Up-Regulation of Interleukin-10 in Splenic Immune Response Induced by Serotype A Pasteurellamultocida
Abstract
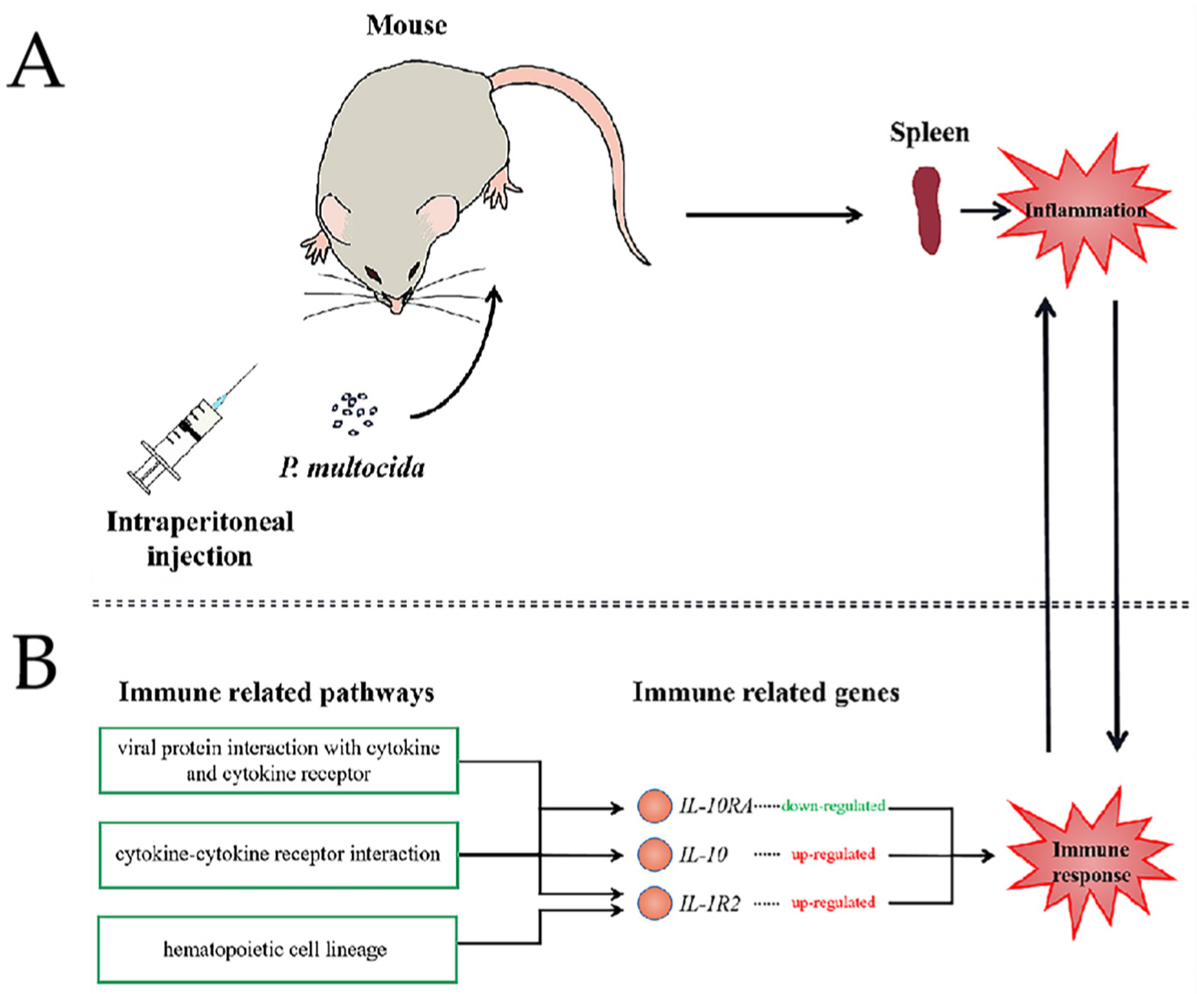
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Experimental Animals
2.3. Pathogenic of P. multocida HN02
2.4. Bacterial Colonization
2.5. Total RNA Extraction, Library Construction, and Sequencing
2.6. Differentially Expressed Genes Analysis
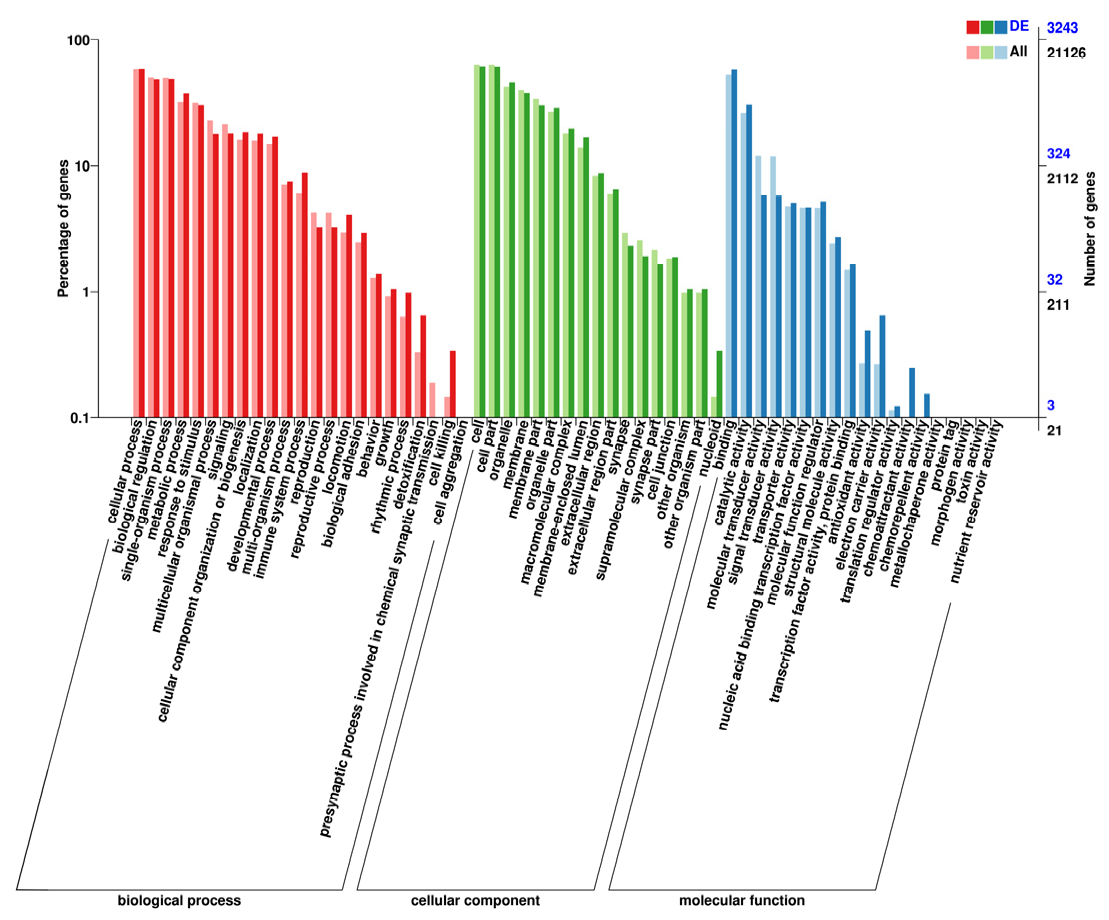
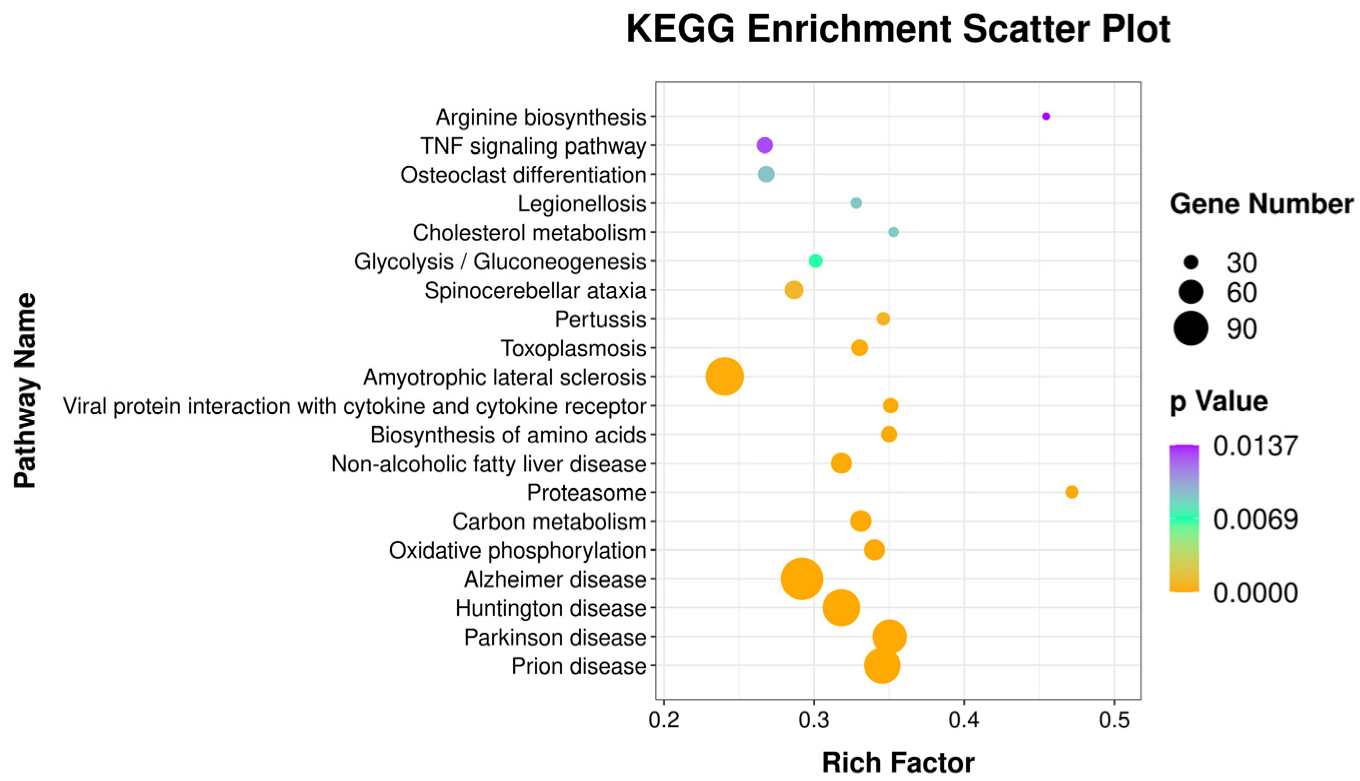
2.7. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analyses
2.8. Quantitative Real-Time PCR
2.9. Statistical Analysis
3. Results
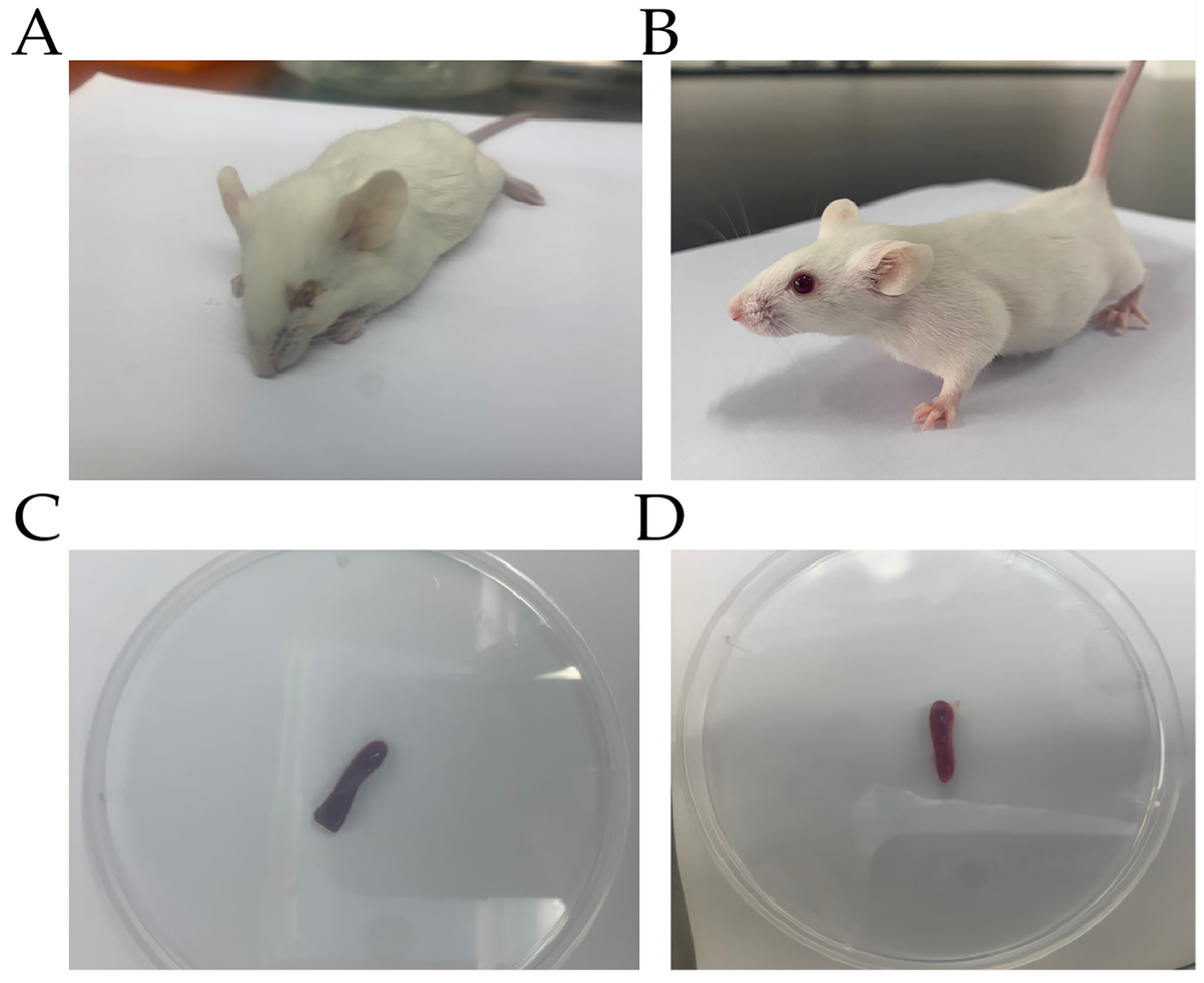
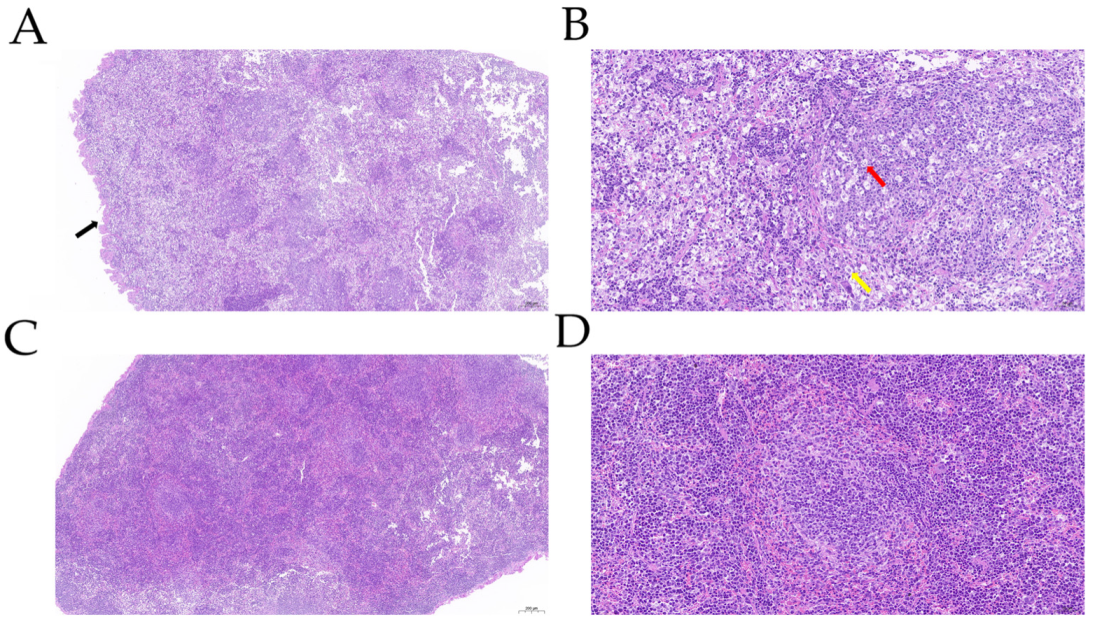
3.1. P. multocida Infection in Mice
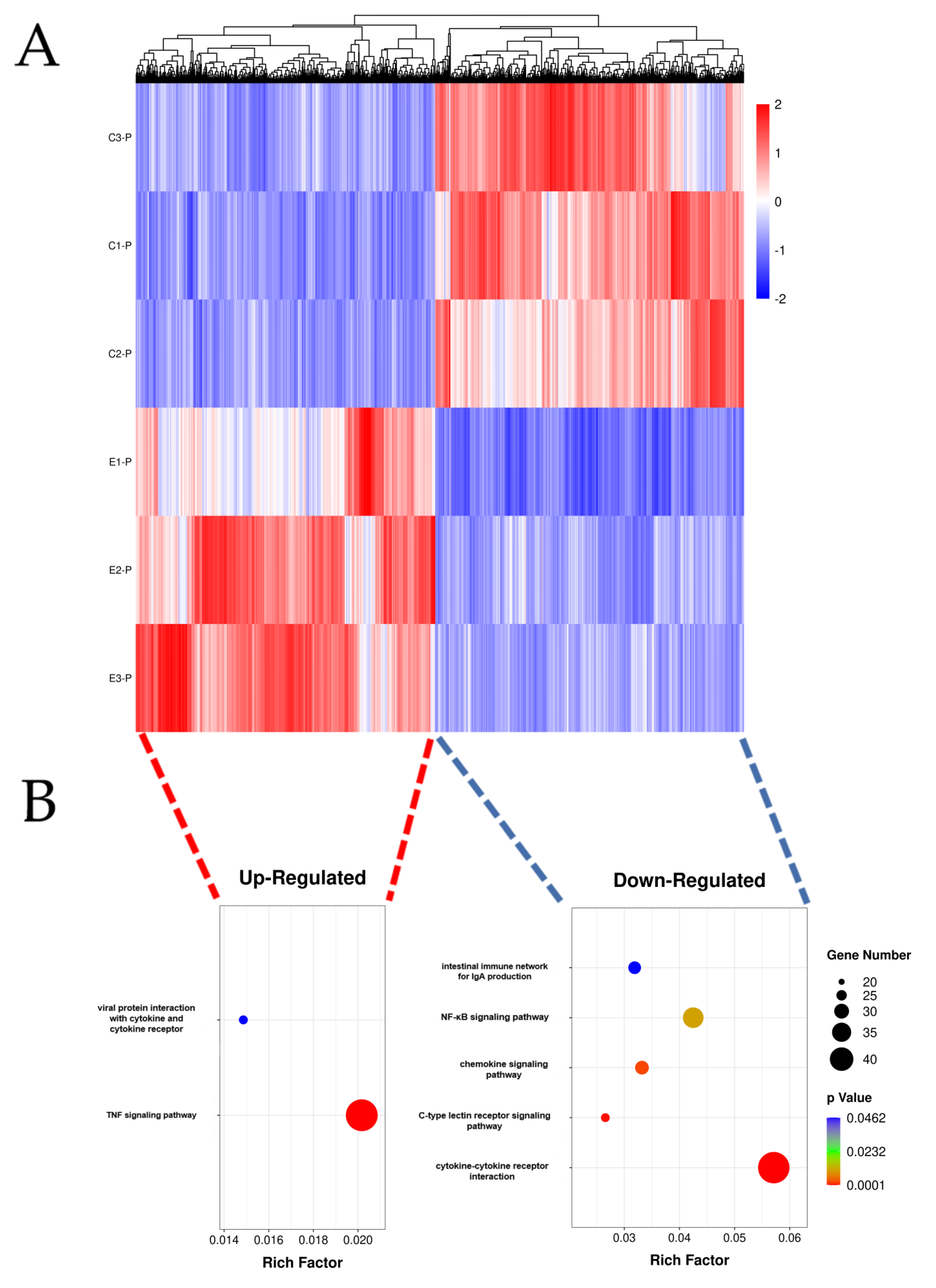
3.2. Transcriptomic Sequencing Analysis
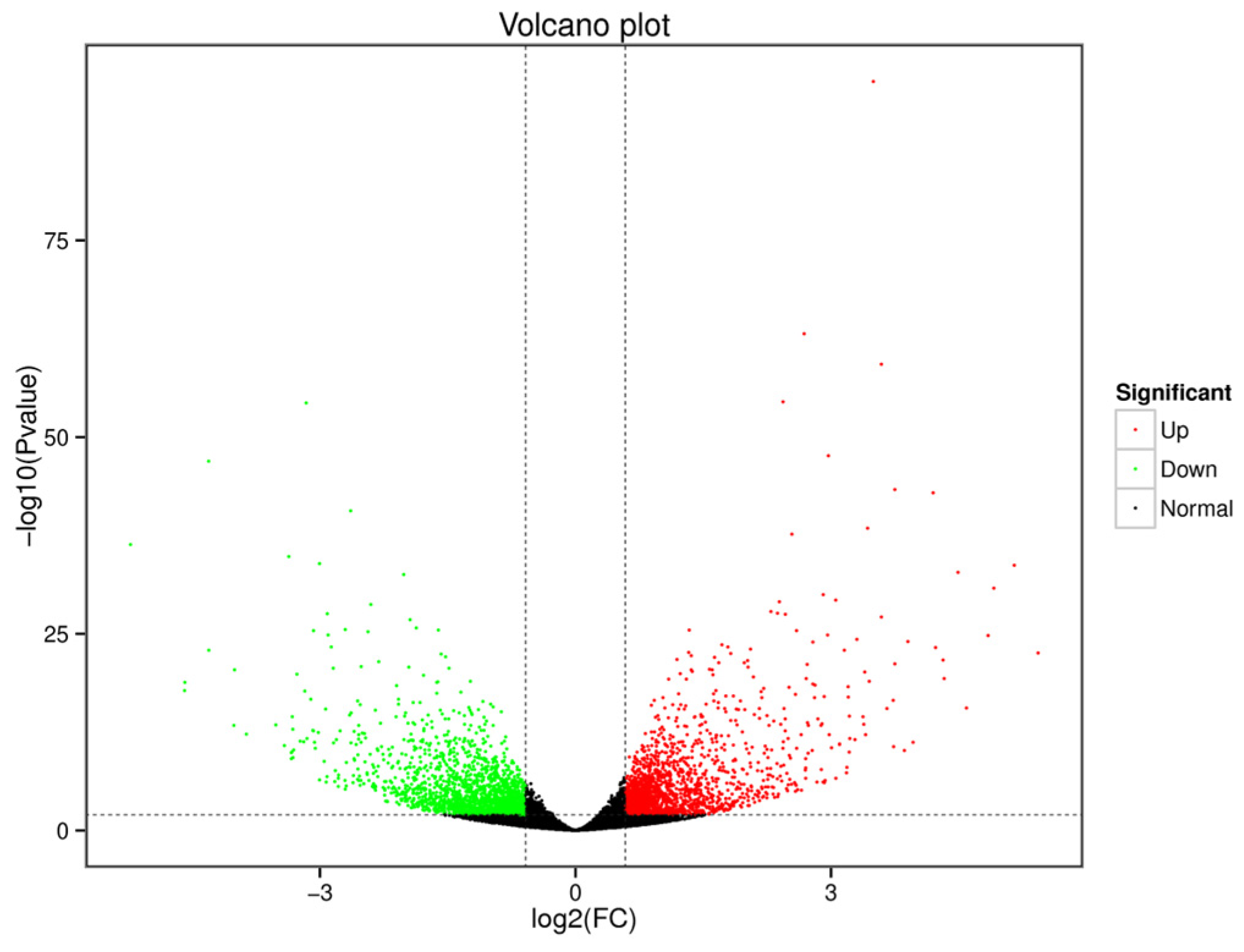
3.3. Identification of DEGs
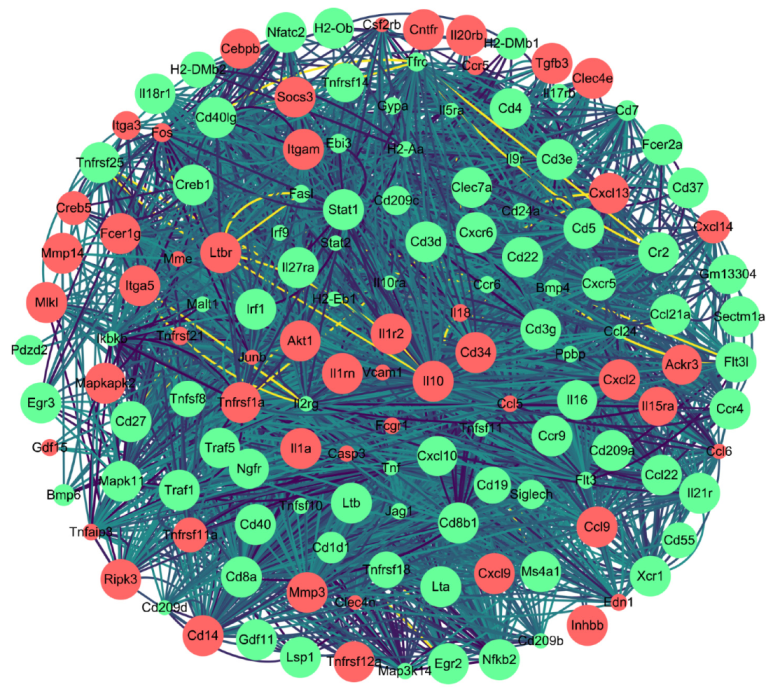
3.4. DEGs Related to Host Immune Response against Infection
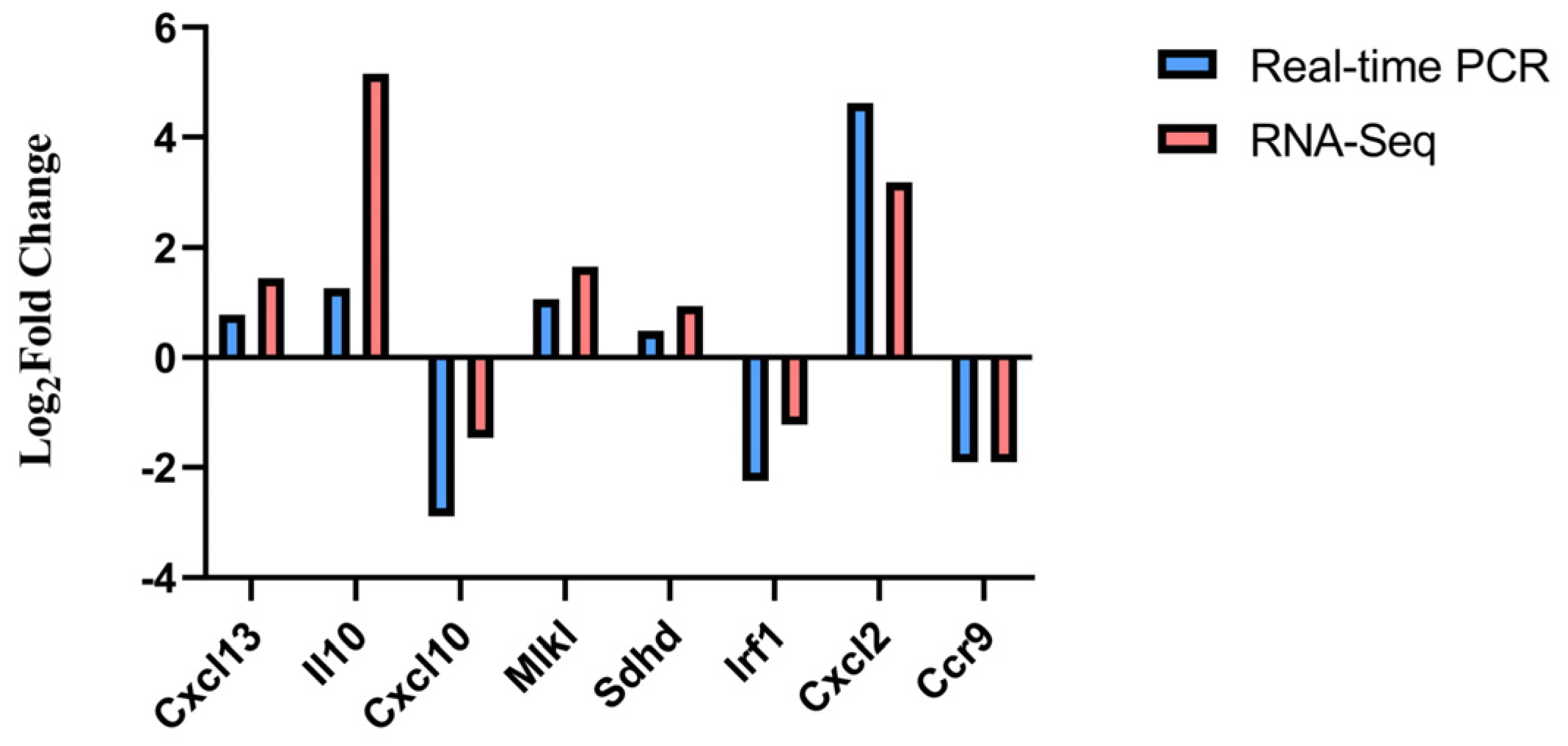
3.5. Experimental Validation of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wilson, B.A.; Ho, M.F. Pasteurella multocida: From Zoonosis to Cellular Microbiology. Clin. Microbiol. Rev. 2013, 26, 631–655. [Google Scholar] [CrossRef] [PubMed]
- Marcantonio, Y.C.; Kulkarni, P.A.; Sachs, S.; Ting, K.; Lee, J.; Mendoza, D. Disseminated Pasteurella multocida infection: Cellulitis, osteomyelitis, and myositis. IDCases 2017, 10, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Townsend, K.M.; Boyce, J.D.; Chung, J.Y.; Frost, A.J.; Alder, B. Genetic Organization of Pasteurella multocida cap Loci and Development of a Multiplex Capsular PCR Typing System. J. Clin. Microbiol. 2001, 39, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, N.D.; Ajam, N.; Blackall, P.J.; Asiah, N.M.; Ramlan, M.; Maria, J.; Thong, K.L. Capsular serotyping of Pasteurella multocida from various animal hosts–a comparison of phenotypic and genotypic methods. Trop. Biomed. 2011, 28, 55–63. [Google Scholar] [PubMed]
