Association of LBX1 Gene Methylation Level with Disease Severity in Patients with Idiopathic Scoliosis: Study on Deep Paravertebral Muscles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Tissue Samples
2.3. Genomic DNA Methylation Analysis
2.4. Statistical Analysis
3. Results
3.1. Patients and Controls
3.2. DNA Methylation at the LBX1 Promoter Regions—A Case-Control Study
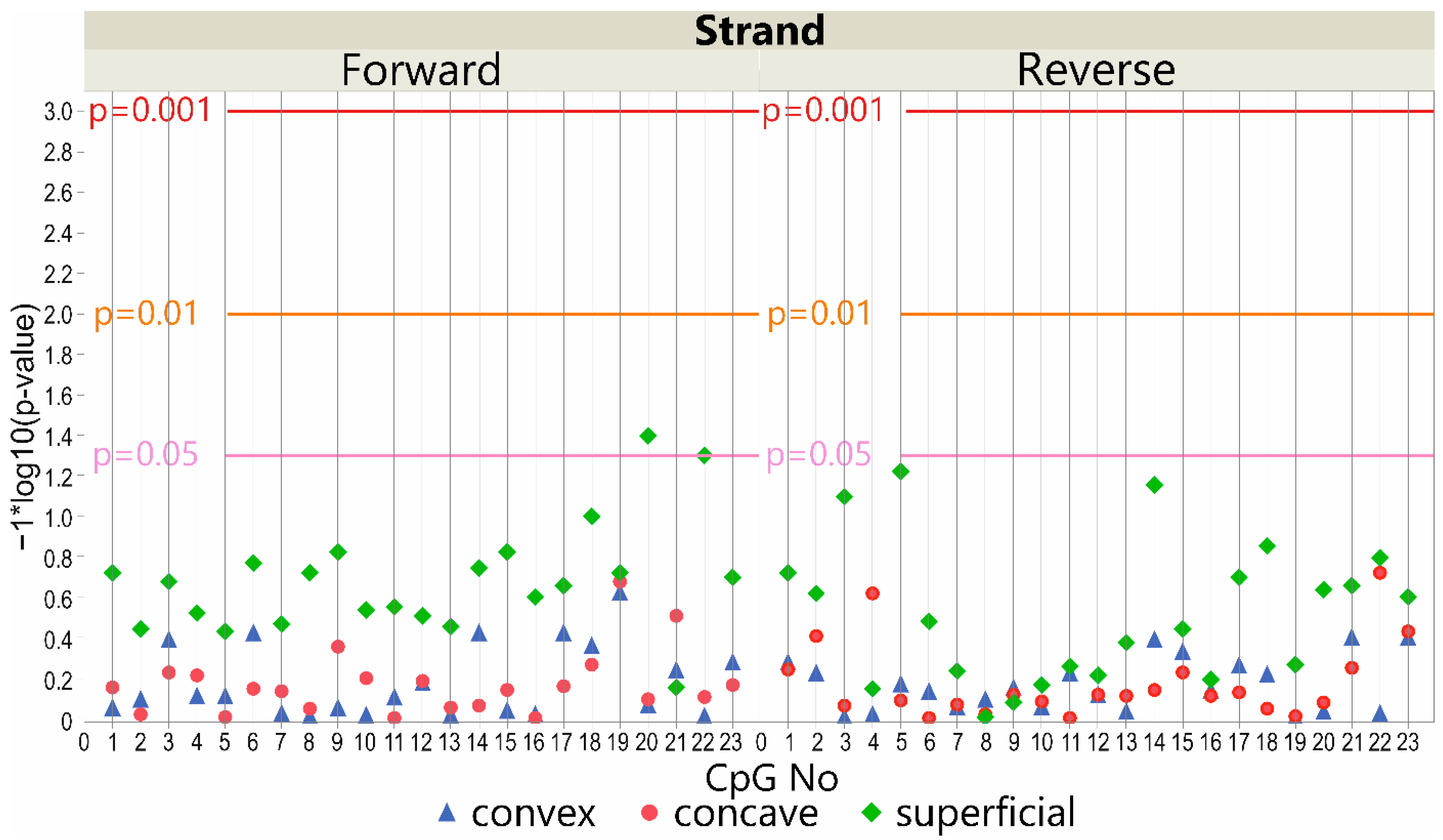
3.3. DNA Methylation at the LBX1 Promoter Regions—Deep Paravertebral Muscles vs. Superficial Muscles
3.3.1. DNA Forward Strand Promoter Region
3.3.2. DNA Reverse Strand Promoter Region
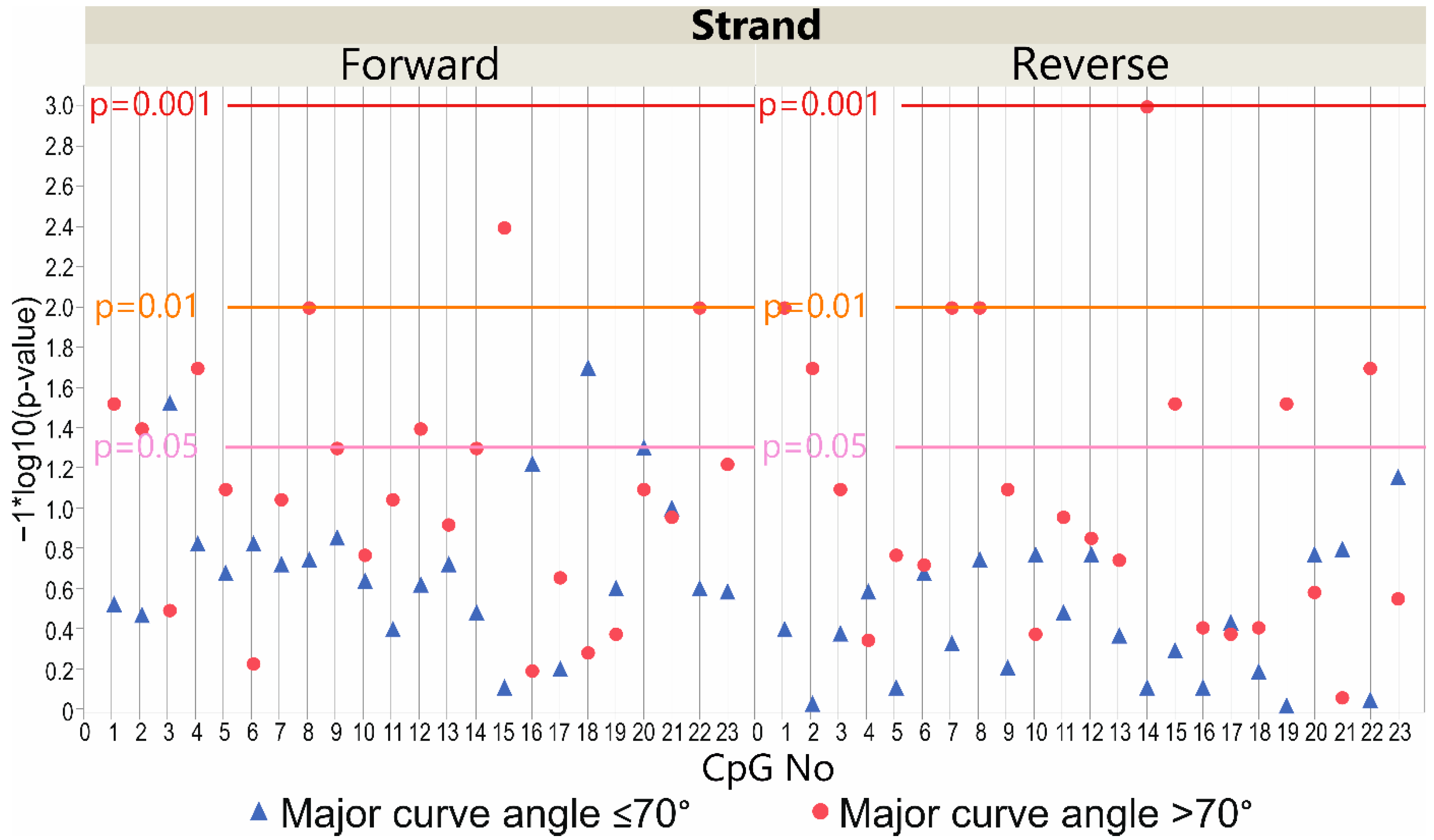
3.4. LBX1 Methylation Status and Major Curve Angle—Case-Only Study
3.4.1. DNA Forward Strand Promoter Region
3.4.2. DNA Reverse Strand Promoter Region
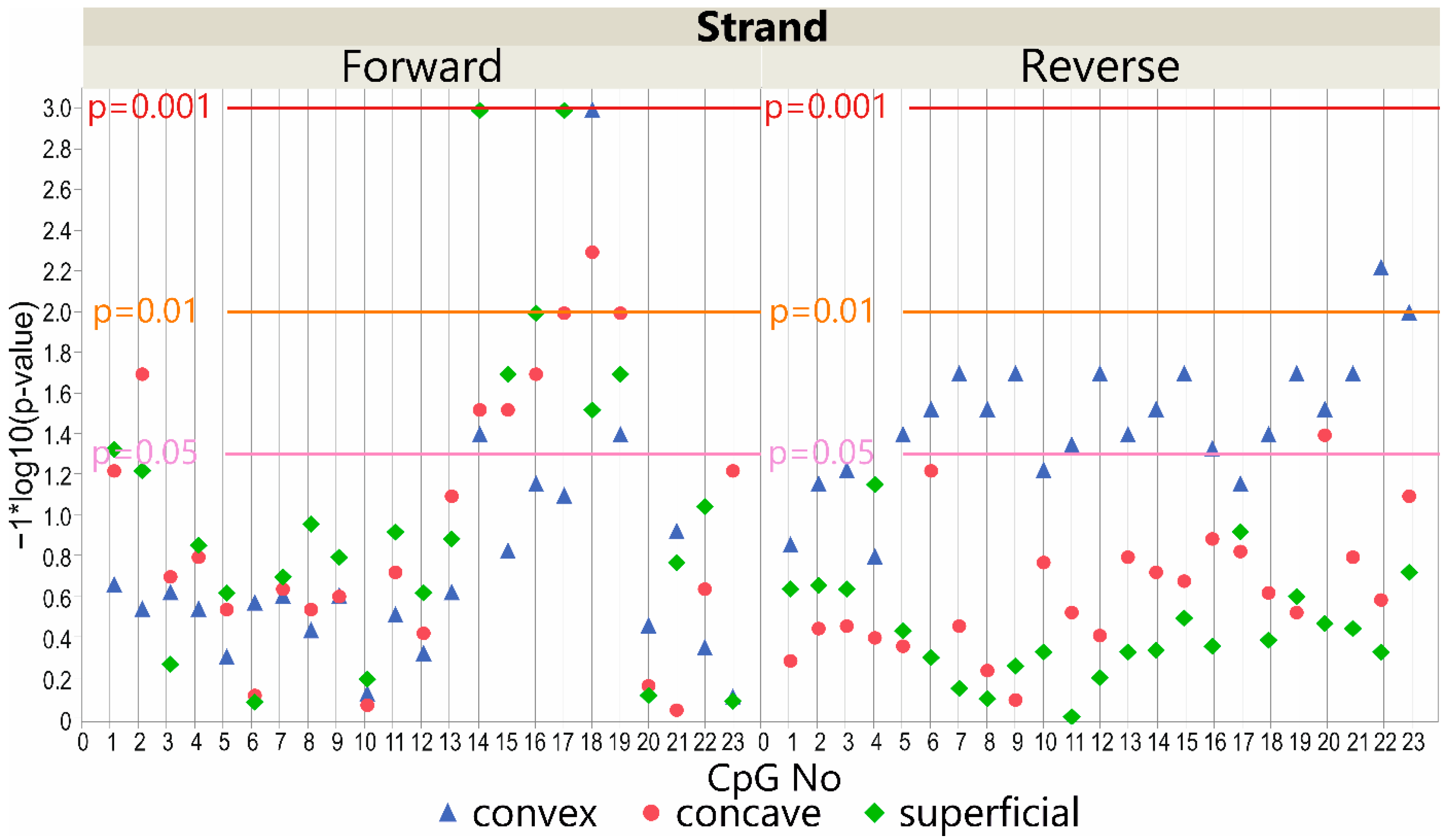
3.5. LBX1 Methylation Status and Major Curve Angle—Deep Paravertebral Muscles vs. Superficial Muscles
3.5.1. DNA Forward Strand Promoter Region
3.5.2. DNA Reverse Strand Promoter Region
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinstein, S.L.; Dolan, L.A.; Cheng, J.C.; Danielsson, A.; Morcuende, J.A. Adolescent idiopathic scoliosis. Lancet 2008, 371, 1527–1537. [Google Scholar] [CrossRef]
- Weinstein, S.L. The Natural History of Adolescent Idiopathic Scoliosis. J. Pediatr. Orthop. 2019, 39, S44–S46. [Google Scholar] [CrossRef]
- Huh, S.; Eun, L.Y.; Kim, N.K.; Jung, J.W.; Choi, J.Y.; Kim, H.S. Cardiopulmonary function and scoliosis severity in idiopathic scoliosis children. Korean J. Pediatr. 2015, 58, 218–223. [Google Scholar] [CrossRef]
- Lowe, T.G.; Edgar, M.; Margulies, J.Y.; Miller, N.H.; Raso, V.J.; Reinker, K.A.; Rivard, C.H. Etiology of idiopathic scoliosis: Current trends in research. J. Bone Jt. Surg.-Ser. A 2000, 82, 1157–1168. [Google Scholar] [CrossRef]
- De Salvatore, S.; Ruzzini, L.; Longo, U.G.; Marino, M.; Greco, A.; Piergentili, I.; Costici, P.F.; Denaro, V. Exploring the association between specific genes and the onset of idiopathic scoliosis: A systematic review. BMC Med. Genom. 2022, 15, 115. [Google Scholar] [CrossRef]
- Khanshour, A.M.; Kou, I.; Fan, Y.; Einarsdottir, E.; Makki, N.; Kidane, Y.H.; Kere, J.; Grauers, A.; Johnson, T.A.; Paria, N.; et al. Genome-wide meta-analysis and replication studies in multiple ethnicities identify novel adolescent idiopathic scoliosis susceptibility loci. Hum. Mol. Genet. 2018, 27, 3986–3998. [Google Scholar] [CrossRef]
