.; Steiner, I.; Kempkensteffen, C.; Stephan, C.; Erbersdobler, A.; Jung, K. Robust MicroRNA Stability in Degraded RNA Preparations from Human Tissue and Cell Samples. Clin. Chem. 2010, 56, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Kou, R.; Lam, H.; Duan, H.; Ye, L.; Jongkam, N.; Chen, W.; Zhang, S.; Li, S. Benefits and Challenges with Applying Unique Molecular Identifiers in next Generation Sequencing to Detect Low Frequency Mutations. PLoS ONE 2016, 11, e0146638. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lönnerberg, P.; Linnarsson, S. Quantitative Single-Cell RNA-Seq with Unique Molecular Identifiers. Nat. Methods 2014, 11, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Jones, D.F.; Fleming, R. Transcriptomic Analysis of Degraded Forensic Body Fluids. Forensic Sci. Int. Genet. 2015, 17, 35–42. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, M.X.; Ye, N.H.; Qiao, W.M.; Gao, B.; Law, W.K.; Tian, Y.; Zhang, D.; Zhang, D.; Liu, T.Y.; et al. Comparative Performance of the BGISEQ-500 and Illumina HiSeq4000 Sequencing Platforms for Transcriptome Analysis in Plants. Plant Methods 2018, 14, 69. [Google Scholar] [CrossRef]
- Fehlmann, T.; Reinheimer, S.; Geng, C.; Su, X.; Drmanac, S.; Alexeev, A.; Zhang, C.; Backes, C.; Ludwig, N.; Hart, M.; et al. CPAS-Based Sequencing on the BGISEQ-500 to Explore Small Non-Coding RNAs. Clin. Epigenetics 2016, 8, 123. [Google Scholar] [CrossRef]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef]