- Chen, Q.L.; Zhang, Z.X.; Chen, S.; Chen, J.; Cheng, Y.W.; Liu, A.; Li, B.; Chen, Z.; Zheng, Y.Y. Genome-Wide Differential Expression Profiling of Pulmonary circRNAs Associated With Immune Reaction to Pasteurella multocida in Goats. Front. Vet. Sci. 2021, 8, 615405. [Google Scholar] [CrossRef]
- He, F.; Zhao, Z.L.; Wu, X.Y.; Duan, L.J.; Li, N.Z.; Fang, R.D. Transcriptomic Analysis of High- and Low-Virulence Bovine Pasteurella multocida in vitro and in vivo. Front.Vet. Sci. 2021, 8, 616774. [Google Scholar] [CrossRef]
- Alexey, V.N.; Tatyana, I.G.; Alexander, G.G.; Vladimir, A.T.; Alexandra, O.S. Prevalence of different OmpH-types among Pasteurella multocida isolated from the lungs of calves with respiratory problems. Microb. Pathog. 2017, 104, 184–189. [Google Scholar]
- Praveena, P.E.; Periasamy, S.; Kumar, A.A.; Singh, N. Cytokine profiles, apoptosis and pathology of experimental Pasteurella multocida serotype A1 infection in mice. Res. Vet. Sci. 2020, 89, 332–339. [Google Scholar] [CrossRef]
- Fang, R.D.; Lei, G.H.; Jiang, J.L.; Du, H.H.; Liu, Y.J.; Lei, Z.H.; Ye, C.; Li, N.Z. High- and low-virulent bovine Pasteurella multocida induced differential NLRP3 inflammasome activation and subsequent IL-1β secretion. Vet. Microbiol. 2020, 243, 108646. [Google Scholar] [CrossRef]
- Parker, T.A.; Cheng, H.W.; Willeford, K.O. Interleukin-6 expression in response to innate immune regulatory factor stimulation. Biomed. Pharmacother. 2011, 65, 90–94. [Google Scholar] [CrossRef]
- Medzhitov, R.; Shevach, E.M.; Trinchieri, G.; Mellor, A.L.; Munn, D.H.; Gordon, S.; Libby, P.; Hansson, G.K.; Shortman, K.; Dong, C.; et al. Highlights of 10 years of immunology in Nature Reviews Immunology. Nat. Rev. Immunol. 2011, 11, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.W.; de Malefyt, R.W.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef]
