Endometrial Cancer-Adjacent Tissues Express Higher Levels of Cancer-Promoting Genes than the Matched Tumors
Abstract
:1. Introduction
2. Materials and methods
2.1. Patients and Samples
2.2. RNA Isolation, Reverse Transcription and Quantitative PCR
2.3. Statistical Analysis
3. Results
3.1. Gene Expression in Tumor-Adjacent Tissues and Tumors in Patients with EC and in Endometrial Tissue in Cancer-Free Patients with Leiomyoma
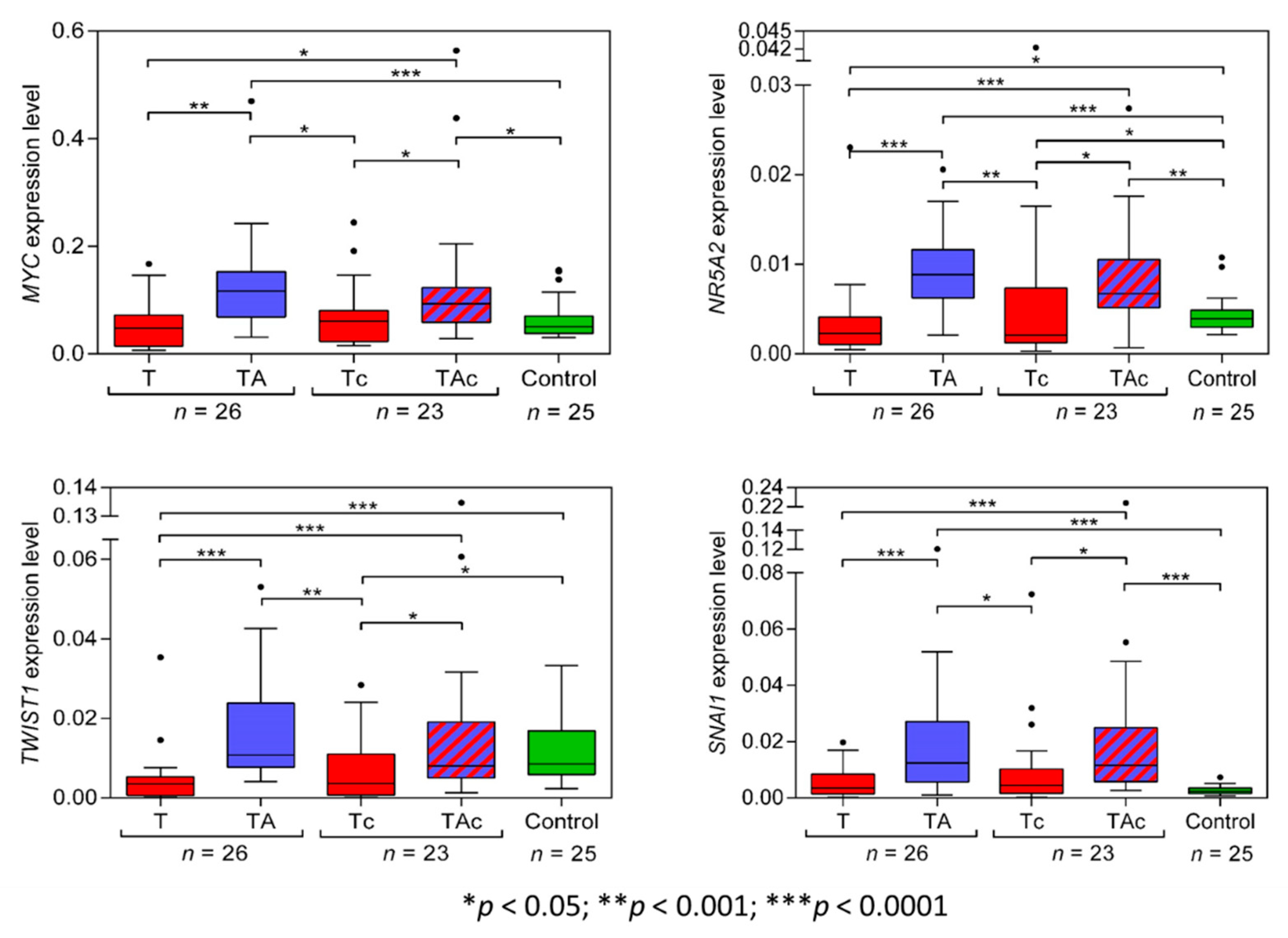
3.1.1. MYC
3.1.2. NR5A2
3.1.3. TWIST1
3.1.4. SNAI1
3.1.5. STK11(LKB1)
3.1.6. CXCR2
3.1.7. HMGA2
3.1.8. LIN28A
3.1.9. POU5F1 Isoform A (OCT4A)
3.1.10. POU5F1 isoform B (OCT4B)
3.1.11. POU5F1 isoform B1 (OCT4B1)
3.1.12. miR-205-5p
3.2. Gene Expression in Relation to Histological Results in Tumor-Adjacent Tissues
3.3. Relationships of Gene Expression with Clinical Data
3.4. Expression Levels of the Analyzed Transcripts in EC Cell Lines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yan, P.S.; Venkataramu, C.; Ibrahim, A.; Liu, J.C.; Shen, R.Z.; Diaz, N.M.; Centeno, B.; Weber, F.; Leu, Y.W.; Shapiro, C.L.; et al. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin. Cancer Res. 2006, 12, 6626–6636. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Gao, Y.; Jones, A.; Ruebner, M.; Beckmann, M.W.; Wachter, D.L.; Fasching, P.A.; Widschwendter, M. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat. Commun. 2016, 7, 10478. [Google Scholar] [CrossRef] [PubMed]
- Reed, M.A.C.; Singhal, R.; Ludwig, C.; Carrigan, J.B.; Ward, D.G.; Taniere, P.; Alderson, D.; Günther, U.L. Metabolomic Evidence for a Field Effect in Histologically Normal and Metaplastic Tissues in Patients with Esophageal Adenocarcinoma. Neoplasia 2017, 19, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Chandran, U.R.; Dhir, R.; Ma, C.; Michalopoulos, G.; Becich, M.; Gilbertson, J. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer 2005, 5, 45. [Google Scholar] [CrossRef]
- Sanz-Pamplona, R.; Berenguer, A.; Cordero, D.; Mollevi, D.G.; Crous-Bou, M.; Sole, X.; Paré-Brunet, L.; Guino, E.; Salazar, R.; Santos, C.; et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol. Cancer 2014, 13, 46. [Google Scholar] [CrossRef]
- Abdalla, M.; Tran-Thanh, D.; Moreno, J.; Iakovlev, V.; Nair, R.; Kanwar, N.; Abdalla, M.; Lee, J.P.Y.; Kwan, J.Y.Y.; Cawthorn, T.R.; et al. Mapping genomic and transcriptomic alterations spatially in epithelial cells adjacent to human breast carcinoma. Nat. Commun. 2017, 8, 1245. [Google Scholar] [CrossRef]
- Casbas-Hernandez, P.; Sun, X.; Roman-Perez, E.; D’Arcy, M.; Sandhu, R.; Hishida, A.; McNaughton, K.K.; Yang, X.R.; Makowski, L.; Sherman, M.E.; et al. Tumor intrinsic subtype is reflected in cancer-adjacent tissue. Cancer Epidemiol. Biomark. Prev. 2015, 24, 406–414. [Google Scholar] [CrossRef]
- Pineda, A.L.; Ogoe, H.A.; Balasubramanian, J.B.; Rangel Escareno, C.; Visweswaran, S.; Herman, J.G.; Gopalakrishnan, V. On Predicting lung cancer subtypes using ‘omic’ data from tumor and tumor-adjacent histologically-normal tissue. BMC Cancer 2016, 16, 184. [Google Scholar] [CrossRef]
- Raudenska, M.; Sztalmachova, M.; Gumulec, J.; Fojtu, M.; Polanska, H.; Balvan, J.; Feith, M.; Binkova, H.; Horakova, Z.; Kostrica, R.; et al. Prognostic significance of the tumour-adjacent tissue in head and neck cancers. Tumour Biol. 2015, 36, 9929–9939. [Google Scholar] [CrossRef]