- Grauers, A.; Einarsdottir, E.; Gerdhem, P. Genetics and pathogenesis of idiopathic scoliosis. Scoliosis Spinal Disord. 2016, 11, 45. [Google Scholar] [CrossRef]
- Gorman, K.F.; Julien, C.; Moreau, A. The genetic epidemiology of idiopathic scoliosis. Eur. Spine J. 2012, 21, 1905–1919. [Google Scholar] [CrossRef]
- Takahashi, Y.; Kou, I.; Takahashi, A.; Johnson, T.A.; Kono, K.; Kawakami, N.; Uno, K.; Ito, M.; Minami, S.; Yanagida, H.; et al. A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat. Genet. 2011, 43, 1237–1240. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Gao, S.-J.; Xu, H.; Liu, Y.; Li, H.-L.; Chen, X.-Y.; Ning, G.-Z.; Feng, S.-Q. The association of rs11190870 near LBX1 with the susceptibility and severity of AIS, a meta-analysis. Int. J. Surg. 2018, 54, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yang, Q.; Liu, Y.; Guan, Y.; Zhan, X.; Xiao, Z.; Wei, Q. Association between ladybird homeobox 1 gene polymorphisms and adolescent idiopathic scoliosis: A MOOSE-compliant meta-analysis. Medicine 2019, 98, e16314. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhang, Y.; Huang, S.; Song, Y. The Susceptibility and Potential Functions of the LBX1 Gene in Adolescent Idiopathic Scoliosis. Front. Genet. 2021, 11, 1802. [Google Scholar] [CrossRef] [PubMed]
- Jennings, W.; Hou, M.; Perterson, D.; Missiuna, P.; Thabane, L.; Tarnopolsky, M.; Samaan, M.C. Paraspinal muscle ladybird homeobox 1 (LBX1) in adolescent idiopathic scoliosis: A cross-sectional study. Spine J. 2019, 19, 1911–1916. [Google Scholar] [CrossRef]
- Brohmann, H.; Jagla, K.; Birchmeier, C. The role of Lbx1 in migration of muscle precursor cells. Development 2000, 127, 437–445. [Google Scholar] [CrossRef]
- Sieber, M.A.; Storm, R.; Martinez-de-la-Torre, M.; Müller, T.; Wende, H.; Reuter, K.; Vasyutina, E.; Birchmeier, C. Lbx1 Acts as a Selector Gene in the Fate Determination of Somatosensory and Viscerosensory Relay Neurons in the Hindbrain. J. Neurosci. 2007, 27, 4902–4909. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Schäfer, K.; Braun, T. The homeobox containing gene Lbx1 is required for correct dorsal-ventral patterning of the neural tube. J. Neurochem. 2002, 82, 774–782. [Google Scholar] [CrossRef]