- Kang, M.J.; Jang, A.R.; Park, J.Y.; Ahn, J.H.; Lee, T.S.; Kim, D.Y.; Lee, M.S.; Hwang, S.; Jeong, Y.J.; Park, J.H. IL-10 Protects Mice From the Lung Infection of Acinetobacter baumannii and Contributes to Bacterial Clearance by Regulating STAT3-Mediated MARCO Expression in Macrophages. Front. Immunol. 2020, 11, 270. [Google Scholar] [CrossRef] [PubMed]
- Aziz, F.; Chakraborty, A.; Liu, K.; Zhang, T.; Li, X.; Du, R.; Monts, J.; Xu, G.; Li, Y.; Bai, R.; et al. Gastric tumorigenesis induced by combining Helicobacter pylori infection and chronic alcohol through IL-10 inhibition. Carcinogenesis 2022, 43, 126–139. [Google Scholar] [CrossRef]
- Khatun, M.M.; Islam, M.A.; Baek, B.K. In Vitro and In Vivo IFN-γ and IL-10 Measurement in Experimental Brucella abortus Biotype 1 Infection in Sprague-Dawley Rats. Vector Borne Zoonotic Dis. 2021, 21, 579–585. [Google Scholar] [CrossRef]
- Guo, Z.J.; Zhao, Y.; Zhang, Z.D.; Li, Y.M. Interleukin-10-Mediated Lymphopenia Caused by Acute Infection with Foot-and-Mouth Disease Virus in Mice. Viruses 2021, 13, 2358. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Lin, H.C. Characterization and function of membrane vesicles in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 2021, 105, 1795–1801. [Google Scholar] [CrossRef]
- Kobayashi, S.D.; Malachowa, N.; DeLeo, F.R. Pathogenesis of Staphylococcus aureus abscesses. Am. J. Pathol. 2015, 185, 1518–1527. [Google Scholar] [CrossRef]
- Lowy, F.D. How Staphylococcus aureus adapts to its host. N. Engl. J. Med. 2011, 364, 1987–1990. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, X.; Zhou, R.; Chen, H.; Wilson, B.A.; Wu, B. Pasteurella multocida: Genotypes and Genomics. Microbiol. Mol. Biol. Rev. 2019, 83, e00014–e00019. [Google Scholar] [CrossRef]
- Miao, Z.; Ding, Y.; Zhao, N.; Chen, X.; Cheng, H.; Wang, J.; Liu, J.; Wang, F. Transcriptome sequencing reveals fibrotic associated-genes involved in bovine mammary fibroblasts with Staphylococcus aureus. Int. J. Biochem. Cell Biol. 2020, 121, 105696. [Google Scholar] [CrossRef]
- Wu, C.; Qin, X.; Li, P.; Pan, T.; Ren, W.; Li, N.; Peng, Y. Transcriptomic Analysis on Responses of Murine Lungs to Pasteurella multocida Infection. Front. Cell Infect. Microbiol. 2017, 7, 251. [Google Scholar] [CrossRef]