- Roman-Perez, E.; Casbas-Hernandez, P.; Pirone, J.R.; Rein, J.; Carey, L.A.; Lubet, R.A.; Mani, S.A.; Amos, K.D.; Troester, M.A. Gene expression in extratumoral microenvironment predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012, 14, R51. [Google Scholar] [CrossRef]
- Grinchuk, O.V.; Yenamandra, S.P.; Iyer, R.; Singh, M.; Lee, H.K.; Lim, K.H.; Chow, P.K.; Kuznetsov, V.A. Tumor-Adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol. Oncol. 2018, 12, 89–113. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Villanueva, A.; Kobayashi, M.; Peix, J.; Chiang, D.Y.; Camargo, A.; Gupta, S.; Moore, J.; Wrobel, M.J.; Lerner, J.; et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 1995–2004. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Stern, D.F.; Zhao, H. Transcriptional Profiles from Paired Normal Samples Offer Complementary Information on Cancer Patient Survival--Evidence from TCGA Pan-Cancer Data. Sci. Rep. 2016, 6, 20567. [Google Scholar] [CrossRef] [PubMed]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Bai, M.K.; Costopoulos, J.S.; Christoforidou, B.P.; Papadimitriou, C.S. Immunohistochemical detection of the c-myc oncogene product in normal, hyperplastic and carcinomatous endometrium. Oncology 1994, 51, 314–319. [Google Scholar] [CrossRef]
- Geisler, J.P.; Geisler, H.E.; Manahan, K.J.; Miller, G.A.; Wiemann, M.C.; Zhou, Z.; Crabtree, W. Nuclear and cytoplasmic c-myc staining in endometrial carcinoma and their relationship to survival. Int. J. Gynecol. Cancer 2004, 14, 133–137. [Google Scholar] [CrossRef]
- Meinsohn, M.C.; Smith, O.E.; Bertolin, K.; Murphy, B.D. The Orphan Nuclear Receptors Steroidogenic Factor-1 and Liver Receptor Homolog-1: Structure, Regulation, and Essential Roles in Mammalian Reproduction. Physiol. Rev. 2019, 99, 1249–1279. [Google Scholar] [CrossRef]
- Michalek, S.; Brunner, T. Nuclear-Mitochondrial crosstalk: On the role of the nuclear receptor liver receptor homolog-1 (NR5A2) in the regulation of mitochondrial metabolism, cell survival, and cancer. IUBMB Life 2021, 73, 592–610. [Google Scholar] [CrossRef]
- Dube, C.; Bergeron, F.; Vaillant, M.J.; Robert, N.M.; Brousseau, C.; Tremblay, J.J. The nuclear receptors SF1 and LRH1 are expressed in endometrial cancer cells and regulate steroidogenic gene transcription by cooperating with AP-1 factors. Cancer Lett. 2009, 275, 127–138. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Seo, J.; Ha, J.; Kang, E.; Cho, S. The role of epithelial-mesenchymal transition-regulating transcription factors in anti-cancer drug resistance. Arch. Pharm. Res. 2021, 44, 281–292. [Google Scholar] [CrossRef]
- Zhang, P.; Hu, P.; Shen, H.; Yu, J.; Liu, Q.; Du, J. Prognostic role of Twist or Snail in various carcinomas: A systematic review and meta-analysis. Eur. J. Clin. Investig. 2014, 44, 1072–1094. [Google Scholar] [CrossRef]
