Identification of Hypothalamic Long Noncoding RNAs Associated with Hypertension and the Behavior/Neurological Phenotype of Hypertensive ISIAH Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA-Seq Analysis
2.3. qPCR for Validation of the RNA-Seq Results
2.4. Functional Annotation of DEGs
2.5. Correlation Analysis
3. Results
3.1. LncRNAs Detected as Expressed by RNA-Seq Analysis
3.2. Correlation of LncRNAs’ and DEGs’ Expression Levels
3.3. Validation of the LncRNAs’ Differential Expression
3.4. Functional Analysis of the DEGs Whose Expression Correlates with the Expression of LncRNAs
3.5. Correlating Expression Levels between LncRNAs and DEGs Associated with the Behavior/Neurological Phenotype
3.6. Correlating Expression Levels between LncRNAs and DEGs Associated with Hypertension and Blood Pressure Regulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef]
- Kumar, M.M.; Goyal, R. LncRNA as a Therapeutic Target for Angiogenesis. Curr. Top. Med. Chem. 2017, 17, 1750–1757. [Google Scholar] [CrossRef]
- Charles Richard, J.L.; Eichhorn, P.J.A. Platforms for Investigating LncRNA Functions. SLAS Technol. 2018, 23, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, M.; Wang, D.Z. Non-Coding RNAs Including miRNAs and lncRNAs in Cardiovascular Biology and Disease. Cells 2014, 3, 883–898. [Google Scholar] [CrossRef]
- Huang, Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell Mol. Med. 2018, 22, 5768–5775. [Google Scholar] [CrossRef]
- Fang, Y.; Xu, Y.; Wang, R.; Hu, L.; Guo, D.; Xue, F.; Guo, W.; Zhang, D.; Hu, J.; Li, Y.; et al. Recent advances on the roles of LncRNAs in cardiovascular disease. J. Cell Mol. Med. 2020, 24, 12246–12257. [Google Scholar] [CrossRef]
- Shen, S.; Jiang, H.; Bei, Y.; Xiao, J.; Li, X. Long Non-Coding RNAs in Cardiac Remodeling. Cell Physiol. Biochem. 2017, 41, 1830–1837. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Zaccagnini, G.; Perfetti, A.; Fuschi, P.; Valaperta, R.; Voellenkle, C.; Castelvecchio, S.; Gaetano, C.; Finato, N.; Beltrami, A.P.; et al. Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 2016, 14, 183. [Google Scholar] [CrossRef]
- Liao, J.; He, Q.; Li, M.; Chen, Y.; Liu, Y.; Wang, J. LncRNA MIAT: Myocardial infarction associated and more. Gene 2016, 578, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, L.; Xu, H.; Liu, Y.; Yang, C.; Cowley, A.W., Jr.; Wang, N.; Liu, P.; Liang, M. Characteristics of long non-coding RNAs in the Brown Norway rat and alterations in the Dahl salt-sensitive rat. Sci. Rep. 2014, 4, 7146. [Google Scholar] [CrossRef]
- Gopalakrishnan, K.; Kumarasamy, S.; Mell, B.; Joe, B. Genome-wide identification of long noncoding RNAs in rat models of cardiovascular and renal disease. Hypertension 2015, 65, 200–210. [Google Scholar] [CrossRef]
- Hou, L.; Lin, Z.; Ni, Y.; Wu, Y.; Chen, D.; Song, L.; Huang, X.; Hu, H.; Yang, D. Microarray expression profiling and gene ontology analysis of long non-coding RNAs in spontaneously hypertensive rats and their potential roles in the pathogenesis of hypertension. Mol. Med. Rep. 2016, 13, 295–300. [Google Scholar] [CrossRef]
- Wu, G.; Jose, P.A.; Zeng, C. Noncoding RNAs in the Regulatory Network of Hypertension. Hypertension 2018, 72, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Leimena, C.; Qiu, H. Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension. Int. J. Mol. Sci. 2018, 19, 927. [Google Scholar] [CrossRef]