- Grauers, A.; Rahman, I.; Gerdhem, P. Heritability of scoliosis. Eur. Spine J. 2012, 21, 1069–1074. [Google Scholar] [CrossRef]
- Balioglu, M.B.; Aydin, C.; Kargin, D.; Albayrak, A.; Atici, Y.; Tas, S.K.; Kaygusuz, M.A. Vitamin-D measurement in patients with adolescent idiopathic scoliosis. J. Pediatr. Orthop. B 2017, 26, 48–52. [Google Scholar] [CrossRef]
- Yang, Z.; Xie, Y.; Chen, J.; Zhang, D.; Yang, C.; Li, M. High selenium may be a risk factor of adolescent idiopathic scoliosis. Med. Hypotheses 2010, 75, 126–127. [Google Scholar] [CrossRef]
- Watanabe, K.; Michikawa, T.; Yonezawa, I.; Takaso, M.; Minami, S.; Soshi, S.; Tsuji, T.; Okada, E.; Abe, K.; Takahashi, M.; et al. Physical Activities and Lifestyle Factors Related to Adolescent Idiopathic Scoliosis. J. Bone Jt. Surg. Am. 2017, 99, 284–294. [Google Scholar] [CrossRef]
- Grauers, A.; Danielsson, A.; Karlsson, M.; Ohlin, A.; Gerdhem, P. Family history and its association to curve size and treatment in 1,463 patients with idiopathic scoliosis. Eur. Spine J. 2013, 22, 2421–2426. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.L.S.; Yeung, H.Y.; Hung, V.W.Y.; Di Liao, C.; Lam, T.P.; Yeung, H.M.; Lee, K.M.; Ng, B.K.W.; Cheng, J.C.Y. Genetic epidemiology and heritability of AIS: A study of 415 Chinese female patients. J. Orthop. Res. 2012, 30, 1464–1469. [Google Scholar] [CrossRef] [PubMed]
- Maqsood, A.; Frome, D.K.; Gibly, R.F.; Larson, J.E.; Patel, N.M.; Sarwark, J.F. IS (Idiopathic Scoliosis) etiology: Multifactorial genetic research continues. A systematic review 1950 to 2017. J. Orthop. 2020, 21, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Faldini, C.; Manzetti, M.; Neri, S.; Barile, F.; Viroli, G.; Geraci, G.; Ursini, F.; Ruffilli, A. Epigenetic and Genetic Factors Related to Curve Progression in Adolescent Idiopathic Scoliosis: A Systematic Scoping Review of the Current Literature. Int. J. Mol. Sci. 2022, 23, 5914. [Google Scholar] [CrossRef]
- Burwell, R.G.; Dangerfield, P.H.; Moulton, A.; Grivas, T.B.; Meredith, D.; Wägele, B.; Altmaier, E.; Deloukas, P.; Erdmann, J.; Grundberg, E.; et al. Adolescent idiopathic scoliosis (AIS), environment, exposome and epigenetics: A molecular perspective of postnatal normal spinal growth and the etiopathogenesis of AIS with consideration of a network approach and possible implications for medical therapy. Scoliosis 2011, 6, 26. [Google Scholar] [CrossRef]