- Zubakov, D.; Boersma, A.W.M.; Choi, Y.; Van Kuijk, P.F.; Wiemer, E.A.C.; Kayser, M. MicroRNA Markers for Forensic Body Fluid Identification Obtained from Microarray Screening and Quantitative RT-PCR Confirmation. Int. J. Legal Med. 2010, 124, 217–226. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Luo, H.; Ye, Y.; Yan, J.; Hou, Y. Screening and Confirmation of MicroRNA Markers for Forensic Body Fluid Identification. Forensic Sci. Int. Genet. 2013, 7, 116–123. [Google Scholar] [CrossRef]
- Hanson, E.K.; Lubenow, H.; Ballantyne, J. Identification of Forensically Relevant Body Fluids Using a Panel of Differentially Expressed MicroRNAs. Anal. Biochem. 2009, 387, 303–314. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Courts, C.; Madea, B. Specific Micro-RNA Signatures for the Detection of Saliva and Blood in Forensic Body-Fluid Identification. J. Forensic Sci. 2011, 56, 1464–1470. [Google Scholar] [CrossRef]
- Omelia, E.J.; Uchimoto, M.L.; Williams, G. Quantitative PCR Analysis of Blood- and Saliva-Specific MicroRNA Markers Following Solid-Phase DNA Extraction. Anal. Biochem. 2013, 435, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-L.; Park, S.-M.; Kwon, O.-H.; Lee, H.-C.; Kim, J.-Y.; Seok, H.H.; Lee, W.S.; Lee, S.-H.; Kim, Y.S.; Woo, K.-M.; et al. Microarray Screening and QRT-PCR Evaluation of MicroRNA Markers for Forensic Body Fluid Identification. Electrophoresis 2014, 35, 3062–3068. [Google Scholar] [CrossRef] [PubMed]
- Tong, D.; Jin, Y.; Xue, T.; Ma, X.; Zhang, J.; Ou, X.; Cheng, J.; Sun, H. Investigation of the Application of MiR10b and MiR135b in the Identification of Semen Stains. PLoS ONE 2015, 10, e0137067. [Google Scholar] [CrossRef]