- Jusic, A.; Devaux, Y. Noncoding RNAs in Hypertension. Hypertension 2019, 74, 477–492. [Google Scholar] [CrossRef]
- Matsui, M.; Corey, D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017, 16, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Luo, T.; Dong, D.; Wu, X.; Wang, Y. Characterization of long non-coding RNAs to reveal potential prognostic biomarkers in hepatocellular carcinoma. Gene 2018, 663, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Deng, C. Identification of a novel four-lncRNA signature as a prognostic indicator in cirrhotic hepatocellular carcinoma. PeerJ 2019, 7, e7413. [Google Scholar] [CrossRef]
- Berthoud, H.R.; Munzberg, H. The lateral hypothalamus as integrator of metabolic and environmental needs: From electrical self-stimulation to opto-genetics. Physiol. Behav. 2011, 104, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Bains, J.S.; Wamsteeker Cusulin, J.I.; Inoue, W. Stress-related synaptic plasticity in the hypothalamus. Nat. Rev. Neurosci. 2015, 16, 377–388. [Google Scholar] [CrossRef]
- Goncharuk, V.D. The hypothalamus and its role in hypertension. Handb. Clin. Neurol. 2021, 182, 333–354. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- De Wardener, H.E. The hypothalamus and hypertension. Physiol. Rev. 2001, 81, 1599–1658. [Google Scholar] [CrossRef]
- Burdakov, D.; Luckman, S.M.; Verkhratsky, A. Glucose-sensing neurons of the hypothalamus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 2227–2235. [Google Scholar] [CrossRef]
- Ferri, S.L.; Flanagan-Cato, L.M. Oxytocin and dendrite remodeling in the hypothalamus. Horm. Behav. 2012, 61, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S. The hypothalamus in anxiety disorders. Handb. Clin. Neurol. 2021, 180, 149–160. [Google Scholar] [CrossRef]
- Markel, A.L.; Redina, O.E.; Gilinsky, M.A.; Dymshits, G.M.; Kalashnikova, E.V.; Khvorostova, Y.V.; Fedoseeva, L.A.; Jacobson, G.S. Neuroendocrine profiling in inherited stress-induced arterial hypertension rat strain with stress-sensitive arterial hypertension. J. Endocrinol. 2007, 195, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Markel, A.L.; Maslova, L.N.; Shishkina, G.T.; Bulygina, V.V.; Machanova, N.A.; Jacobson, G.S. Developmental influences on blood pressure regulation in ISIAH rats. In Development of the Hypertensive Phenotype: Basic and Clinical Studies; McCarty, R., Blizard, D.A., Chevalier, R.L., Birkenhager, W.H., Reid, J.L., Eds.; Handbook of Hypertension; Elsevier: Amsterdam, The Netherlands, 1999; Volume 19, pp. 493–526. [Google Scholar]
- Redina, O.E.; Smolenskaya, S.E.; Polityko, Y.K.; Ershov, N.I.; Gilinsky, M.A.; Markel, A.L. Hypothalamic Norepinephrine Concentration and Heart Mass in Hypertensive ISIAH Rats Are Associated with a Genetic Locus on Chromosome 18. J. Pers Med. 2021, 11, 67. [Google Scholar] [CrossRef]
- Tanaka, M.; Yoshida, M.; Emoto, H.; Ishii, H. Noradrenaline systems in the hypothalamus, amygdala and locus coeruleus are involved in the provocation of anxiety: Basic studies. Eur. J. Pharmacol. 2000, 405, 397–406. [Google Scholar] [CrossRef]