- Meng, Y.; Lin, T.; Liang, S.; Gao, R.; Jiang, H.; Shao, W.; Yang, F.; Zhou, X. Value of DNA methylation in predicting curve progression in patients with adolescent idiopathic scoliosis. EBioMedicine 2018, 36, 489–496. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Landen, S.; Jacques, M.; Hiam, D.; Alvarez-Romero, J.; Harvey, N.R.; Haupt, L.M.; Griffiths, L.R.; Ashton, K.J.; Lamon, S.; Voisin, S.; et al. Skeletal muscle methylome and transcriptome integration reveals profound sex differences related to muscle function and substrate metabolism. Clin. Epigenet. 2021, 13, 202. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Yan, J.; Chen, M.; Zhu, M.; Tang, Y.; Liu, S.; Tang, Z. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 2021, 49, 1313–1329. [Google Scholar] [CrossRef]
- Liu, G.; Wang, L.; Wang, X.; Yan, Z.; Yang, X.; Lin, M.; Liu, S.; Zuo, Y.; Niu, Y.; Zhao, S.; et al. Whole-Genome Methylation Analysis of Phenotype Discordant Monozygotic Twins Reveals Novel Epigenetic Perturbation Contributing to the Pathogenesis of Adolescent Idiopathic Scoliosis. Front. Bioeng. Biotechnol. 2019, 7, 364. [Google Scholar] [CrossRef]
- Mao, S.-H.; Qian, B.-P.; Shi, B.; Zhu, Z.-Z.; Qiu, Y. Quantitative evaluation of the relationship between COMP promoter methylation and the susceptibility and curve progression of adolescent idiopathic scoliosis. Eur. Spine J. 2018, 27, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Xu, L.; Mao, S.; Xu, L.; Liu, Z.; Sun, X.; Zhu, Z.; Qiu, Y. Abnormal PITX1 gene methylation in adolescent idiopathic scoliosis: A pilot study. BMC Musculoskelet. Disord. 2018, 19, 138. [Google Scholar] [CrossRef]
- Shi, B.; Mao, S.; Xu, L.; Li, Y.; Sun, X.; Liu, Z.; Zhu, Z.; Qiu, Y. Quantitation Analysis of PCDH10 Methylation In Adolescent Idiopathic Scoliosis Using Pyrosequencing Study. Spine 2020, 45, E373–E378. [Google Scholar] [CrossRef]
- Cobb, J.R. Outline for the Study of Scoliosis. In Instructional Course Lectures; Edwards: Ann Arbor, MI, USA, 1948; Volume 5, pp. 261–275. [Google Scholar]
- Risser, J.C.; Brand, R.A. The iliac apophysis: An invaluable sign in the management of scoliosis. Clin. Orthop. Relat. Res. 2010, 468, 646–653. [Google Scholar] [CrossRef]