- Kashimura, M. The human spleen as the center of the blood defense system. Int. J. Hematol. 2020, 112, 147–158. [Google Scholar] [CrossRef]
- Zhang, W.; Jiao, Z.; Huang, H.; Wu, Y.; Wu, H.; Liu, Z.; Zhang, Z.; An, Q.; Cheng, Y.; Chen, S.; et al. Effects of Pasteurella multocida on Histopathology, miRNA and mRNA Expression Dynamics in Lung of Goats. Animals 2022, 12, 1529. [Google Scholar] [CrossRef]
- Uenoyama, K.; Ueno, Y.; Tosaki, K.; Abeto, Y.; Ito, H.; Katsuda, K.; Shibahara, T. Immunohistochemical and molecular analysis of Pasteurella multocida in a rabbit with suppurative pleuropneumonia. J. Vet. Med. Sci. 2020, 82, 89–93. [Google Scholar] [CrossRef]
- Lalić, I.M.; Bichele, R.; Repar, A.; Despotović, S.Z.; Petričević, S.; Laan, M.; Peterson, P.; Westermann, J.; Milićević, Ž.; Mirkov, I.; et al. Lipopolysaccharide induces tumor necrosis factor receptor-1 independent relocation of lymphocytes from the red pulp of the mouse spleen. Ann. Anat. 2018, 216, 125–134. [Google Scholar] [CrossRef]
- DeAngelis, P.L. Microbial glycosaminoglycan glycosyltransferases. Glycobiology 2002, 12, 9R–16R. [Google Scholar] [CrossRef]
- Townley, R.A.; Bülow, H.E. Deciphering functional glycosaminoglycan motifs in development. Curr. Opin. Struct. Biol. 2018, 50, 144–154. [Google Scholar] [CrossRef]
- Cress, B.F.; Englaender, J.A.; He, W.; Kasper, D.; Linhardt, R.J.; Koffas, M.A. Masquerading microbial pathogens: Capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol. Rev. 2014, 38, 660–697. [Google Scholar] [CrossRef]
- DeAngelis, P.L. Evolution of glycosaminoglycans and their glycosyltransferases: Implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat. Rec. 2002, 268, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, S.K.; Thies, E.S. Influence of encapsulation on phagocytosis of Pasteurella multocida by bovine neutrophils. Infect. Immun. 1979, 26, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Harper, M.; John, M.; Edmunds, M.; Wright, A.; Ford, M.; Turni, C.; Blackall, P.J.; Cox, A.; Adler, B.; Boyce, J.D. Protective efficacy afforded by live Pasteurella multocida vaccines in chickens is independent of lipopolysaccharide outer core structure. Vaccine 2016, 34, 1696–1703. [Google Scholar] [CrossRef]