- Redina, O.E.; Smolenskaya, S.E.; Maslova, L.N.; Sakharov, D.G.; Markel, A.L. The characteristics of motor activity in ISIAH rats in an open field test are controlled by genes on chromosomes 2 and 16. Neurosci. Behav. Physiol. 2009, 39, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Meshkov, I.O.; Alekhina, T.A.; Moreva, T.A.; Markel, A.L. Behavioral characterictics of ISIAH rat strain. Zh. Vyssh. Nerv. Deiat. Im. I. P. Pavlova 2012, 62, 233–242. [Google Scholar]
- Klimov, L.O.; Ershov, N.I.; Efimov, V.M.; Markel, A.L.; Redina, O.E. Genome-wide transcriptome analysis of hypothalamus in rats with inherited stress-induced arterial hypertension. BMC Genet. 2016, 17 (Suppl. 1), 13. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Fedoseeva, L.A.; Ryazanova, M.A.; Ershov, N.I.; Markel, A.L.; Redina, O.E. Comparative transcriptional profiling of renal cortex in rats with inherited stress-induced arterial hypertension and normotensive Wistar Albino Glaxo rats. BMC Genet. 2016, 17 (Suppl. 1), 12. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Smith, J.R.; Hayman, G.T.; Wang, S.J.; Laulederkind, S.J.F.; Hoffman, M.J.; Kaldunski, M.L.; Tutaj, M.; Thota, J.; Nalabolu, H.S.; Ellanki, S.L.R.; et al. The Year of the Rat: The Rat Genome Database at 20: A multi-species knowledgebase and analysis platform. Nucleic Acids Res. 2020, 48, D731–D742. [Google Scholar] [CrossRef] [PubMed]
- Ravasi, T.; Suzuki, H.; Cannistraci, C.V.; Katayama, S.; Bajic, V.B.; Tan, K.; Akalin, A.; Schmeier, S.; Kanamori-Katayama, M.; Bertin, N.; et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 2010, 140, 744–752. [Google Scholar] [CrossRef]
- Polunin, D.; Shtaiger, I.; Efimov, V. JACOBI4 software for multivariate analysis of biological data. bioRxiv 2019, 803684. [Google Scholar] [CrossRef]
- Wang, H.; Iacoangeli, A.; Lin, D.; Williams, K.; Denman, R.B.; Hellen, C.U.; Tiedge, H. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 2005, 171, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Zong, W.; Gao, Q.; Chen, S.; Chen, L.; Zeng, G.; Huang, W.; Li, Z.; Zeng, C.; Xie, Y.; et al. The Expression Alteration of BC1 RNA and its Interaction with Eukaryotic Translation Initiation Factor eIF4A Post-Status Epilepticus. Neurochem. Res. 2018, 43, 1328–1338. [Google Scholar] [CrossRef]
- Lewejohann, L.; Skryabin, B.V.; Sachser, N.; Prehn, C.; Heiduschka, P.; Thanos, S.; Jordan, U.; Dell’Omo, G.; Vyssotski, A.L.; Pleskacheva, M.G.; et al. Role of a neuronal small non-messenger RNA: Behavioural alterations in BC1 RNA-deleted mice. Behav. Brain Res. 2004, 154, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fuscoe, J.C.; Zhao, C.; Guo, C.; Jia, M.; Qing, T.; Bannon, D.I.; Lancashire, L.; Bao, W.; Du, T.; et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014, 5, 3230. [Google Scholar] [CrossRef]
- Comprido, D. Regulation of Heterogeneous Nuclear Ribonucleoprotein K (hnRNP K) by BDNF: Changes in mRNA Binding as Determined by Microarray Analysis. Master’s Thesis, University of Coimbra, Coimbra, Portugal, 2011. [Google Scholar]
- Abdo, S.; Lo, C.S.; Chenier, I.; Shamsuyarova, A.; Filep, J.G.; Ingelfinger, J.R.; Zhang, S.L.; Chan, J.S. Heterogeneous nuclear ribonucleoproteins F and K mediate insulin inhibition of renal angiotensinogen gene expression and prevention of hypertension and kidney injury in diabetic mice. Diabetologia 2013, 56, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Sun, W.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, H. Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine-Related Cancers. Onco. Targets Ther. 2020, 13, 7699–7717. [Google Scholar] [CrossRef]