- Janusz, P.; Chmielewska, M.; Andrusiewicz, M.; Kotwicka, M.; Kotwicki, T. Methylation of Estrogen Receptor 1 Gene in the Paraspinal Muscles of Girls with Idiopathic Scoliosis and Its Association with Disease Severity. Genes 2021, 12, 790. [Google Scholar] [CrossRef]
- Cheung, K.M.C.; Wang, T.; Qiu, G.X.; Luk, K.D.K. Recent advances in the aetiology of adolescent idiopathic scoliosis. Int. Orthop. 2008, 32, 729–734. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, S.R.; Qiu, G.X.; Zhang, J.G.; Zhuang, Q.Y.; Wang, N.N. Research progress on the etiology and pathogenesis of adolescent idiopathic scoliosis. Chin. Med. J. 2020, 133, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Fadzan, M.; Bettany-Saltikov, J. Etiological Theories of Adolescent Idiopathic Scoliosis: Past and Present. Open Orthop. J. 2017, 11, 1466–1489. [Google Scholar] [CrossRef]
- Roaf, R. The basic anatomy of scoliosis. J. Bone Jt. Surg. 1966, 48, 786–792. [Google Scholar] [CrossRef]
- Guo, X.; Chau, W.W.; Chan, Y.L.; Cheng, J.C.Y. Relative anterior spinal overgrowth in adolescent idiopathic scoliosis. Results of disproportionate endochondral-membranous bone growth. J. Bone Jt. Surg. Br. 2003, 85, 1026–1031. [Google Scholar] [CrossRef]
- Chu, W.C.W.; Lam, W.W.M.; Chan, Y.L.; Ng, B.K.W.; Lam, T.P.; Lee, K.M.; Guo, X.; Cheng, J.C.Y. Relative shortening and functional tethering of spinal cord in adolescent idiopathic scoliosis? Study with multiplanar reformat magnetic resonance imaging and somatosensory evoked potential. Spine 2006, 31, E19–E25. [Google Scholar] [CrossRef] [PubMed]
- Roye, B.D.; Wright, M.L.; Matsumoto, H.; Yorgova, P.; McCalla, D.; Hyman, J.E.; Roye, D.P.; Shah, S.A.; Vitale, M.G. An Independent Evaluation of the Validity of a DNA-Based Prognostic Test for Adolescent Idiopathic Scoliosis. J. Bone Jt. Surg. Am. 2015, 97, 1994–1998. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, S.L.; Zavala, D.C.; Ponseti, I.V. Idiopathic scoliosis. Long-term follow-up and prognosis in untreated patients. J. Bone Jt. Surg.-Ser. A 1981, 63, 702–712. [Google Scholar] [CrossRef]
- Nepple, J.J.; Lenke, L.G. Severe idiopathic scoliosis with respiratory insufficiency treated with preoperative traction and staged anteroposterior spinal fusion with a 2-level apical vertebrectomy. Spine J. 2009, 9, e9–e13. [Google Scholar] [CrossRef]
- Asher, M.A.; Burton, D.C. Adolescent idiopathic scoliosis: Natural history and long term treatment effects. Scoliosis 2006, 1, 2. [Google Scholar] [CrossRef]