- Da Correia, J.S.; Soldau, K.; Christen, U.; Tobias, P.S.; Ulevitch, R.J. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex. transfer from CD14 to TLR4 and MD-2. J. Biol. Chem. 2001, 276, 21129–21135. [Google Scholar] [CrossRef]
- Li, B.; Chen, Q.L.; Cheng, Y.; Chen, J.; Zhang, Z.; An, Q.; Huang, H.; Liu, Z.; Zhai, Z.; Man, C.; et al. Transcriptome Analysis of Mouse Spleen After Infection with Goat-derived Pasteurella multocida Type D. China Anim. Husb. Vet. Med. 2021, 48, 4391–4402. [Google Scholar]
- An, Q.; Chen, S.; Zhang, L.; Zhang, Z.; Cheng, Y.; Wu, H.; Liu, A.; Chen, Z.; Li, B.; Chen, J.; et al. The mRNA and miRNA profiles of goat bronchial epithelial cells stimulated by Pasteurella multocida strains of serotype A and D. PeerJ 2022, 10, e13047. [Google Scholar] [CrossRef]
- Garlanda, C.; Riva, F.; Bonavita, E.; Mantovani, A. Negative regulatory receptors of the IL-1 family. Semin. Immunol. 2013, 25, 408–415. [Google Scholar] [CrossRef]
- Colotta, F.; Re, F.; Muzio, M.; Bertini, R.; Polentarutti, N.; Sironi, M.; Giri, J.G.; Dower, S.K.; Sims, J.E.; Mantovani, A. Interleukin-1 type II receptor: A decoy target for IL-1 that is regulated by IL-4. Science 1993, 261, 472–475. [Google Scholar] [CrossRef]
- Re, F.; Sironi, M.; Muzio, M.; Matteucci, C.; Introna, M.; Orlando, S.; Penton-Rol, G.; Dower, S.K.; Sims, J.E.; Colotta, F.; et al. Inhibition of interleukin-1 responsiveness by type II receptor gene transfer: A surface “receptor” with anti-interleukin-1 function. J. Exp. Med. 1996, 183, 1841–1850. [Google Scholar] [CrossRef]
- Spriggs, M.K.; Nevens, P.J.; Grabstein, K.; Dower, S.K.; Cosman, D.; Armitage, R.J.; McMahan, C.J.; Sims, J.E. Molecular characterization of the interleukin-1 receptor (IL-1R) on monocytes and polymorphonuclear cells. Cytokine 1992, 4, 90–95. [Google Scholar] [CrossRef]