- Chu, Q.; Gu, X.; Zheng, Q.; Guo, Z.; Shan, D.; Wang, J.; Zhu, H. Long noncoding RNA SNHG4: A novel target in human diseases. Cancer Cell Int. 2021, 21, 583. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, W.C.; Liang, Z.D.; Yin, X.R.; Ji, Z.R.; Chen, X.H.; Wei, M.J.; Pei, L. LncRNA SNHG4 Attenuates Inflammatory Responses by Sponging miR-449c-5p and Up-Regulating STAT6 in Microglial During Cerebral Ischemia-Reperfusion Injury. Drug Des. Devel. Ther. 2020, 14, 3683–3695. [Google Scholar] [CrossRef]
- Li, H.; Wu, Y.; Suo, G.; Shen, F.; Zhen, Y.; Chen, X.; Lv, H. Profiling neuron-autonomous lncRNA changes upon ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2018, 495, 104–109. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Z.; Ren, S.; Li, K.; Ning, Q.; Jiang, X. Aberrant expression of long noncoding RNAs in the serum and myocardium of spontaneous hypertensive rats. Mol. Biol. Rep. 2019, 46, 6399–6404. [Google Scholar] [CrossRef]
- Jiang, X.Y.; Ning, Q.L. Expression profiling of long noncoding RNAs and the dynamic changes of lncRNA-NR024118 and Cdkn1c in angiotensin II-treated cardiac fibroblasts. Int. J. Clin. Exp. Pathol. 2014, 7, 1325–1336. [Google Scholar] [PubMed]
- Jiang, X.; Zhang, F.; Ning, Q. Losartan reverses the down-expression of long noncoding RNA-NR024118 and Cdkn1c induced by angiotensin II in adult rat cardiac fibroblasts. Pathol. Biol. 2015, 63, 122–125. [Google Scholar] [CrossRef]
- Yao, C.; Wang, Y.; Zhang, H.; Feng, W.; Wang, Q.; Shen, D.; Qian, T.; Liu, F.; Mao, S.; Gu, X.; et al. lncRNA TNXA-PS1 Modulates Schwann Cells by Functioning as a Competing Endogenous RNA Following Nerve Injury. J. Neurosci. 2018, 38, 6574–6585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Le, T.D.; Liu, L.; Li, J. Inferring and analyzing module-specific lncRNA-mRNA causal regulatory networks in human cancer. Brief. Bioinform. 2019, 20, 1403–1419. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.C.; Chiang, E.F.; Tong, S.K.; Lai, W.; Hsu, N.C.; Wang, L.C.; Chung, B.C. Regulation of steroidogenesis in transgenic mice and zebrafish. Mol. Cell Endocrinol. 2001, 171, 9–14. [Google Scholar] [CrossRef]
- Geerling, J.C.; Loewy, A.D. Aldosterone in the brain. Am. J. Physiol. Renal. Physiol. 2009, 297, F559–F576. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sanchez, E.P.; Ahmad, N.; Romero, D.G.; Gomez-Sanchez, C.E. Is aldosterone synthesized within the rat brain? Am. J. Physiol. Endocrinol. Metab. 2005, 288, E342–E346. [Google Scholar] [CrossRef]
- Gomez-Sanchez, E.P.; Gomez-Sanchez, C.M.; Plonczynski, M.; Gomez-Sanchez, C.E. Aldosterone synthesis in the brain contributes to Dahl salt-sensitive rat hypertension. Exp. Physiol. 2010, 95, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Oki, K.; Gomez-Sanchez, E.P.; Gomez-Sanchez, C.E. Role of mineralocorticoid action in the brain in salt-sensitive hypertension. Clin. Exp. Pharmacol. Physiol. 2012, 39, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Urade, Y.; Kimura, H.; Eguchi, N.; Nishikawa, A.; Hayaishi, O. Lipocalin-type prostaglandin D synthase (beta-trace) is a newly recognized type of retinoid transporter. J. Biol. Chem. 1997, 272, 15789–15795. [Google Scholar] [CrossRef] [PubMed]