- Weinstein, S.L.; Ponseti, I. V Curve progression in idiopathic scoliosis. J. Bone Jt. Surg. Am. 1983, 65, 447–455. [Google Scholar] [CrossRef]
- Luhmann, S.J.; Lenke, L.G.; Kim, Y.J.; Bridwell, K.H.; Schootman, M. Thoracic adolescent idiopathic scoliosis curves between 70° and 100°: Is anterior release necessary? Spine 2005, 30, 2061–2067. [Google Scholar] [CrossRef]
- Suk, S.I.; Kim, J.H.; Cho, K.J.; Kim, S.S.; Lee, J.J.; Han, Y.T. Is anterior release necessary in severe scoliosis treated by posterior segmental pedicle screw fixation? Eur. Spine J. 2007, 16, 1359–1365. [Google Scholar] [CrossRef]
- Solla, F.; Clement, J.L.; Doria, C.; Bertoncelli, C.; Rosello, O.; Rampal, V. Adolescent idiopathic scoliosis exceeding 70°: A single unit surgical experience. Minerva Ortop. Traumatol. 2018, 69, 69–77. [Google Scholar] [CrossRef]
- Wajchenberg, M.; Martins, D.E.; De Paiva Luciano, R.; Puertas, E.B.; Del Curto, D.; Schmidt, B.; De Souza Oliveira, A.B.; Faloppa, F. Histochemical analysis of paraspinal rotator muscles from patients with adolescent idiopathic scoliosis. Medicine 2015, 94, e598. [Google Scholar] [CrossRef] [Green Version]
- Newton Ede, M.M.P.; Jones, S.W. Adolescent idiopathic scoliosis: Evidence for intrinsic factors driving aetiology and progression. Int. Orthop. 2016, 40, 2075–2080. [Google Scholar] [CrossRef] [PubMed]
- Mannion, A.F.; Meier, M.; Grob, D.; Muntener, M. Paraspinal muscle fibre type alterations associated with scoliosis: An old problem revisited with new evidence. Eur. Spine J. 1998, 7, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Meng, Y.; Jin, X.; Zhang, C.; Zhao, J.; Wang, C.; Gao, R.; Zhou, X. Volumetric and fatty infiltration imbalance of deep paravertebral muscles in adolescent idiopathic scoliosis. Med. Sci. Monit. 2017, 23, 2089–2095. [Google Scholar] [CrossRef]
- Avikainen, V.; Rezasoltani, A.; Kauhanen, H. Asymmetry of paraspinal EMG-time characteristics in idiopathic scoliosis. J. Spinal Disord. 1999, 12, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Veldhuizen, A.G.; Halbertsma, J.P.K.; Maurits, N.M.; Sluiter, W.J.; Cool, J.C.; Van Horn, J.R. The Relation Between Electromyography and Growth Velocity of the Spine in the Evaluation of Curve Progression in Idiopathic Scoliosis. Spine 2004, 29, 1011–1016. [Google Scholar] [CrossRef]