- Sirker, M.; Fimmers, R.; Schneider, P.M.; Gomes, I. Evaluating the Forensic Application of 19 Target MicroRNAs as Biomarkers in Body Fluid and Tissue Identification. Forensic Sci. Int. Genet. 2017, 27, 41–49. [Google Scholar] [CrossRef]
- Korma, W.; Mihret, A.; Tarekegn, A.; Chang, Y.; Hwang, D.; Tessema, T.S.; Lee, H. Identification of Circulating MiR-22-3p and MiR-93-5p as Stable Endogenous Control in Tuberculosis Study. Diagnostics 2020, 10, 868. [Google Scholar] [CrossRef]
- Ragni, E.; De Luca, P.; Marmotti, A.; de Girolamo, L. MiR-26a-5p Is a Stable Reference Gene for MiRNA Studies in Chondrocytes from Developing Human Cartilage. Cells 2019, 8, 631. [Google Scholar] [CrossRef]
- Peltier, H.J.; Latham, G.J. Normalization of MicroRNA Expression Levels in Quantitative RT-PCR Assays: Identification of Suitable Reference RNA Targets in Normal and Cancerous Human Solid Tissues. RNA 2008, 14, 844–852. [Google Scholar] [CrossRef]
- Fujimoto, S.; Manabe, S.; Morimoto, C.; Ozeki, M.; Hamano, Y.; Tamaki, K. Optimal Small-Molecular Reference RNA for RT-qPCR-Based Body Fluid Identification. Forensic Sci. Int. Genet. 2018, 37, 135–142. [Google Scholar] [CrossRef]
- Sirker, M.; Liang, W.; Zhang, L.; Fimmers, R.; Rothschild, M.A.; Gomes, I.; Schneider, P.M. Impact of Using Validated or Standard Reference Genes for MiRNA qPCR Data Normalization in Cell Type Identification. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e199–e201. [Google Scholar] [CrossRef]