- Kyo, S.; Sakaguchi, J.; Ohno, S.; Mizumoto, Y.; Maida, Y.; Hashimoto, M.; Nakamura, M.; Takakura, M.; Nakajima, M.; Masutomi, K.; et al. High Twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum. Pathol. 2006, 37, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Abouhashem, N.S.; Ibrahim, D.A.; Mohamed, A.M. Prognostic implications of epithelial to mesenchymal transition related proteins (E-cadherin, Snail) and hypoxia inducible factor 1alpha in endometrioid endometrial carcinoma. Ann. Diagn. Pathol. 2016, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, Q.; Li, N.; Bai, X.; Wang, F.; Li, B. TWIST1 expression and clinical significance in type I endometrial cancer and premalignant lesions: A retrospective clinical study. Medicine 2020, 99, e23397. [Google Scholar] [CrossRef] [PubMed]
- Blechschmidt, K.; Kremmer, E.; Hollweck, R.; Mylonas, I.; Hofler, H.; Kremer, M.; Becker, K.F. The E-cadherin repressor snail plays a role in tumor progression of endometrioid adenocarcinomas. Diagn. Mol. Pathol. 2007, 16, 222–228. [Google Scholar] [CrossRef]
- Herrmann, J.L.; Byekova, Y.; Elmets, C.A.; Athar, M. Liver kinase B1 (LKB1) in the pathogenesis of epithelial cancers. Cancer Lett. 2011, 306, 1–9. [Google Scholar] [CrossRef]
- Yang, G.; Rosen, D.G.; Liu, G.; Yang, F.; Guo, X.; Xiao, X.; Xue, F.; Mercado-Uribe, I.; Huang, J.; Lin, S.H.; et al. CXCR2 promotes ovarian cancer growth through dysregulated cell cycle, diminished apoptosis, and enhanced angiogenesis. Clin. Cancer Res. 2010, 16, 3875–3886. [Google Scholar] [CrossRef]
- Matsuo, Y.; Ochi, N.; Sawai, H.; Yasuda, A.; Takahashi, H.; Funahashi, H.; Takeyama, H.; Tong, Z.; Guha, S. CXCL8/IL-8 and CXCL12/SDF-1alpha co-operatively promote invasiveness and angiogenesis in pancreatic cancer. Int. J. Cancer 2009, 124, 853–861. [Google Scholar] [CrossRef]
- Gabellini, C.; Trisciuoglio, D.; Desideri, M.; Candiloro, A.; Ragazzoni, Y.; Orlandi, A.; Zupi, G.; Del Bufalo, D. Functional activity of CXCL8 receptors, CXCR1 and CXCR2, on human malignant melanoma progression. Eur. J. Cancer 2009, 45, 2618–2627. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, L.; Xu, J.; Zhang, X.; Wang, B. Enhanced expression and clinical significance of chemokine receptor CXCR2 in hepatocellular carcinoma. J. Surg. Res. 2011, 166, 241–246. [Google Scholar] [CrossRef]
- Yung, M.M.; Tang, H.W.; Cai, P.C.; Leung, T.H.; Ngu, S.F.; Chan, K.K.; Xu, D.; Yang, H.; Ngan, H.Y.; Chan, D.W. GRO-α and IL-8 enhance ovarian cancer metastatic potential via the CXCR2-mediated TAK1/NFkappaB signaling cascade. Theranostics 2018, 8, 1270–1285. [Google Scholar] [CrossRef] [PubMed]
- Henriques, T.B.; Dos Santos, D.Z.; Dos Santos Guimaraes, I.; Tessarollo, N.G.; Lyra-Junior, P.C.M.; Mesquita, P.; Pádua, D.; Amaral, A.L.; Cavadas, B.; Pereira, L.; et al. Inhibition of CXCR2 plays a pivotal role in re-sensitizing ovarian cancer to cisplatin treatment. Aging 2021, 13, 13405–13420. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Purcell, C.; Seaton, A.; Oladipo, O.; Maxwell, P.J.; O’Sullivan, J.M.; Wilson, R.H.; Johnston, P.G.; Waugh, D.J. Chemotherapy-Induced CXC-chemokine/CXC-chemokine receptor signaling in metastatic prostate cancer cells confers resistance to oxaliplatin through potentiation of nuclear factor-kappaB transcription and evasion of apoptosis. J. Pharmacol. Exp. Ther. 2008, 327, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.W.D.; Coulter, J.A.; Ong, C.W.; Maxwell, P.J.; Walker, S.; Butterworth, K.T.; Lyubomska, O.; Berlingeri, S.; Gallagher, R.; O’Sullivan, J.M. Clinical and functional characterization of CXCR1/CXCR2 biology in the relapse and radiotherapy resistance of primary PTEN-deficient prostate carcinoma. NAR Cancer. 2020, 2, zcaa012. [Google Scholar] [CrossRef]