- Re, F.; Muzio, M.; de Rossi, M.; Polentarutti, N.; Giri, J.G.; Mantovani, A.; Colotta., F. The type II “receptor” as a decoy target for interleukin 1 in polymorphonuclear leukocytes: Characterization of induction by dexamethasone and ligand binding properties of the released decoy receptor. J. Exp. Med. 1994, 179, 739–743. [Google Scholar] [CrossRef]
- Colotta, F.; Re, F.; Muzio, M.; Polentarutti, N.; Minty, A.; Caput, D.; Ferrara, P.; Mantovani, A. Interleukin-13 induces expression and release of interleukin-1 decoy receptor in human polymorphonuclear cells. J. Biol. Chem. 1994, 269, 12403–12406. [Google Scholar] [CrossRef]
- Dawson, J.; Engelhardt, P.; Kastelic, T.; Cheneval, D.; MacKenzie, A.; Ramage, P. Effects of soluble interleukin-1 type II receptor on rabbit antigen-induced arthritis: Clinical, biochemical and histological assessment. Rheumatology 1999, 38, 401–406. [Google Scholar] [CrossRef]
- Simeoni, E.; Dudler, J.; Fleury, S.; Li, J.; Pagnotta, M.; Pascual, M.; von Segesser, L.K.; Vassalli, G. Gene transfer of a soluble IL-1 type 2 receptor-Ig fusion protein improves cardiac allograft survival in rats. Eur. J. Cardio Thorac. Surg. 2007, 31, 222–228. [Google Scholar] [CrossRef]
- Szot, P.; Franklin, A.; Figlewicz, D.P.; Beuca, T.P.; Bullock, K.; Hansen, K.; Banks, W.A.; Raskind, M.A.; Peskind, E.R. Multiple lipopolysaccharide (LPS) injections alter interleukin 6 (IL-6), IL-7, IL-10 and IL-6 and IL-7 receptor mRNA in CNS and spleen. Neuroscience 2017, 355, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Kastelein, R.A.; Hunter, C.A. IL-10 enhances NK cell proliferation, cytotoxicity and production of IFN-gamma when combined with IL-18. Eur. J. Immunol. 1999, 29, 2658–2665. [Google Scholar] [CrossRef]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Gallucci, S.; Lolkema, M.; Matzinger, P. Natural adjuvants: Endogenous activators of dendritic cells. Nat. Med. 1999, 5, 1249–1255. [Google Scholar] [CrossRef]