- Sauer, E.; Madea, B.; Courts, C. An Evidence Based Strategy for Normalization of Quantitative PCR Data from MiRNA Expression Analysis in Forensically Relevant Body Fluids. Forensic Sci. Int. Genet. 2014, 11, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.; Manabe, S.; Morimoto, C.; Ozeki, M.; Hamano, Y.; Hirai, E.; Kotani, H.; Tamaki, K. Distinct Spectrum of MicroRNA Expression in Forensically Relevant Body Fluids and Probabilistic Discriminant Approach. Sci. Rep. 2019, 9, 14332. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Wei, T.; Janson, P.C.J.; Sääf, A.; Lundeberg, L.; Tengvall-Linder, M.; Norstedt, G.; Alenius, H.; Homey, B.; Scheynius, A.; et al. MicroRNAs: Novel Regulators Involved in the Pathogenesis of Psoriasis? PLoS ONE 2007, 2, e610. [Google Scholar] [CrossRef]
- Bamberg, M.; Bruder, M.; Dierig, L.; Kunz, S.N.; Schwender, M.; Wiegand, P. Best of Both: A Simultaneous Analysis of MRNA and MiRNA Markers for Body Fluid Identification. Forensic Sci. Int. Genet. 2022, 59, 102707. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Z.; Tao, R.; He, G.; Liu, J.; Li, C.; Hou, Y. The Potential Use of Piwi-Interacting RNA Biomarkers in Forensic Body Fluid Identification: A Proof-of-Principle Study. Forensic Sci. Int. Genet. 2019, 39, 129–135. [Google Scholar] [CrossRef]
- Mccann, K.L.; Kavari, S.L.; Burkholder, A.B.; Phillips, B.T.; Hall, T.M.T. H/ACA SnoRNA Levels Are Regulated during Stem Cell Differentiation. Nucleic Acids Res. 2020, 48, 8686–8703. [Google Scholar] [CrossRef]
- Bratkovič, T.; Bozič, J.; Rogelj, B. Functional Diversity of Small Nucleolar RNAs. Nucleic Acids Res. 2020, 48, 1627–1651. [Google Scholar] [CrossRef]