- Kassim, S.K.; El-Salahy, E.M.; Fayed, S.T.; Helal, S.A.; Helal, T.; Azzam, E.-D.; Khalifa, A. Vascular endothelial growth factor and interleukin-8 are associated with poor prognosis in epithelial ovarian cancer patients. Clin. Biochem. 2004, 37, 363–369. [Google Scholar] [CrossRef]
- Wei, L.; Liu, Y.; Ma, Y.; Ding, C.; Zhang, H.; Lu, Z.; Gu, Z.; Zhu, C. C-X-C chemokine receptor 2 correlates with unfavorable prognosis and facilitates malignant cell activities via activating JAK2/STAT3 pathway in non-small cell lung cancer. Cell Cycle. 2019, 18, 3456–3471. [Google Scholar] [CrossRef]
- Zhao, J.; Ou, B.; Feng, H.; Wang, P.; Yin, S.; Zhu, C.; Wang, S.; Chen, C.; Zheng, M.; Zong, Y.; et al. Overexpression of CXCR2 predicts poor prognosis in patients with colorectal cancer. Oncotarget 2017, 8, 28442–28454. [Google Scholar] [CrossRef]
- Sui, P.; Hu, P.; Zhang, T.; Zhang, X.; Liu, Q.; Du, J. High expression of CXCR-2 correlates with lymph node metastasis and predicts unfavorable prognosis in resected esophageal carcinoma. Med. Oncol. 2014, 31, 809. [Google Scholar] [CrossRef]
- Mehravar, M.; Ghaemimanesh, F.; Poursani, E.M. An Overview on the Complexity of OCT4: At the Level of DNA, RNA and Protein. Stem Cell Rev. Rep. 2021, 17, 1121–1136. [Google Scholar] [CrossRef]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. The Role Specific Mechanism of OCT4 in Cancer Stem Cells: A Review. Int. J. Stem Cells 2020, 13, 312–325. [Google Scholar] [CrossRef]
- Saha, S.K.; Jeong, Y.; Cho, S.; Cho, S.G. Systematic expression alteration analysis of master reprogramming factor OCT4 and its three pseudogenes in human cancer and their prognostic outcomes. Sci. Rep. 2018, 8, 14806. [Google Scholar] [CrossRef] [PubMed]
- Mohiuddin, I.S.; Wei, S.J.; Kang, M.H. Role of OCT4 in cancer stem-like cells and chemotherapy resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165432. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Stender, J.D.; Joshi, S.; Wu, G.; Katzenellenbogen, B.S. OCT-4: A novel estrogen receptor-α collaborator that promotes tamoxifen resistance in breast cancer cells. Oncogene 2016, 35, 5722–5734. [Google Scholar] [CrossRef] [PubMed]
- Tsialikas, J.; Romer-Seibert, J. LIN28: Roles and regulation in development and beyond. Development 2015, 142, 2397–2404. [Google Scholar] [CrossRef]
- Balzeau, J.; Menezes, M.R.; Cao, S.; Hagan, J.P. The LIN28/let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31. [Google Scholar] [CrossRef]
- Donkers, H.; Bekkers, R.; Galaal, K. Diagnostic value of microRNA panel in endometrial cancer: A systematic review. Oncotarget 2020, 11, 2010–2023. [Google Scholar] [CrossRef]
- McCluggage, W.G.; Connolly, L.E.; McBride, H.A.; Kalloger, S.; Gilks, C.B. HMGA2 is commonly expressed in uterine serous carcinomas and is a useful adjunct to diagnosis. Histopathology 2012, 60, 547–553. [Google Scholar] [CrossRef]
- Wei, L.; Liu, X.; Zhang, W.; Wei, Y.; Li, Y.; Zhang, Q.; Dong, R.; Kwon, J.S.; Liu, Z.; Zheng, W.; et al. Overexpression and oncogenic function of HMGA2 in endometrial serous carcinogenesis. Am. J. Cancer Res. 2016, 6, 249–259. [Google Scholar]