- Tokudome, S.; Sano, M.; Shinmura, K.; Matsuhashi, T.; Morizane, S.; Moriyama, H.; Tamaki, K.; Hayashida, K.; Nakanishi, H.; Yoshikawa, N.; et al. Glucocorticoid protects rodent hearts from ischemia/reperfusion injury by activating lipocalin-type prostaglandin D synthase-derived PGD2 biosynthesis. J. Clin. Investig. 2009, 119, 1477–1488. [Google Scholar] [CrossRef]
- Joo, M.; Sadikot, R.T. PGD synthase and PGD2 in immune resposne. Mediators Inflamm. 2012, 2012, 503128. [Google Scholar] [CrossRef] [PubMed]
- Saleem, S.; Zhuang, H.; de Brum-Fernandes, A.J.; Maruyama, T.; Narumiya, S.; Dore, S. PGD(2) DP1 receptor protects brain from ischemia-reperfusion injury. Eur. J. Neurosci. 2007, 26, 73–78. [Google Scholar] [CrossRef]
- Ragolia, L.; Hall, C.E.; Palaia, T. Lipocalin-type prostaglandin D(2) synthase stimulates glucose transport via enhanced GLUT4 translocation. Prostaglandins Other Lipid Mediat. 2008, 87, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Marin-Mendez, J.J.; Patino-Garcia, A.; Segura, V.; Ortuno, F.; Galvez, M.D.; Soutullo, C.A. Differential expression of prostaglandin D2 synthase (PTGDS) in patients with attention deficit-hyperactivity disorder and bipolar disorder. J. Affect. Disord. 2012, 138, 479–484. [Google Scholar] [CrossRef]
- Ouhaddi, Y.; Najar, M.; Pare, F.; Lussier, B.; Urade, Y.; Benderdour, M.; Pelletier, J.P.; Martel-Pelletier, J.; Fahmi, H. L-PGDS deficiency accelerated the development of naturally occurring age-related osteoarthritis. Aging 2020, 12, 24778–24797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Srivastava, A.; Palaia, T.; Hall, C.; Lee, J.; Stevenson, M.; Zhao, C.L.; Ragolia, L. Lipocalin-type prostaglandin D2 synthase deletion induces dyslipidemia and non-alcoholic fatty liver disease. Prostaglandins Other Lipid Mediat. 2020, 149, 106429. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Yun, S.J.; Ha, J.M.; Jin, S.Y.; Ha, H.K.; Song, S.H.; Kim, C.D.; Bae, S.S. Prostaglandin D2 stimulates phenotypic changes in vascular smooth muscle cells. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Spielewoy, C.; Roubert, C.; Hamon, M.; Nosten-Bertrand, M.; Betancur, C.; Giros, B. Behavioural disturbances associated with hyperdopaminergia in dopamine-transporter knockout mice. Behav. Pharmacol. 2000, 11, 279–290. [Google Scholar] [CrossRef]
- Cui, C.; Booker, T.K.; Allen, R.S.; Grady, S.R.; Whiteaker, P.; Marks, M.J.; Salminen, O.; Tritto, T.; Butt, C.M.; Allen, W.R.; et al. The beta3 nicotinic receptor subunit: A component of alpha-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J. Neurosci. 2003, 23, 11045–11053. [Google Scholar] [CrossRef] [PubMed]
- Booker, T.K.; Butt, C.M.; Wehner, J.M.; Heinemann, S.F.; Collins, A.C. Decreased anxiety-like behavior in beta3 nicotinic receptor subunit knockout mice. Pharmacol. Biochem. Behav. 2007, 87, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Durand, D.; Pampillo, M.; Caruso, C.; Lasaga, M. Role of metabotropic glutamate receptors in the control of neuroendocrine function. Neuropharmacology 2008, 55, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Lane, T.A.; Boerner, T.; Bannerman, D.M.; Kew, J.N.; Tunbridge, E.M.; Sharp, T.; Harrison, P.J. Decreased striatal dopamine in group II metabotropic glutamate receptor (mGlu2/mGlu3) double knockout mice. BMC Neurosci. 2013, 14, 102. [Google Scholar] [CrossRef] [PubMed]