- Wang, S.; Tao, R.; Ming, T.; Wang, M.; Liu, J.; He, G.; Zou, X.; Wang, Z.; Hou, Y. Expression Profile Analysis and Stability Evaluation of 18 Small RNAs in the Chinese Han Population. Electrophoresis 2020, 41, 2021–2028. [Google Scholar] [CrossRef]
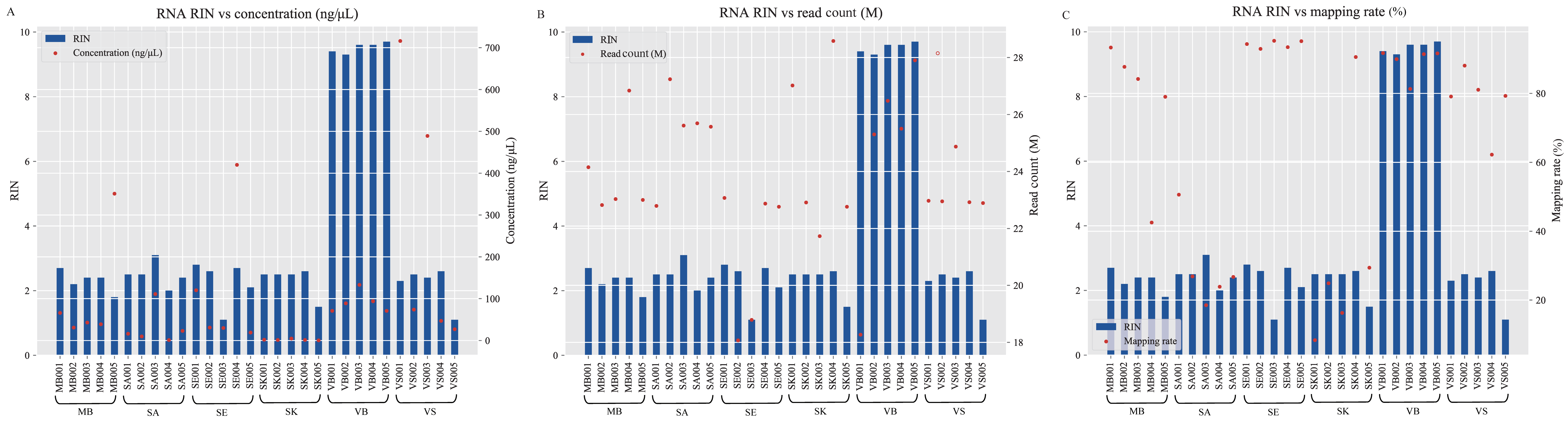
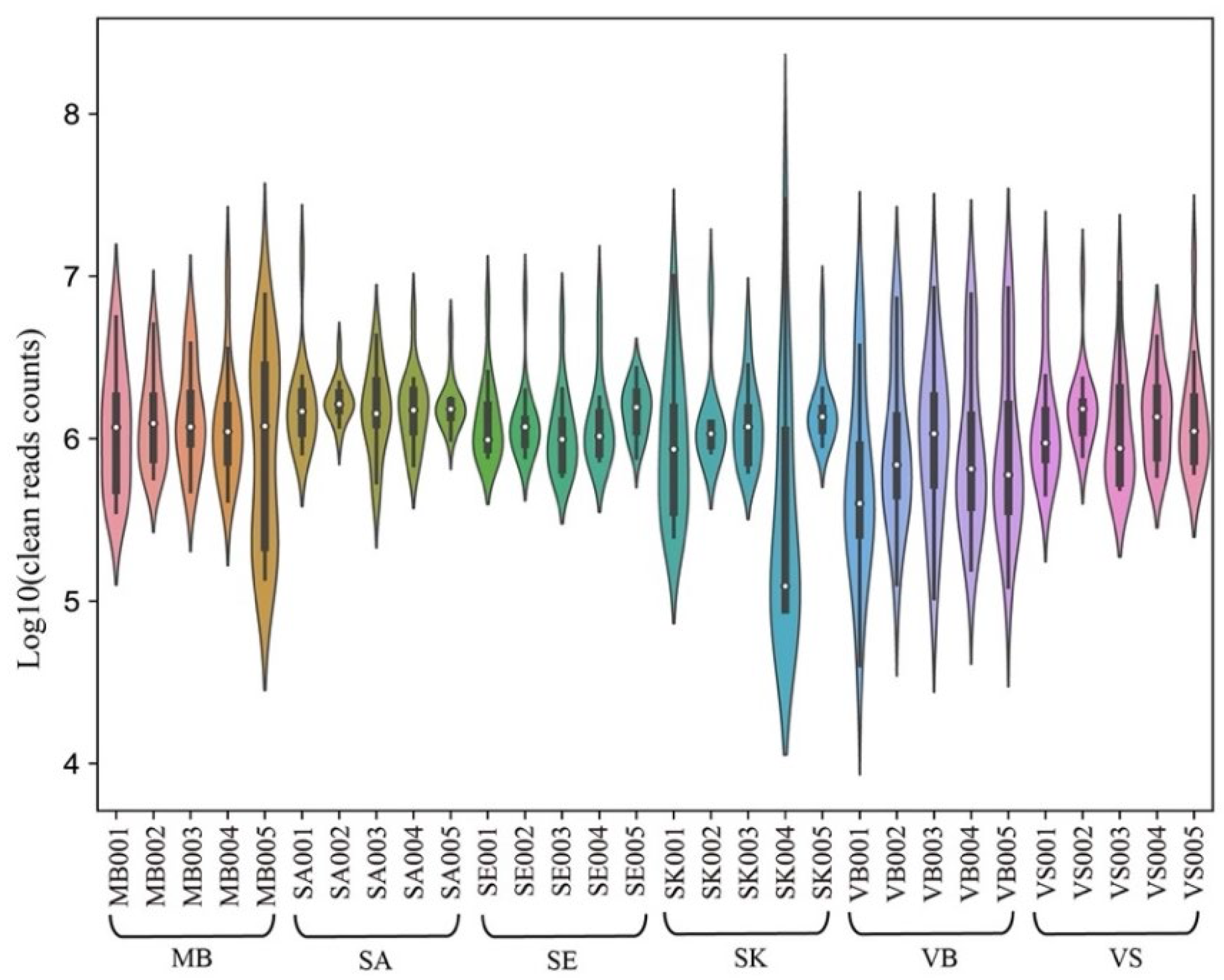
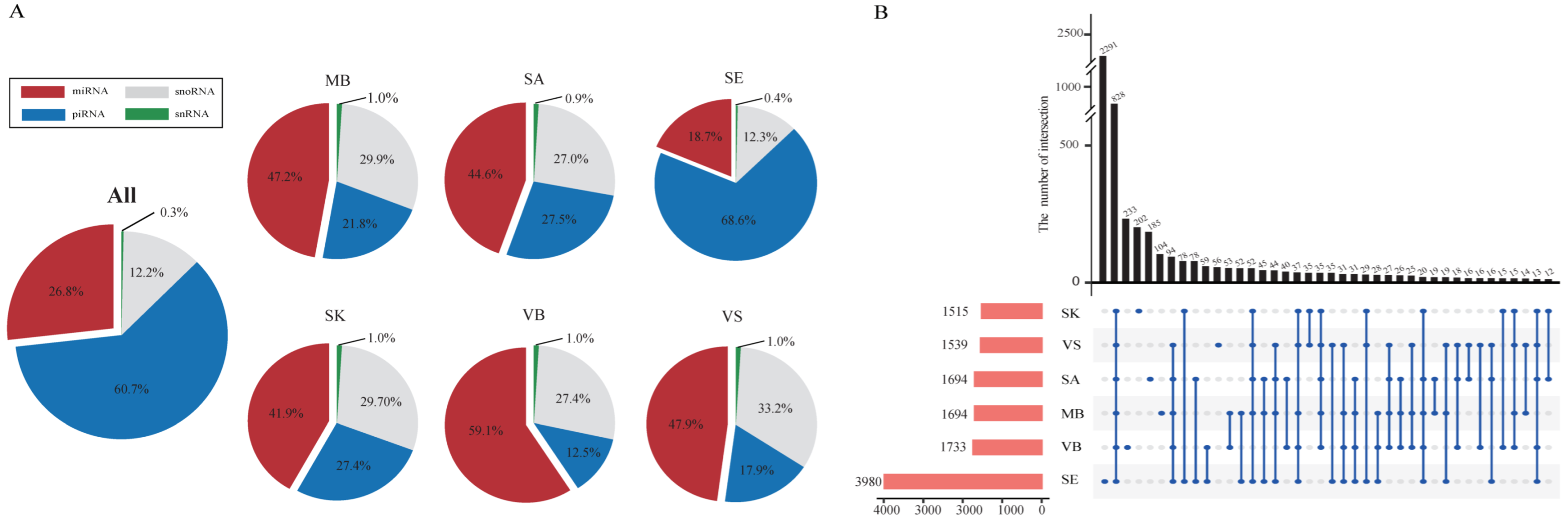
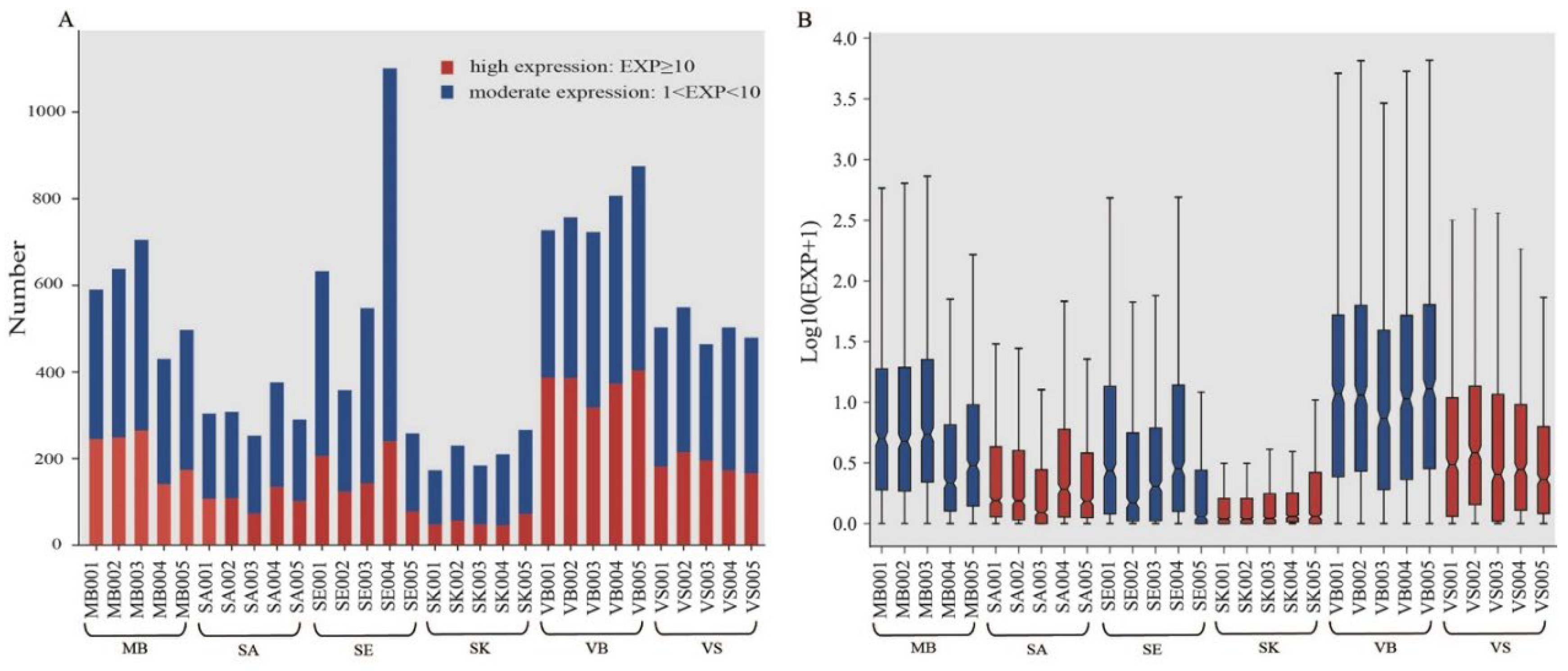


| Group | Mean Concentration (ng/μL) | Mean RIN | Mean Q20 (%) | Mean Read Count (M) | Mean Mapping (%) |
|---|---|---|---|---|---|
| MB | 106.00 | 2.30 | 98.78 | 23.97 | 77.34 |
| SA | 32.23 | 2.50 | 98.78 | 25.38 | 29.31 |
| SE | 124.00 | 2.30 | 98.80 | 21.11 | 94.21 |
| SK | 1.90 | 2.32 | 98.76 | 24.60 | 33.90 |
| VB | 91.60 | 9.52 | 98.82 | 24.69 | 89.18 |
| VS | 270.60 | 2.18 | 98.92 | 23.32 | 77.94 |
| Group | From | Markers |
|---|---|---|
| MB | Published data | miR-144 [25,48], miR-214 [26,49], miR-185 [25,48], miR-412 [50], miR-451 [50], miR-1246 [9] |
| This study | miR-144-3p, miR-144-5p, miR-214-3p, miR-214-5p, miR-3120-3p, novel-miR254-3p | |
| SA | Published data | miR-26a [51], miR-96 [51], miR-135b [51], miR-182 [51], miR-200c [27,52], miR-203 [9,27,52], miR-205 [27,50,52,53], miR-208b [48], miR-381 [51], miR-431 [51], miR-450b-5p [51], miR-518c [48], miR-583 [48], miR-622 [51], miR-658 [50], miR-1228 [51], miR-223 [27,54], miR-145 [51,54], miR-141 [27,51], miR-375 [27], miR-34a [27], let-7c [27], miR-22 [27], miR-27a,b [27],miR-23a,b [27], miR-125b [27],miR-99a [27], miR-29a,b [27], miR-210 [27] |
| This study | - | |
| SE | Published data | miR-891a [9,25,48,49], miR-888 [49], miR-10a [48], miR-10b [48,50,55], miR-17 [51], miR-29b-2 [51], miR-135b [48,50,55], miR-340 [51], miR-380 [51], miR-507 [48], miR-508-5p [51], miR-644 [51], miR-943 [48], miR-2392 [48,54], miR-3197 [54], miR-26b [9] |