- Ma, J.; Li, D.; Kong, F.F.; Yang, D.; Yang, H.; Ma, X.X. miR-302a-5p/367-3p-HMGA2 axis regulates malignant processes during endometrial cancer development. J. Exp. Clin. Cancer Res. 2018, 37, 19. [Google Scholar] [CrossRef]
- Montserrat, N.; Mozos, A.; Llobet, D.; Dolcet, X.; Pons, C.; de Herreros, A.G.; Matias-Guiu, X.; Prat, J. Epithelial to mesenchymal transition in early stage endometrioid endometrial carcinoma. Hum. Pathol. 2012, 43, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Palumbo Junior, A.; de Sousa, V.P.L.; Esposito, F.; De Martino, M.; Forzati, F.; Moreira, F.C.B.; Simão, T.A.; Nasciutti, L.E.; Fusco, A.; Ribeiro Pinto, L.F.; et al. Overexpression of HMGA1 Figures as a Potential Prognostic Factor in Endometrioid Endometrial Carcinoma (EEC). Genes 2019, 10, 372. [Google Scholar] [CrossRef]
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: Present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Spinelli, C.; Reis-Sobreiro, M.; Cavallini, L.; You, S.; Zandian, M.; Li, X.; Mishra, R.; Chiarugi, P.; Adam, R.M.; et al. MYC Mediates Large Oncosome-Induced Fibroblast Reprogramming in Prostate Cancer. Cancer Res. 2017, 77, 2306–2317. [Google Scholar] [CrossRef]
- Balaj, L.; Lessard, R.; Dai, L.; Cho, Y.J.; Pomeroy, S.L.; Breakefield, X.O.; Skog, J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011, 2, 180. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wu, X.; Zhou, W.; Fong, M.Y.; Cao, M.; Liu, J.; Liu, X.; Chen, C.H.; Fadare, O.; Pizzo, D.P.; et al. Cancer-Cell-Secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat. Cell. Biol. 2018, 20, 597–609. [Google Scholar] [CrossRef]
- Johnston, L.A. Socializing with MYC: Cell competition in development and as a model for premalignant cancer. Cold Spring Harb. Perspect. Med. 2014, 4, a014274. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, D.P.; Southwick, H.W.; Smejkal, W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer 1953, 6, 963–968. [Google Scholar] [CrossRef]
- Di Giacomo, S.; Sollazzo, M.; de Biase, D.; Ragazzi, M.; Bellosta, P.; Pession, A.; Grifoni, D. Human Cancer Cells Signal Their Competitive Fitness Through MYC Activity. Sci. Rep. 2017, 7, 12568. [Google Scholar] [CrossRef]
- Raeder, M.B.; Birkeland, E.; Trovik, J.; Krakstad, C.; Shehata, S.; Schumacher, S.; Zack, T.I.; Krohn, A.; Werner, H.M.; Moody, S.E.; et al. Integrated genomic analysis of the 8q24 amplification in endometrial cancers identifies ATAD2 as essential to MYC-dependent cancers. PLoS ONE 2013, 8, e54873. [Google Scholar] [CrossRef]
- Zhou, J.; Suzuki, T.; Kovacic, A.; Saito, R.; Miki, Y.; Ishida, T.; Moriya, T.; Simpson, E.R.; Sasano, H.; Clyne, C.D. Interactions between prostaglandin E(2), liver receptor homologue-1, and aromatase in breast cancer. Cancer Res. 2005, 65, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Annicotte, J.S.; Chavey, C.; Servant, N.; Teyssier, J.; Bardin, A.; Licznar, A.; Badia, E.; Pujol, P.; Vignon, F.; Maudelonde, T.; et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene 2005, 24, 8167–8175. [Google Scholar] [CrossRef]