- Highland, J.N.; Zanos, P.; Georgiou, P.; Gould, T.D. Group II metabotropic glutamate receptor blockade promotes stress resilience in mice. Neuropsychopharmacology 2019, 44, 1788–1796. [Google Scholar] [CrossRef]
- Zarrindast, M.R.; Khakpai, F. The Modulatory Role of Dopamine in Anxiety-like Behavior. Arch. Iran. Med. 2015, 18, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Juarez Olguin, H.; Calderon Guzman, D.; Hernandez Garcia, E.; Barragan Mejia, G. The Role of Dopamine and Its Dysfunction as a Consequence of Oxidative Stress. Oxid Med. Cell Longev. 2016, 2016, 9730467. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Fujita, M.; Ito, Y.; Okada, T.; Kusuoka, H.; Nishimura, T. Brain dopamine transporter in spontaneously hypertensive rats. J. Nucl. Med. 1997, 38, 470–474. [Google Scholar]
- Viggiano, D.; Vallone, D.; Sadile, A. Dysfunctions in dopamine systems and ADHD: Evidence from animals and modeling. Neural. Plast. 2004, 11, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.T.; Zhen, J.; Reith, M.E. Importance of cholesterol in dopamine transporter function. J. Neurochem. 2012, 123, 700–715. [Google Scholar] [CrossRef]
- Threlfell, S.; Mohammadi, A.S.; Ryan, B.J.; Connor-Robson, N.; Platt, N.J.; Anand, R.; Serres, F.; Sharp, T.; Bengoa-Vergniory, N.; Wade-Martins, R.; et al. Striatal Dopamine Transporter Function Is Facilitated by Converging Biology of alpha-Synuclein and Cholesterol. Front. Cell Neurosci. 2021, 15, 658244. [Google Scholar] [CrossRef] [PubMed]
- Wheatcroft, S.B.; Kearney, M.T.; Shah, A.M.; Ezzat, V.A.; Miell, J.R.; Modo, M.; Williams, S.C.; Cawthorn, W.P.; Medina-Gomez, G.; Vidal-Puig, A.; et al. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes 2007, 56, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Boughanem, H.; Yubero-Serrano, E.M.; Lopez-Miranda, J.; Tinahones, F.J.; Macias-Gonzalez, M. Potential Role of Insulin Growth-Factor-Binding Protein 2 as Therapeutic Target for Obesity-Related Insulin Resistance. Int. J. Mol. Sci. 2021, 22, 1133. [Google Scholar] [CrossRef]
- Liu, Y.; Song, C.; Shen, F.; Zhang, J.; Song, S.W. IGFBP2 promotes immunosuppression associated with its mesenchymal induction and FcgammaRIIB phosphorylation in glioblastoma. PLoS ONE 2019, 14, e0222999. [Google Scholar] [CrossRef]
- Khan, S. IGFBP-2 Signaling in the Brain: From Brain Development to Higher Order Brain Functions. Front. Endocrinol. 2019, 10, 822. [Google Scholar] [CrossRef] [PubMed]
- Bhat, P.V.; Manolescu, D.C. Role of vitamin A in determining nephron mass and possible relationship to hypertension. J. Nutr. 2008, 138, 1407–1410. [Google Scholar] [CrossRef]
- Guo, X.; Li, Z.; Zhou, Y.; Yu, S.; Yang, H.; Zheng, L.; Liu, Y.; Sun, Y. Metabolic Profile for Prediction of Ischemic Stroke in Chinese Hypertensive Population. J. Stroke Cerebrovasc. Dis. 2019, 28, 1062–1069. [Google Scholar] [CrossRef]
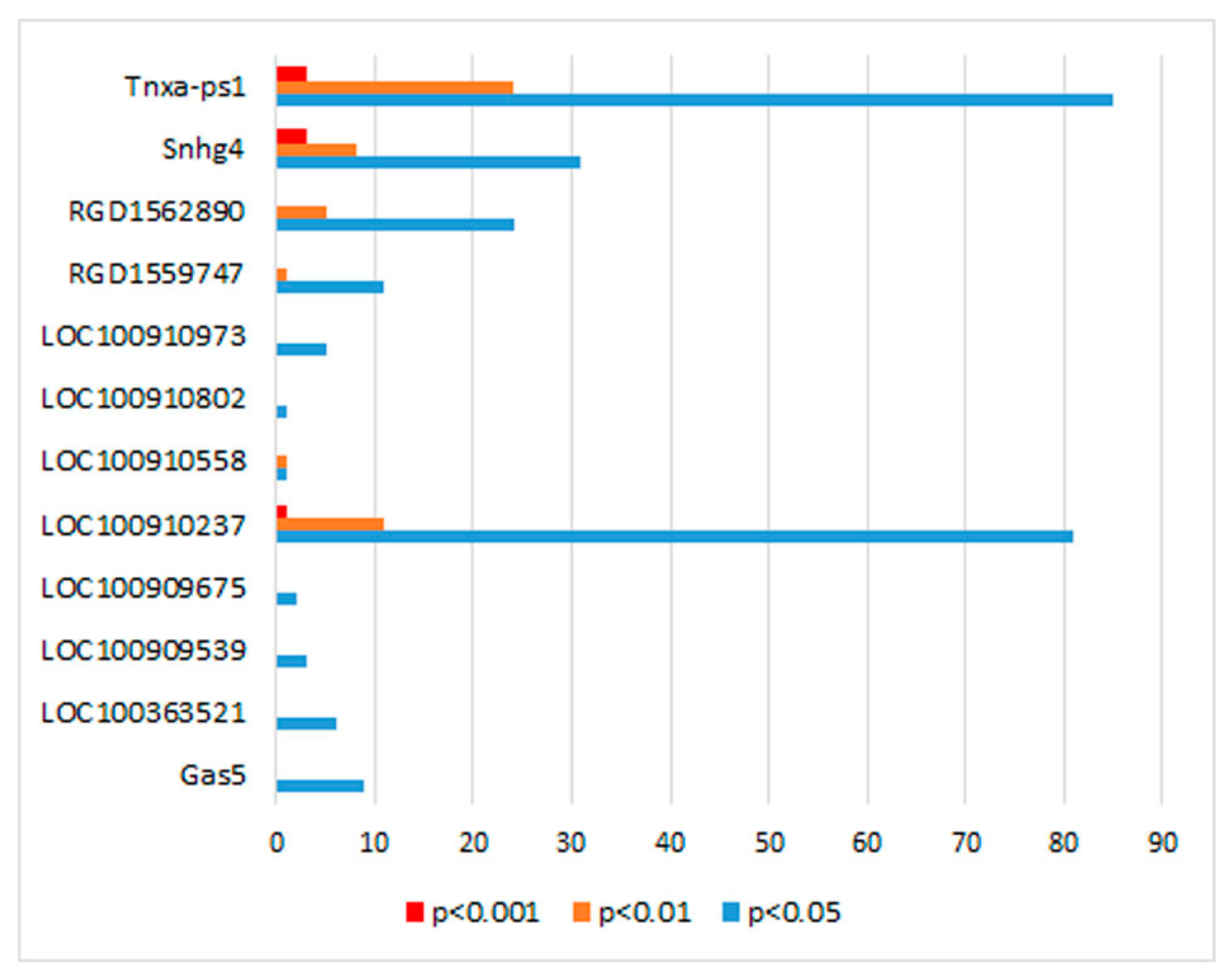
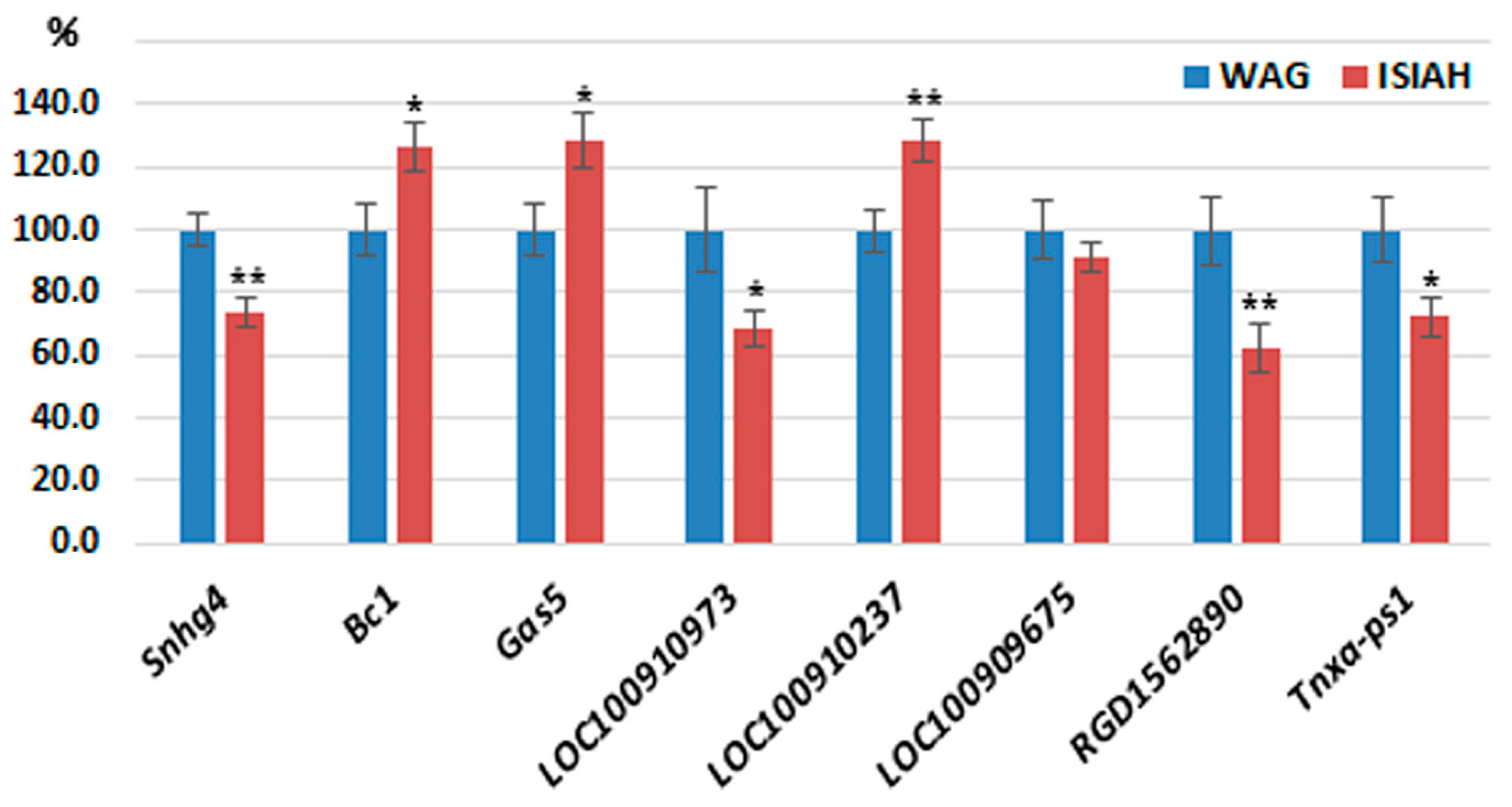


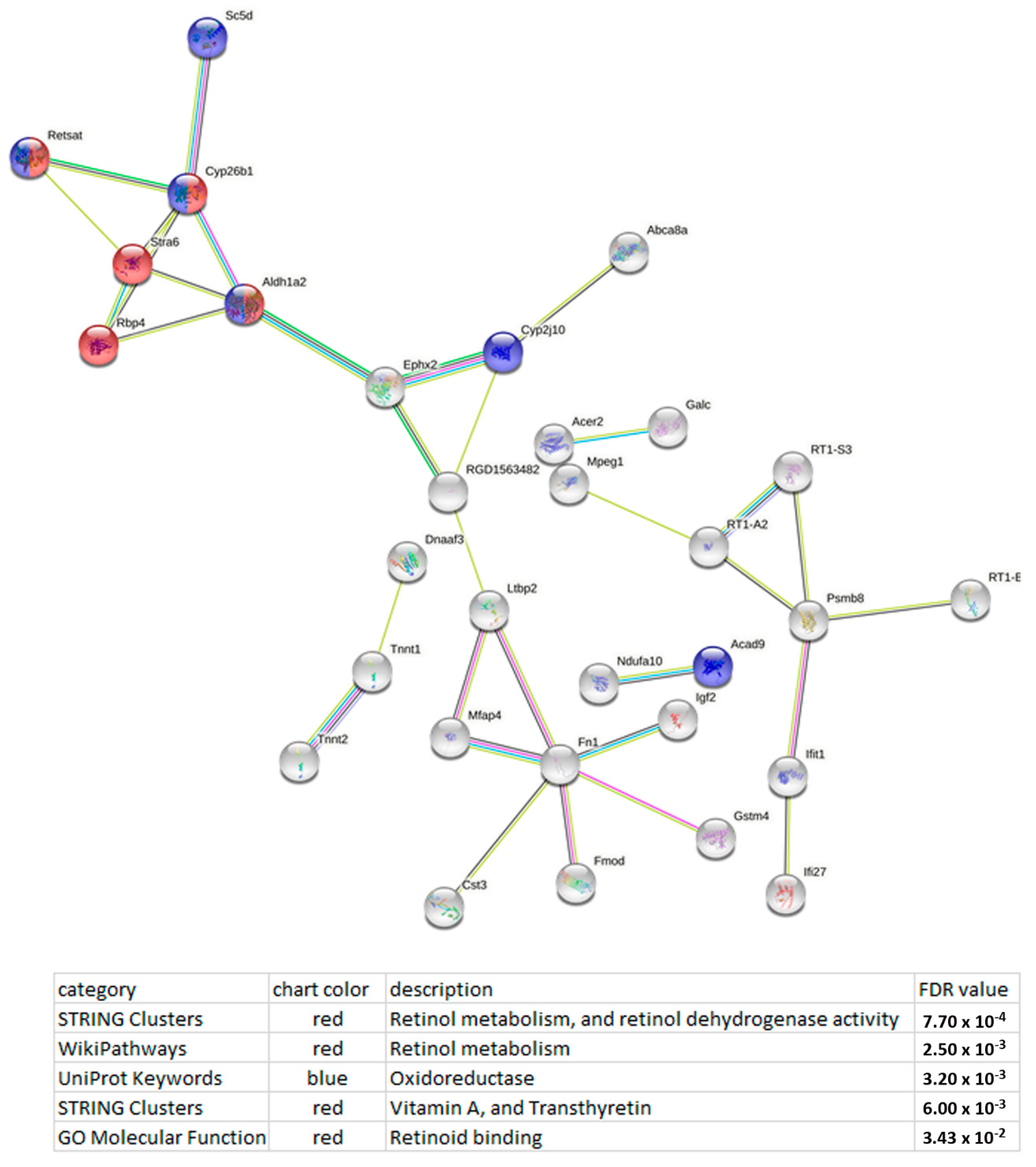
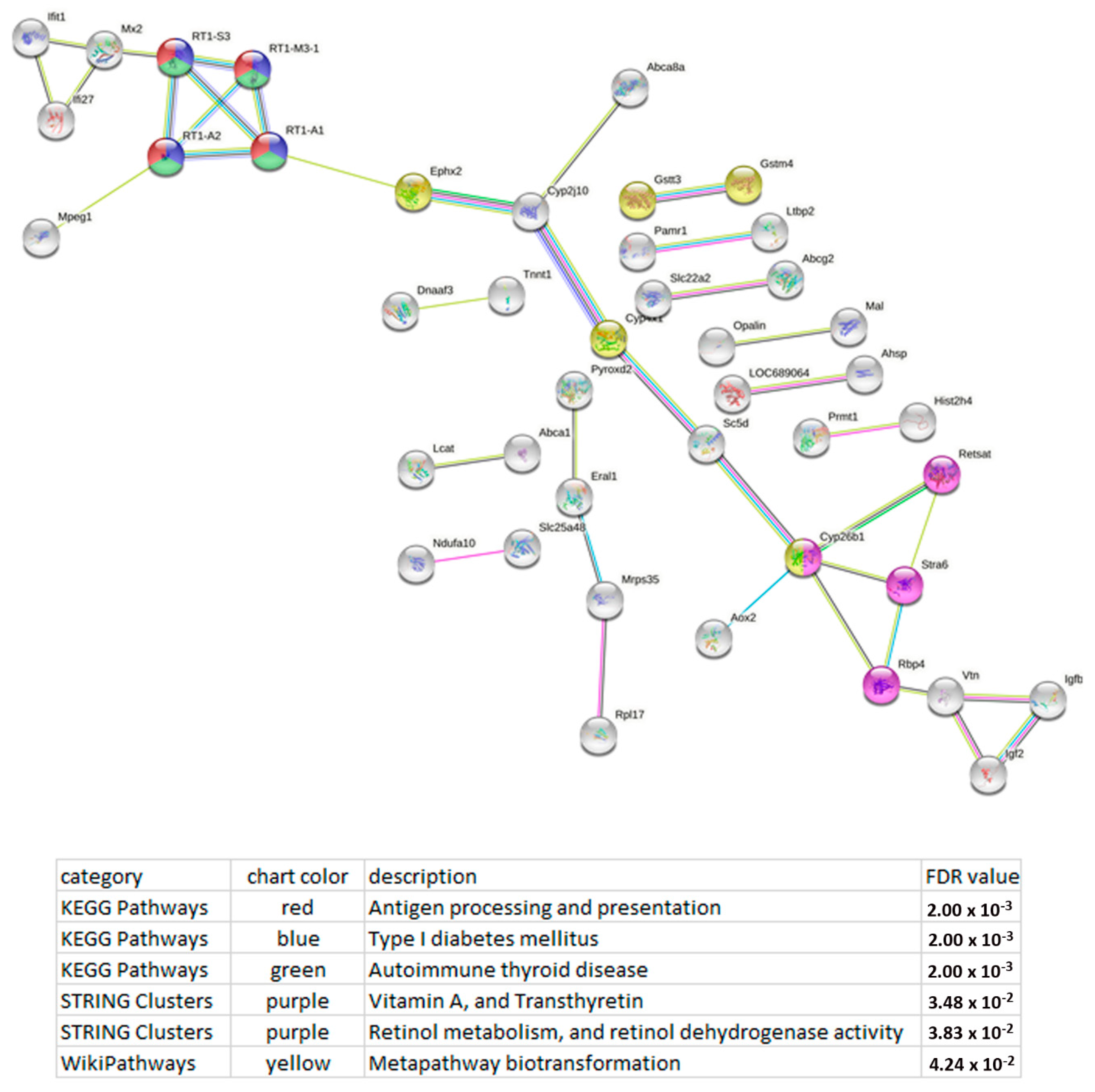
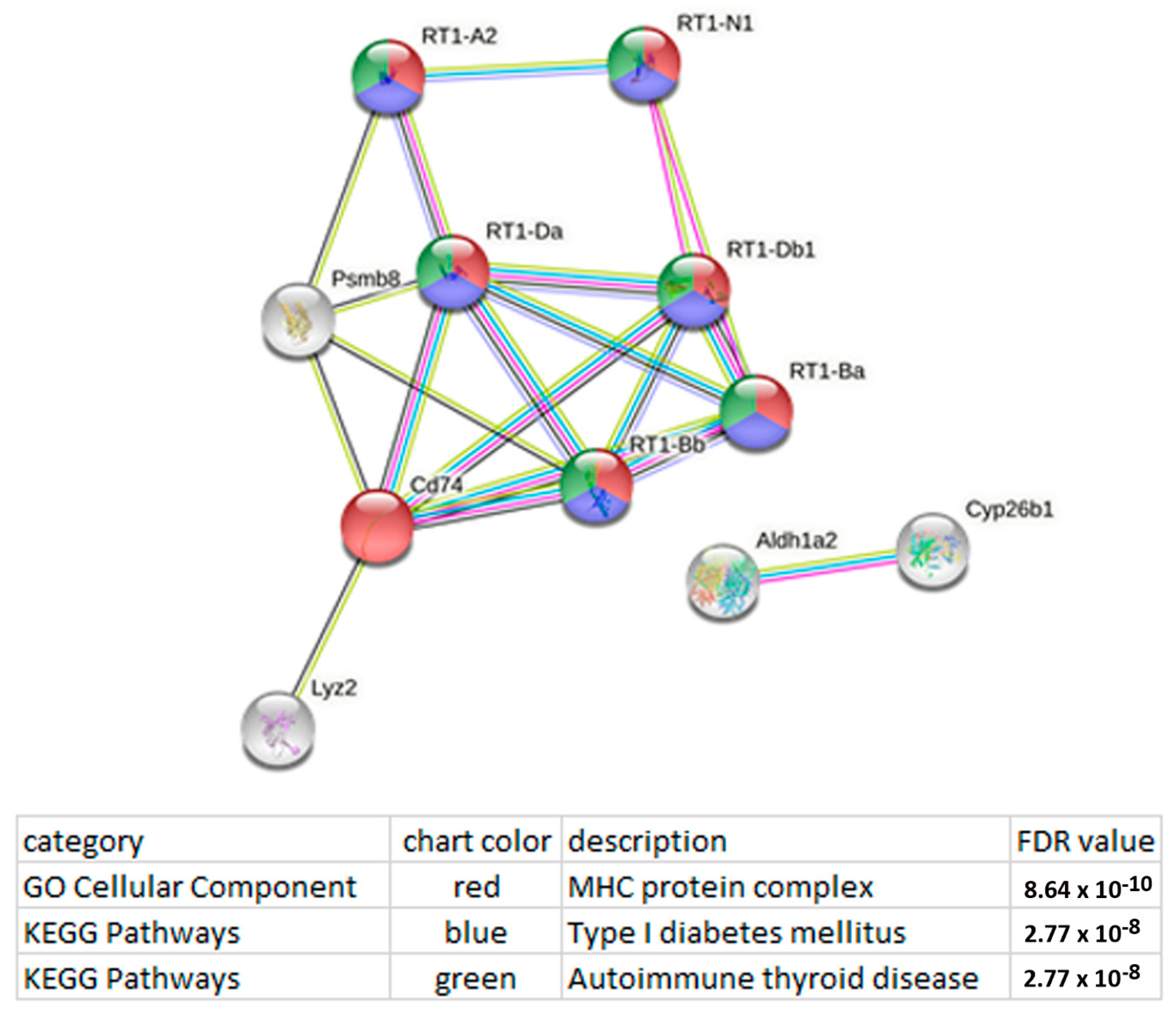
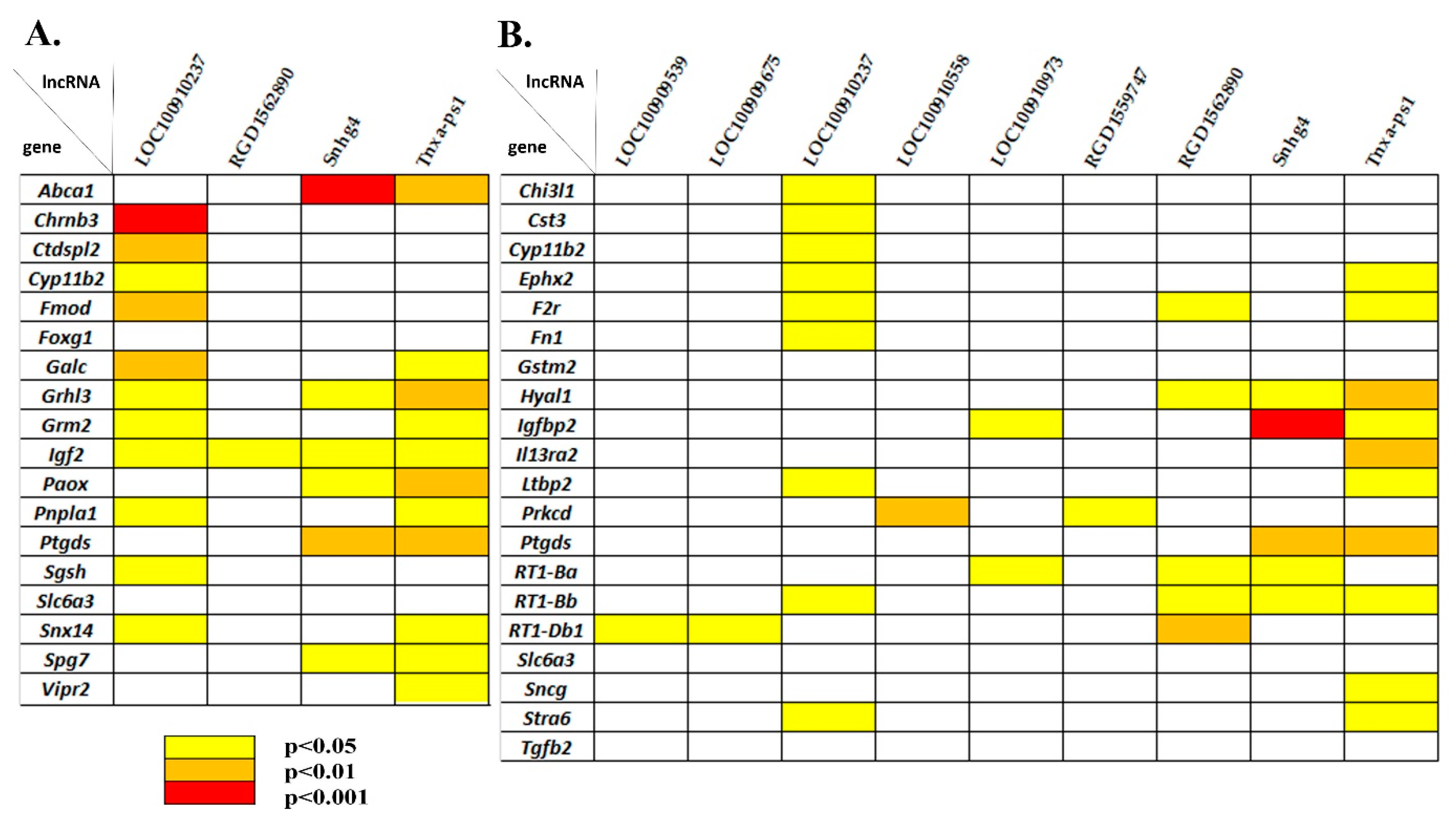