| This study | miR-891a-5p, miR-888-5p, miR-12136, miR-122-5p, miR-202-5p, miR-4477b, miR-509-3-5p, miR-890, piR-001089, piR-004800, piR-005660, piR-005816, piR-006557, piR-012901, piR-013521, piR-015150, piR-019823, piR-020247, piR-023378, RF00285, RF01135, RF01151, RF01273, RF01276, RF01297 | |
| SK | Published data | miR-203a-3p [39], miR-205-5p [39], miR-139 [56], miR-494 [56], miR-3169 [56] |
| This study | RF00335, RF00343, RF00472, RF01218, RF01499, RF01501, RF01509, RF01514, RF01846, RF01860 | |
| VB | Published data | miR-484 [54], miR-126 [52], miR-16 [48,49,50,51], miR-20a [48], miR-106a [48], miR-150 [52], miR-185 [48], miR-451 [27,48,50,52,53], miR-182 [54], miR-144-3p [17,27], miR-200b [9],miR-486 [27,49], miR-16 [27], miR-126 [27] |
| This study | miR-484, miR-126-5p, let-7d-3p, miR-1260b, miR-1277-3p, miR-1284, miR-139-3p, miR-3173-5p, miR-337-3p, miR-340-3p, miR-4433b-5p, miR-4435, miR-5010-3p, miR-5193, miR-5581-3p, miR-584-5p, miR-664b-3p, miR-671-3p, miR-6842-3p, piR-000441, piR-000586, piR-000805, piR-013306, piR-013350, piR-017061, piR-019676, piR-020813, RF00154, RF00158, RF00231, RF00266, RF00283, RF00416, RF00420, RF00493, RF00548, RF00570, RF00592, RF00609, RF00610, RF00611 | |
| VS | Published data | miR-124a [50,52], miR-372 [50,52], miR-617 [48], miR-654-5q [54],miR-155-5p [25], miR-1260b [27,49], miR-654-5p [27] |
| This study | miR-193b-3p, miR-203a-3p, miR-203b-5p, piR-015026, piR-020388, RF00093, RF01306, RF01641 | |