- Konieczny, M.R.; Senyurt, H.; Krauspe, R. Epidemiology of adolescent idiopathic scoliosis. J. Child. Orthop. 2013, 7, 3–9. [Google Scholar] [CrossRef]
- Wu, N.; Ming, X.; Xiao, J.; Wu, Z.; Chen, X.; Shinawi, M.; Shen, Y.; Yu, G.; Liu, J.; Xie, H.; et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015, 372, 341–350. [Google Scholar] [CrossRef]
- Takeda, K.; Kou, I.; Kawakami, N.; Iida, A.; Nakajima, M.; Ogura, Y.; Imagawa, E.; Miyake, N.; Matsumoto, N.; Yasuhiko, Y.; et al. Compound Heterozygosity for Null Mutations and a Common Hypomorphic Risk Haplotype in TBX6 Causes Congenital Scoliosis. Hum. Mutat. 2017, 38, 317–323. [Google Scholar] [CrossRef]
- Sparrow, D.B.; Chapman, G.; Smith, A.J.; Mattar, M.Z.; Major, J.A.; O’Reilly, V.C.; Saga, Y.; Zackai, E.H.; Dormans, J.P.; Alman, B.A.; et al. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell 2012, 149, 295–306. [Google Scholar] [CrossRef]
- Lin, M.; Zhao, S.; Liu, G.; Huang, Y.; Yu, C.; Zhao, Y.; Wang, L.; Zhang, Y.; Yan, Z.; Wang, S.; et al. Identification of novel FBN1 variations implicated in congenital scoliosis. J. Hum. Genet. 2020, 65, 221–230. [Google Scholar] [CrossRef] [Green Version]
- Giampietro, P.F.; Raggio, C.L.; Reynolds, C.E.; Shukla, S.K.; McPherson, E.; Ghebranious, N.; Jacobsen, F.S.; Kumar, V.; Faciszewski, T.; Pauli, R.M.; et al. An analysis of PAX1 in the development of vertebral malformations. Clin. Genet. 2005, 68, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, T.; Nashabat, M.; Alghanem, B.; Alhallaj, A.S.; Boudjelal, M.; Umair, M.; Alarifi, S.; Alfares, A.; Al Mohrij, S.A.; Alfadhel, M. Delta Like-1 Gene Mutation: A Novel Cause of Congenital Vertebral Malformation. Front. Genet. 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Feng, Z.; Dai, Z.; Lee, W.Y.W.; Wu, Z.; Liu, Z.; Sun, X.; Tang, N.; Cheng, J.C.Y.; Qiu, Y.; et al. A Functional SNP in the Promoter of LBX1 Is Associated With the Development of Adolescent Idiopathic Scoliosis Through Involvement in the Myogenesis of Paraspinal Muscles. Front. Cell Dev. Biol. 2021, 9, 777890. [Google Scholar]