| Gene ID | LncRNA | Chr.:Megabases | Expression Level in | log2 (Fold Change) of ISIAH/WAG | p-Value | |
|---|---|---|---|---|---|---|
| ISIAH, RPKM | WAG, RPKM | |||||
| 100568448 | Snhg4 * | 18:28.07 | 13.7 | 40.6 | −1.56 | 0.0057 |
| 29294 | Bc1 | 1:224.94 | 127,304.0 | 91,730.1 | 0.47 | 0.0128 |
| 81714 | Gas5 * | 13:83.75 | 15.3 | 21.4 | −0.49 | 0.0409 |
| 100910973 | LOC100910973 * | 1:234.18 | 24.5 | 30.9 | −0.34 | 0.0676 |
| 100910237 | LOC100910237 * | 1:24.23 | 6.1 | 4.1 | 0.59 | 0.0701 |
| 100909675 | LOC100909675 * | 4:7.83 | 1.5 | 1.2 | 0.29 | 0.3363 |
| 100363521 | LOC100363521 * | 4:43.75 | 31.7 | 34.9 | −0.14 | 0.4049 |
| 294945 | RGD1562890 * | 2:124.12 | 2.8 | 3.6 | −0.37 | 0.4079 |
| 100910802 | LOC100910802 * | 12:24.79 | 1.9 | 1.5 | 0.32 | 0.4600 |
| 29536 | Rmrp | 15:31.57 | 20.0 | 16.4 | 0.28 | 0.4703 |
| 312310 | RGD1559747 * | 4:142.93 | 7.6 | 7.0 | 0.12 | 0.4978 |
| 100909539 | LOC100909539 * | 2:105.05 | 12.2 | 13.7 | −0.16 | 0.5426 |
| 25602 | Tnxa-ps1 * | 20:6.40 | 8.1 | 4.5 | 0.83 | 0.7158 |
| 680254 | LOC680254 | 13:79.98 | 5.7 | 5.4 | 0.08 | 0.7843 |
| 309109 | Tmem80 | 1:221.20 | 17.2 | 16.6 | 0.06 | 0.7931 |
| 100910558 | LOC100910558 * | 5:104.68 | 2.2 | 2.1 | 0.03 | 0.8448 |
| 29206 | Ybx1-ps3 | 10:40.22 | 23.4 | 23.8 | −0.03 | 0.8541 |
| 500568 | RGD1564482 | 5:158.41 | 1.3 | 1.4 | −0.15 | 0.8674 |
| 259227 | Vof16 | 8:44.27 | 3.0 | 3.1 | −0.04 | 0.8873 |
| 360865 | Prrc2c | 13:85.44 | 29.5 | 29.1 | 0.02 | 0.8921 |
| 686120 | LOC686120 | 12:26.72 | 1.4 | 1.4 | 0.01 | 0.9253 |
| 100911177 | LOC100911177 | 3:169.95 | 89.9 | 89.7 | 0.00 | 0.9913 |
| Gene | Chr.:Megabases | Expression Level in | log2 (Fold Change) of ISIAH/WAG | q-Value | Definition | |
|---|---|---|---|---|---|---|
| WAG, RPKM | ISIAH, RPKM | |||||
| Abca1∆ | 5:74.0 | 7.47 | 11.71 | 0.6 | 5.94 × 10−3 | ATP-binding cassette subfamily A member 1 |
| Chrnb3∆¶ | 16:68.5 | 3.12 | 1.31 | −1.2 | 5.94 × 10−3 | cholinergic receptor nicotinic beta 3 subunit |
| Ctdspl2 | 3:120.5 | 4.97 | 7.81 | 0.7 | 2.47 × 10−2 | CTD small phosphatase-like 2 |
| Cyp11b2¥ | 7:116.2 | 0.90 | 2.07 | 1.2 | 1.52 × 10−2 | cytochrome P450, family 11, subfamily b, polypeptide 2 |
| Fmod∆ | 13: 55.9 | 22.13 | 12.70 | −0.8 | 5.94 × 10−3 | fibromodulin |
| Foxg1∆¶€ | 6:79.5 | 5.45 | 2.94 | −0.9 | 5.94 × 10−3 | forkhead box G1 |
| Galc∆¶ | 6:131.4 | 10.27 | 6.30 | −0.7 | 1.52 × 10−2 | galactosylceramidase |
| Grhl3 *∆€ | 5:157.7 | 1.05 | 2.68 | 1.4 | 5.94 × 10−3 | grainyhead-like transcription factor 3 |
| Grm2©∆¶ | 8:114.7 | 10.93 | 2.73 | −2.0 | 5.94 × 10−3 | glutamate metabotropic receptor 2 |
| Igf2∆ | 1:222.7 | 201.43 | 94.08 | −1.1 | 5.94 × 10−3 | insulin-like growth factor 2 |
| Paox∆¶ | 1:219.5 | 7.27 | 13.01 | 0.8 | 5.94 × 10−3 | polyamine oxidase |
| Pnpla1∆ | 20:8.4 | 7.33 | 1.12 | −2.7 | 5.94 × 10−3 | patatin-like phospholipase domain-containing 1 |
| Ptgds¥ | 3:2.7 | 3548.34 | 2356.79 | −0.6 | 1.09 × 10−2 | prostaglandin D2 synthase |
| Sgsh | 10:108.1 | 2.49 | 4.90 | 1.0 | 1.99 × 10−2 | N-sulfoglucosamine sulfohydrolase |
| Slc6a3 *©§#∆¤¶¥ | 1:33.7 | 6.86 | 2.65 | −1.4 | 5.94 × 10−3 | solute carrier family 6 member 3 |
| Snx14©§# | 8:95.5 | 21.12 | 11.94 | −0.8 | 5.94 × 10−3 | sorting nexin 14 |
| Spg7∆ | 19:66.6 | 17.80 | 26.18 | 0.6 | 2.83 × 10−2 | SPG7 matrix AAA peptidase subunit, paraplegin |