- Sauter, B.; Albert, M.L.; Francisco, L.; Larsson, M.; Somersan, S.; Bhardwaj, N. Consequences of cell death: Exposure to necrotic tumor cells, but not primary tissue cells or apoptotic cells, induces the maturation of immunostimulatory dendritic cells. J. Exp. Med. 2000, 191, 423–434. [Google Scholar] [CrossRef]
- Corinti, S.; Albanesi, C.; la Sala, A.; Pastore, S.; Girolomoni, G. Regulatory activity of autocrine IL-10 on dendritic cell functions. J. Immunol. 2001, 166, 4312–4318. [Google Scholar] [CrossRef]
- Sellati, T.J.; Sahay, B. Cells of Innate Immunity: Mechanisms of Activation. In Pathobiology of Human Disease: A Dynamic Encyclopedia of Disease Mechanisms; Elsevier: Amsterdam, The Netherlands, 2014; pp. 258–274. [Google Scholar] [CrossRef]
- Wajant, H.; Pfizenmaier, K.; Scheurich, P. Tumor necrosis factor signaling. Cell Death Differ. 2003, 10, 45–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalliolias, G.D.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Waqas, Y.; Yang, P.; Sun, X.; Liu, Y.; Ahmed, N.; Chen, B.; Li, Q.; Hu, L.; Huang, Y.; et al. Cytological study on the regulation of lymphocyte homing in the chicken spleen during LPS stimulation. Oncotarget 2017, 8, 7405–7419. [Google Scholar] [CrossRef]
- Scholten, D.J.; Canals, M.; Maussang, D.; Roumen, L.; Smit, M.J.; Wijtmans, M.; de Graaf, C.; Vischer, H.F.; Leurs, R. Pharmacological modulation of chemokine receptor function. Br. J. Pharmacol. 2012, 165, 1617–1643. [Google Scholar] [CrossRef]
- Kufareva, I.; Salanga, C.L.; Handel, T.M. Chemokine and chemokine receptor structure and interactions: Implications for therapeutic strategies. Immunol. Cell Biol. 2015, 93, 372–383. [Google Scholar] [CrossRef] [Green Version]


| Name of the Primers | Primer Sequences (5′-3′) | Product Length (bp) |
|---|---|---|
| Il-10-F | ACTTGGGTTGCCAAGCCTTA | 161 |
| Il-10-R | GAGAAATCGATGACAGCGCC | |
| Mlkl-F | AGATCCCATTTGAAGGCTGTGA | 174 |
| Mlkl-R | CTCATGGGCACGACACTCAT | |
| Sdhd-F | CCAAGCCACCACTCTGGTTC | 134 |
| Sdhd-R | GCAGCCAGAGAGTAGTCCAC | |