- Xu, L.; Dai, Z.; Xia, C.; Wu, Z.; Feng, Z.; Sun, X.; Liu, Z.; Qiu, Y.; Cheng, J.C.Y.; Zhu, Z. Asymmetric Expression of Wnt/B-catenin Pathway in AIS: Primary or Secondary to the Curve? Spine 2020, 45, E677–E683. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, Z.; Cheng, K.L.; Zhang, J.; Xu, L.; Lam, T.P.; Hung, A.L.H.; Cheng, J.C.Y.; Qiu, Y.; Lee, W.Y.W. Role of differentially expressed LBX1 in Adolescent Idiopathic Scoliosis (AIS) paraspinal muscle phenotypes and muscle-bone crosstalk through modulating myoblasts. Stud. Health Technol. Inform. 2021, 280, 14–17. [Google Scholar]

| Primer | Sequence | Length (nt) | Tm (°C) | GC (%) | PCR Product Size | |
|---|---|---|---|---|---|---|
| LBX1 Forward | B→ PCR | TTTAGGTAGTGGGGTGAG | 18 | 55.8 | 50.0 | 256 bp |
| ← PCR | CCCCAACTATTTATAAATTACATTAACTAC | 30 | 51.9 | 26.7 | ||
| ← SEQ | ATAAATTACATTAACTACTCCTT | 23 | 44.0 | 21.7 | - | |
| LBX1 Reverse | → PCR | GTAGTGGGGTGAGGGGTAA | 19 | 60.3 | 57.9 | 333 bp |
| ← B PCR | ACATTAACTACTCCTTTATTACACC | 25 | 57.2 | 32.0 | ||
| → SEQ | GAGGGGTAAGAGGGT | 15 | 50.8 | 60.0 | - |
| Component | Initial Concentration | Volume Added | Final Concentration | Mixture Volume |
|---|---|---|---|---|
| ZymoTaqTM Premix | 2× | 5 µL | 1× | 10 µL |
| → PCR | 10 µM | 1 µL | 1 µM | |
| ← PCR | 10 µM | 1 µL | 1 µM | |
| DNA | 100 ng/µL | 0.2 µL | 2 ng/µL | |
| Nuclease-free water | 2.8 µL | |||
| Thermal profile of the reactions | ||||
| Number of cycles | Step | Duration, temperature | ||
| 1 | Initial denaturation | 10 min, 95 °C | ||
| 37 | Denaturation | 30 s, 95 °C | ||
| Annealing | 30 s, 54 °C A, 58 °C B | |||
| Extension | 60 s, 72 °C | |||
| 1 | Final extension | 7 min, 72 °C | ||
| 1 | Hold | ∞, 4 °C | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janusz, P.; Tokłowicz, M.; Andrusiewicz, M.; Kotwicka, M.; Kotwicki, T. Association of LBX1 Gene Methylation Level with Disease Severity in Patients with Idiopathic Scoliosis: Study on Deep Paravertebral Muscles. Genes 2022, 13, 1556. https://doi.org/10.3390/genes13091556
Janusz P, Tokłowicz M, Andrusiewicz M, Kotwicka M, Kotwicki T. Association of LBX1 Gene Methylation Level with Disease Severity in Patients with Idiopathic Scoliosis: Study on Deep Paravertebral Muscles. Genes. 2022; 13(9):1556. https://doi.org/10.3390/genes13091556
Chicago/Turabian StyleJanusz, Piotr, Małgorzata Tokłowicz, Mirosław Andrusiewicz, Małgorzata Kotwicka, and Tomasz Kotwicki. 2022. "Association of LBX1 Gene Methylation Level with Disease Severity in Patients with Idiopathic Scoliosis: Study on Deep Paravertebral Muscles" Genes 13, no. 9: 1556. https://doi.org/10.3390/genes13091556