- Chand, A.L.; Herridge, K.A.; Howard, T.L.; Simpson, E.R.; Clyne, C.D. Tissue-Specific regulation of aromatase promoter II by the orphan nuclear receptor LRH-1 in breast adipose stromal fibroblasts. Steroids 2011, 76, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Simpson, E.R.; Mahendroo, M.S.; Means, G.D.; Kilgore, M.W.; Hinshelwood, M.M.; Graham-Lorence, S.; Amarneh, B.; Ito, Y.; Fisher, C.R.; Michael, M.D.; et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr. Rev. 1994, 15, 342–355. [Google Scholar] [PubMed]
- Smuc, T.; Rizner, T.L. Aberrant pre-receptor regulation of estrogen and progesterone action in endometrial cancer. Mol. Cell. Endocrinol. 2009, 301, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Wang, Y.; Liang, W.; Liu, L.; Pan, N.; Deng, H.; Li, L.; Zou, C.; Chan, F.L.; Zhou, Y. LRH-1 drives hepatocellular carcinoma partially through induction of c-myc and cyclin E1, and suppression of p21. Cancer Manag. Res. 2018, 10, 2389–2400. [Google Scholar] [CrossRef] [PubMed]
- Lessl, M.; Klotzbuecher, M.; Schoen, S.; Reles, A.; Stockemann, K.; Fuhrmann, U. Comparative messenger ribonucleic acid analysis of immediate early genes and sex steroid receptors in human leiomyoma and healthy myometrium. J. Clin. Endocrinol. Metab. 1997, 82, 2596–2600. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, K.A.; Heaphy, C.M.; Mai, M.; Vargas, K.M.; Jones, A.C.; Vo, P.; Butler, K.S.; Joste, N.E.; Bisoffi, M.; Griffith, J.K. Markers of fibrosis and epithelial to mesenchymal transition demonstrate field cancerization in histologically normal tissue adjacent to breast tumors. Int. J. Cancer. 2011, 129, 1310–1321. [Google Scholar] [CrossRef]
- Contreras, C.M.; Gurumurthy, S.; Haynie, J.M.; Shirley, L.J.; Akbay, E.A.; Wingo, S.N.; Schorge, J.O.; Broaddus, R.R.; Wong, K.K.; Bardeesy, N.; et al. Loss of Lkb1 provokes highly invasive endometrial adenocarcinomas. Cancer Res. 2008, 68, 759–766. [Google Scholar] [CrossRef]
- Co, N.N.; Iglesias, D.; Celestino, J.; Kwan, S.Y.; Mok, S.C.; Schmandt, R.; Lu, K.H. Loss of LKB1 in high-grade endometrial carcinoma: LKB1 is a novel transcriptional target of p53. Cancer 2014, 120, 3457–3468. [Google Scholar] [CrossRef] [PubMed]
- Pena, C.G.; Nakada, Y.; Saatcioglu, H.D.; Aloisio, G.M.; Cuevas, I.; Zhang, S.; Miller, D.S.; Lea, J.S.; Wong, K.K.; DeBerardinis, R.J.; et al. LKB1 loss promotes endometrial cancer progression via CCL2-dependent macrophage recruitment. J. Clin. Investig. 2015, 125, 4063–4076. [Google Scholar] [CrossRef] [PubMed]
- Karaayvaz, M.; Zhang, C.; Liang, S.; Shroyer, K.R.; Ju, J. Prognostic significance of miR-205 in endometrial cancer. PLoS ONE 2012, 7, e35158. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int. J. Cancer. 2013, 132, 1633–1645. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, Y.P.; Huang, G.R.; Gong, B.L.; Yang, B.; Zhang, D.X.; Hu, P.; Xu, S.R. Expression of the stem cell marker, Nanog, in human endometrial adenocarcinoma. Int. J. Gynecol. Pathol. 2011, 30, 262–270. [Google Scholar] [CrossRef]
- Paul, E.N.; Burns, G.W.; Carpenter, T.J.; Grey, J.A.; Fazleabas, A.T.; Teixeira, J.M. Transcriptome Analyses of Myometrium from Fibroid Patients Reveals Phenotypic Differences Compared to Non-Diseased Myometrium. Int. J. Mol. Sci. 2021, 22, 3618. [Google Scholar] [CrossRef]
- Maclean, A.; Kamal, A.; Adishesh, M.; Alnafakh, R.; Tempest, N.; Hapangama, D.K. Human Uterine Biopsy: Research Value and Common Pitfalls. Int. J. Reprod. Med. 2020, 2020, 9275360. [Google Scholar] [CrossRef]