| Irf1-F | ACTCGAATGCGGATGAGACC | 93 |
| Irf1-R | CTGCTTTGTATCGGCTTTATTGA | |
| Cxcl10-F | CCACGTGTTGAGATCATTGCC | 184 |
| Cxcl10-R | GAGGCTCTCTGCTGTCCATC | |
| Cxcl13-F | CTCCAGGCCACGGTATTCTG | 118 |
| Cxcl13-R | CCAGGGGGCGTAACTTGAAT | |
| Ccr9-F | TGGAGGCTGGTCTGCATTATC | 95 |
| Ccr9-R | CATGCCAGGAATAAGGCTTGTG | |
| Cxcl2-F | CCCAGACAGAAGTCATAGCCAC | 162 |
| Cxcl2-R | TGGTTCTTCCGTTGAGGGAC |
| Animal Number | Weight (g) | Bacterial Loading (CFU × 106/g) | Average Bacterial Loading (CFU × 106/g) |
|---|---|---|---|
| Challenge group-1 | 0.10 | 4.60 ± 0.46 | 3.07 ± 1.09 |
| Challenge group-2 | 0.08 | 1.63 ± 0.54 | |
| Challenge group-3 | 0.06 | 3.30 ± 0.29 | |
| Challenge group-4 | 0.07 | 4.00 ± 1.94 | |
| Challenge group-5 | 0.10 | 1.80 ± 0.61 |
| Sample ID | Clean Reads | Clean Bases | GC (%) | Q30 (%) |
|---|---|---|---|---|
| C1-P | 23,759,030 | 7,104,206,622 | 49.81 | 93.93 |
| C2-P | 27,750,220 | 8,310,925,154 | 50.62 | 94.24 |
| C3-P | 23,587,851 | 7,057,533,332 | 49.86 | 94.24 |
| E1-P | 30,059,831 | 9,003,329,146 | 50.05 | 94.06 |
| E2-P | 25,039,488 | 7,491,410,612 | 50.01 | 93.92 |
| E3-P | 28,211,476 | 8,439,900,430 | 50.51 | 94.18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; He, M.; Cheng, Y.; Jiang, J.; Yang, W.; Zhang, Z.; An, Q.; Chen, S.; Man, C.; Du, L.; et al. Up-Regulation of Interleukin-10 in Splenic Immune Response Induced by Serotype A Pasteurellamultocida. Genes 2022, 13, 1586. https://doi.org/10.3390/genes13091586
Li H, He M, Cheng Y, Jiang J, Yang W, Zhang Z, An Q, Chen S, Man C, Du L, et al. Up-Regulation of Interleukin-10 in Splenic Immune Response Induced by Serotype A Pasteurellamultocida. Genes. 2022; 13(9):1586. https://doi.org/10.3390/genes13091586
Chicago/Turabian StyleLi, Haoyang, Meirong He, Yiwen Cheng, Junming Jiang, Weijie Yang, Zhenxing Zhang, Qi An, Si Chen, Churiga Man, Li Du, and et al. 2022. "Up-Regulation of Interleukin-10 in Splenic Immune Response Induced by Serotype A Pasteurellamultocida" Genes 13, no. 9: 1586. https://doi.org/10.3390/genes13091586