| Vipr2∆ | 6:152.9 | 1.68 | 2.92 | 0.8 | 4.38 × 10−2 | vasoactive intestinal peptide receptor 2 |
| Gene | Chr.:Megabases | Expression Level in | log2 (Fold Change) of ISIAH/WAG | q-Value | Definition | |
|---|---|---|---|---|---|---|
| WAG, RPKM | ISIAH, RPKM | |||||
| Chi3l1 | 13:56.1 | 48.06 | 19.85 | −1.3 | 5.94 × 10−3 | chitinase 3-like 1 |
| Cst3 | 3:149.6 | 3929.44 | 2609.54 | −0.6 | 1.09 × 10−2 | cystatin C |
| Cyp11b2 * | 7:116.2 | 0.90 | 2.07 | 1.2 | 1.52 × 10−2 | cytochrome P450, family 11, subfamily b, polypeptide 2 |
| Ephx2 | 15:48.8 | 0.63 | 13.56 | 4.4 | 5.94 × 10−3 | epoxide hydrolase 2 |
| F2r | 2:45.3 | 12.80 | 18.83 | 0.6 | 3.53 × 10−2 | coagulation factor II (thrombin) receptor |
| Fn1 | 9:78.7 | 24.70 | 12.54 | −1.0 | 5.94 × 10−3 | fibronectin 1 |
| Gstm2 | 2:230.2 | 30.69 | 18.40 | −0.7 | 1.09 × 10−2 | glutathione S-transferase mu 2 |
| Hyal1 | 8:115.7 | 8.44 | 13.64 | 0.7 | 3.79 × 10−2 | hyaluronidase 1 |
| Igfbp2 | 9:79.9 | 95.58 | 49.64 | −0.9 | 5.94 × 10−3 | insulin-like growth factor-binding protein 2 |
| Il13ra2 | X:118.6 | 0.56 | 1.49 | 1.4 | 2.83 × 10−2 | interleukin 13 receptor subunit alpha 2 |
| Ltbp2 | 6:116.9 | 2.65 | 0.36 | −2.9 | 5.94 × 10−3 | latent transforming growth factor beta-binding protein 2 |
| Prkcd | 16:6.6 | 9.96 | 15.32 | 0.6 | 4.98 × 10−2 | protein kinase C, delta |
| Ptgds * | 3:2.7 | 3548.34 | 2356.79 | −0.6 | 1.09 × 10−2 | prostaglandin D2 synthase |
| RT1-Ba | 20:6.1 | 9.75 | 5.48 | −0.8 | 4.72 × 10−2 | RT1 class II, locus Ba |
| RT1-Bb | 20:6.1 | 6.56 | 1.69 | −2.0 | 5.94 × 10−3 | RT1 class II, locus Bb |
| RT1-Db1 | 20:6.2 | 11.30 | 6.65 | −0.8 | 5.94 × 10−3 | RT1 class II, locus Db1 |
| Slc6a3 * | 1:33.7 | 6.86 | 2.65 | −1.4 | 5.94 × 10−3 | solute carrier family 6 member 3 |
| Sncg | 16:9.1 | 64.11 | 35.35 | −0.9 | 5.94 × 10−3 | synuclein, gamma |
| Stra6 | 8:62.7 | 8.73 | 14.19 | 0.7 | 5.94 × 10−3 | signaling receptor and transporter of retinol STRA6 |
| Tgfb2 | 13:109.7 | 6.73 | 12.03 | 0.8 | 5.94 × 10−3 | transforming growth factor beta 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedoseeva, L.A.; Ershov, N.I.; Sidorenko, I.A.; Markel, A.L.; Redina, O.E. Identification of Hypothalamic Long Noncoding RNAs Associated with Hypertension and the Behavior/Neurological Phenotype of Hypertensive ISIAH Rats. Genes 2022, 13, 1598. https://doi.org/10.3390/genes13091598
Fedoseeva LA, Ershov NI, Sidorenko IA, Markel AL, Redina OE. Identification of Hypothalamic Long Noncoding RNAs Associated with Hypertension and the Behavior/Neurological Phenotype of Hypertensive ISIAH Rats. Genes. 2022; 13(9):1598. https://doi.org/10.3390/genes13091598
Chicago/Turabian StyleFedoseeva, Larisa A., Nikita I. Ershov, Ivan A. Sidorenko, Arcady L. Markel, and Olga E. Redina. 2022. "Identification of Hypothalamic Long Noncoding RNAs Associated with Hypertension and the Behavior/Neurological Phenotype of Hypertensive ISIAH Rats" Genes 13, no. 9: 1598. https://doi.org/10.3390/genes13091598
APA StyleFedoseeva, L. A., Ershov, N. I., Sidorenko, I. A., Markel, A. L., & Redina, O. E. (2022). Identification of Hypothalamic Long Noncoding RNAs Associated with Hypertension and the Behavior/Neurological Phenotype of Hypertensive ISIAH Rats. Genes, 13(9), 1598. https://doi.org/10.3390/genes13091598