- Troester, M.A.; Lee, M.H.; Carter, M.; Fan, C.; Cowan, D.W.; Perez, E.R.; Pirone, J.R.; Perou, C.M.; Jerry, D.J.; Schneider, S.S. Activation of host wound responses in breast cancer microenvironment. Clin. Cancer Res. 2009, 15, 7020–7028. [Google Scholar] [CrossRef]
- Ge, L.; Meng, W.; Zhou, H.; Bhowmick, N. Could stroma contribute to field cancerization? Med. Hypotheses 2010, 75, 26–31. [Google Scholar] [CrossRef]
- Chatterjee, S.; Basak, P.; Buchel, E.; Safneck, J.; Murphy, L.C.; Mowat, M.; Kung, S.K.; Eirew, P.; Eaves, C.J.; Raouf, A. Breast Cancers Activate Stromal Fibroblast-Induced Suppression of Progenitors in Adjacent Normal Tissue. Stem Cell Rep. 2018, 10, 196–211. [Google Scholar] [CrossRef]
- Amirrad, F.; Pytak, P.A.; Sadeghiani-Pelar, N.; Nguyen, J.P.T.; Cauble, E.L.; Jones, A.C.; Bisoffi, M. Prostate field cancerization and exosomes: Association between CD9, early growth response 1 and fatty acid synthase. Int. J. Oncol. 2020, 56, 957–968. [Google Scholar] [CrossRef] [PubMed]


| Patients with Cancer Cell-Free Tumor-Adjacent Tissues (TA), n = 26 | |||||
| No. | Patient No. | Age | Histologic Type | Grade | Stage * |
| 1 | 12 | 57 | Endometrioid adenocarcinoma | 3 | IA |
| 2 | 23 | 70 | Endometrioid adenocarcinoma | 2 | IA |
| 3 | 24 | 57 | Endometrioid adenocarcinoma | 2/1 | IA |
| 4 | 25 | 84 | Endometrioid adenocarcinoma | 1 | IIIC |
| 5 | 26 | 72 | Endometrioid adenocarcinoma | 3 | IIIC |
| 6 | 29 | 58 | Clear cell adenocarcinoma | 3 | IB |
| 7 | 32 | 47 | Clear cell adenocarcinoma | 2 | IB |
| 8 | 33 | 60 | Endometrioid adenocarcinoma | 1 | IB |
| 9 | 35 | 61 | Endometrioid adenocarcinoma | 1 | IA |
| 10 | 36 | 70 | Clear cell adenocarcinoma | 2 | IIIC |
| 11 | 41 | 70 | Serous carcinoma | 1 | II |
| 12 | 42 | 73 | Endometrioid adenocarcinoma | 1 | IIA |
| 13 | 44 | 67 | Endometrioid adenocarcinoma | 1 | IA |
| 14 | 52 | 66 | Carcinosarcoma | 3 | IB |
| 15 | 54 | 75 | Endometrioid adenocarcinoma | 2 | IB |
| 16 | 56 | 50 | Endometrioid adenocarcinoma | 1 | IA |
| 17 | 58 | 57 | Endometrioid adenocarcinoma | 2 | II |
| 18 | 59 | 73 | Endometrioid adenocarcinoma | 2 | IB |
| 19 | 60 | 69 | Endometrioid adenocarcinoma | 2 | IA |
| 20 | 61 | 57 | Endometrioid adenocarcinoma | 2 | IA |
| 21 | 62 | 68 | Clear cell adenocarcinoma | - | IB |
| 22 | 63 | 53 | Endometrioid adenocarcinoma | 2 | IA |
| 23 | 67 | 68 | Endometrioid adenocarcinoma | 3 | IA |
| 24 | 69 | 75 | Endometrioid adenocarcinoma | 1 | II |
| 25 | 71 | 53 | Endometrioid adenocarcinoma | 3 | III |
| 26 | 73 | 65 | Endometrioid adenocarcinoma | 1 | IB |
| Patients with tumor-adjacent samples containing cancer cells (TAc), n = 23 | |||||
| No. | Patient no. | Age | Histologic type | Grade | Stage |
| 1 | 11 | 53 | Endometrioid adenocarcinoma | 1 | IB |
| 2 | 16 | 55 | Endometrioid adenocarcinoma | 1 | IB |
| 3 | 17 | 54 | Endometrioid adenocarcinoma | 2 | IB |
| 4 | 18 | 75 | Carcinosarcoma | 3 | I |
| 5 | 19 | 72 | Carcinosarcoma | 3 | I |
| 6 | 20 | 61 | Carcinosarcoma | 2/3 | I |
| 7 | 22 | 63 | Endometrioid adenocarcinoma | 1 | III |
| 8 | 27 | 50 | Endometrioid adenocarcinoma | 2 | I |
| 9 | 31 | 53 | Endometrioid adenocarcinoma | 2 | IB |
| 10 | 34 | 85 | Endometrioid adenocarcinoma | 1 | II |