| Reference gene | Published data | U6 [15], miR-451a [9], miR-22-3p [57], miR-26a-5p [58], let-7a [59], let-7g [9], let-7i [9], miR-484 [56,60], 5S-rRNA [60], miR-92a-3p [60], miR92 [61], miR374 [61], miR-26b [56], miR-92 [61], miR-93 [59,62], miR-191 [59,62], miR-374 [61], miR-423 [61], RNU6b [50], RNU19 [62], RNU24 [59], RNU38B [62], RNU43 [62], RNU48 [59], RNU49 [62], RNU66 [62] |
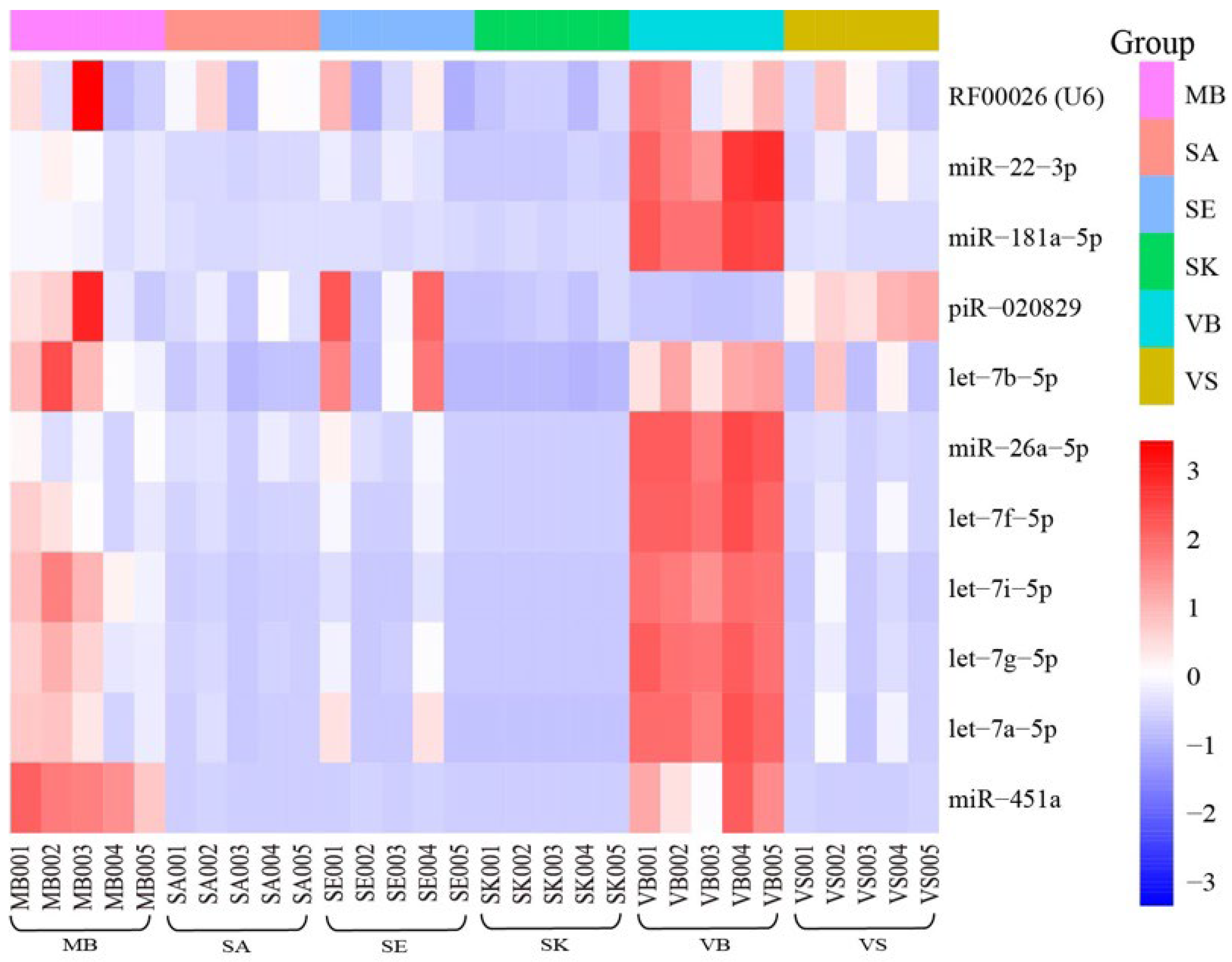
| This study | RF00026(U6), miR-451a, miR-22-3p, miR-26a-5p, let-7a-5p let-7g-5p, let-7i-5p, let-7f-5p, let-7b-5p, miR-181a-5p, piR-020829 |
| Group | Small RNA | Specificity | Expression Level | ||||||
|---|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | I | II | III | IV | ||
| MB | miR-144-3p | 1 | 4 | + | |||||
| miR-214-3p | 5 | + | |||||||
| miR-214-5p | 5 | + | |||||||
| miR-3120-3p | 5 | ++ | |||||||
| miR-144-5p | 5 | ++ | |||||||
| novel-miR254-3p | 5 | + | |||||||
| SE | miR-12136 | 2 | 3 | + | |||||
| piR_005660 | 5 | + | |||||||
| miR-890 | 5 | + | |||||||
| piR-015150 | 1 | 1 | 3 | + | |||||
| miR-888-5p | 5 | + | |||||||
| piR-019823 | 1 | 4 | +++ | ||||||
| piR_020247 | 5 | ++ | |||||||
| RF01297 | 5 | ++ | |||||||
| RF01151 | 1 | 2 | 2 | + | |||||
| miR-122-5p | 5 | + | |||||||
| SK | RF00335 | 5 | + | ||||||
| RF01846 | 5 | + | |||||||
| RF01860 | 5 | ++ | |||||||
| RF01499 | 5 | ++ | |||||||
| RF01509 | 1 | 4 | + | ||||||
| RF01501 | 5 | +++ | |||||||
| RF01218 | 1 | 4 | ++ | ||||||
| RF01514 | 5 | ++ | |||||||
| RF00343 | 5 | ++ | |||||||
| RF00472 | 1 | 4 | ++ | ||||||
| VB | miR-584-5p | 5 | + | ||||||
| miR-484 | 5 | +++ | |||||||
| let-7d-3p | 5 | +++ | |||||||
| miR-337-3p | 5 | ++ | |||||||
| miR-126-5p | 5 | ++ | |||||||
| RF00231 | 2 | 3 | + | ||||||
| RF00610 | 2 | 3 | + | ||||||
| miR-1284 | 5 | + | |||||||
| piR-013306 | 2 | 3 | + | ||||||
| piR-000805 | 1 | 4 | +++ | ||||||
| VS | miR-193b-3p | 1 | 1 | 3 | ++ | ||||
| miR-203a-3p | 1 | 1 | 3 | ++ | |||||
| RF01306 | 2 | 3 | + | ||||||
| RF00093 | 2 | 3 | + | ||||||
| piR-020388 | 4 | 1 | + | ||||||
| piR-015026 | 3 | 2 | + | ||||||
| RF01641 | 2 | 3 | +++ | ||||||
| miR-203b-5p | 1 | 1 | 3 | ++ | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wang, Q.; Wang, N.; Zang, Y.; Wu, R.; Sun, H. A Comprehensive Characterization of Small RNA Profiles by Massively Parallel Sequencing in Six Forensic Body Fluids/Tissue. Genes 2022, 13, 1530. https://doi.org/10.3390/genes13091530
Liu Z, Wang Q, Wang N, Zang Y, Wu R, Sun H. A Comprehensive Characterization of Small RNA Profiles by Massively Parallel Sequencing in Six Forensic Body Fluids/Tissue. Genes. 2022; 13(9):1530. https://doi.org/10.3390/genes13091530
Chicago/Turabian StyleLiu, Zhiyong, Qiangwei Wang, Nana Wang, Yu Zang, Riga Wu, and Hongyu Sun. 2022. "A Comprehensive Characterization of Small RNA Profiles by Massively Parallel Sequencing in Six Forensic Body Fluids/Tissue" Genes 13, no. 9: 1530. https://doi.org/10.3390/genes13091530
APA StyleLiu, Z., Wang, Q., Wang, N., Zang, Y., Wu, R., & Sun, H. (2022). A Comprehensive Characterization of Small RNA Profiles by Massively Parallel Sequencing in Six Forensic Body Fluids/Tissue. Genes, 13(9), 1530. https://doi.org/10.3390/genes13091530