| 11 | 38 | 52 | Endometrioid adenocarcinoma | 1 | II |
| 12 | 39 | 60 | Endometrioid adenocarcinoma | 1 | IIA |
| 13 | 40 | 74 | Endometrioid adenocarcinoma | 2 | II |
| 14 | 46 | 75 | Endometrioid adenocarcinoma partim mucinosum | 1 | II |
| 15 | 47 | 80 | Endometrioid adenocarcinoma | 1 | II |
| 16 | 48 | 61 | Endometrioid adenocarcinoma | 1 | II |
| 17 | 50 | 78 | Endometrioid adenocarcinoma | 1 | IB |
| 18 | 53 | 74 | Endometrioid adenocarcinoma | 2 | IB |
| 19 | 57 | 63 | Endometrioid adenocarcinoma | 1 | IB |
| 20 | 64 | 54 | Endometrioid adenocarcinoma | 2 | IB |
| 21 | 65 | 72 | Carcinosarcoma | 3 | II |
| 22 | 66 | 73 | Endometrioid adenocarcinoma | 2 | IB |
| 23 | 68 | 76 | Endometrioid adenocarcinoma | 2 | IB |
| p-Values (Green Marks Statistical Significance) | ||||||||||||
| MYC | NR5A2 | TWIST1 | SNAI1 | STK11 | CXCR2 | HMGA2 | LIN28A | POU5F1 isoform A | POU5F1 isoform B | POU5F1 isoform B1 | miR-205-5p | |
| T vs TA | 0.0002 | <0.0001 | <0.0001 | <0.0001 | 0.0002 | 0.2079 | 0.0493 | 0.0039 | 0.0068 | 0.001 | 0.2914 | <0.0001 |
| Tc vs TAc | 0.0031 | 0.0101 | 0.0011 | 0.0027 | 0.0196 | 0.2467 | 0.1186 | >0.9999 | 0.6875 | 0.601 | 0.1207 | <0.0001 |
| T vs Control | 0.2053 | 0.0058 | <0.0001 | 0.1857 | 0.0234 | 0.016 | 0.0421 | 0.0017 | 0.2174 | 0.6774 | 0.8409 | <0.0001 |
| TA vs Control | 0.0001 | <0.0001 | 0.0947 | <0.0001 | 0.1506 | 0.0002 | 0.5311 | >0.9999 | <0.0001 | 0.4174 | 0.6432 | 0.1921 |
| Tc vs Control | 0.9511 | 0.0382 | 0.0107 | 0.0948 | 0.0868 | 0.0078 | <0.0001 | 0.0455 | 0.0132 | 0.4241 | 0.8659 | <0.0001 |
| TAc vs Control | 0.0079 | 0.0002 | 0.6637 | <0.0001 | >0.9999 | <0.0001 | 0.0053 | 0.2243 | 0.0038 | 0.9224 | 0.3426 | <0.0001 |
| T vs Tc | 0.1913 | 0.684 | 0.4591 | 0.4804 | 0.6407 | 0.7283 | 0.1715 | 0.2039 | 0.1643 | 0.9921 | 0.8194 | 0.5314 |
| TA vs TAc | 0.2356 | 0.1652 | 0.0526 | 0.8973 | 0.1845 | 0.2689 | 0.0946 | 0.2151 | 0.3834 | 0.3658 | 0.7125 | 0.0001 |
| T vs TAc | 0.0015 | <0.0001 | 0.0001 | 0.0001 | 0.0059 | 0.0132 | 0.8817 | 0.0361 | 0.047 | 0.4579 | 0.4176 | 0.0003 |
| TA vs Tc | 0.0022 | 0.0002 | 0.0003 | 0.0037 | 0.001 | 0.4681 | 0.0034 | 0.0418 | 0.1391 | 0.0902 | 0.6152 | <0.0001 |
| Medians of 2−ΔCt values | ||||||||||||
| MYC | NR5A2 | TWIST1 | SNAI1 | STK11 | CXCR2 | HMGA2 | LIN28A | POU5F1isoform A | POU5F1isoform B | POU5F1isoform B1 | miR-205-5p | |
| T | 0.04787 | 0.0023 | 0.00357 | 0.0035 | 0.02007 | 0.00034 | 0.00566 | 0 | 0 | 3.26 × 10−5 | 2.73 × 10−5 | 0.06781 |
| TA | 0.11679 | 0.00888 | 0.01078 | 0.0124 | 0.03672 | 0.00037 | 0.00401 | 0 | 0 | 1.28 × 10−5 | 3.13 × 10−5 | 0.00269 |
| Tc | 0.06117 | 0.00213 | 0.00362 | 0.00444 | 0.02114 | 0.00029 | 0.00819 | 0 | 0 | 5.63 × 10−5 | 3.33 × 10−5 | 0.09382 |
| TAc | 0.09343 | 0.00675 | 0.00802 | 0.01158 | 0.03373 | 0.00084 | 0.00609 | 0 | 0 | 1.66 × 10−5 | 2.22 × 10−5 | 0.01471 |
| Control | 0.05087 | 0.00395 | 0.00853 | 0.00225 | 0.03665 | 0.00015 | 0.00414 | 0 | 7.10 × 10−5 | 2.67 × 10−5 | 3.44 × 10−5 | 0.00199 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulinczak, M.; Sromek, M.; Panek, G.; Zakrzewska, K.; Lotocka, R.; Szafron, L.M.; Chechlinska, M.; Siwicki, J.K. Endometrial Cancer-Adjacent Tissues Express Higher Levels of Cancer-Promoting Genes than the Matched Tumors. Genes 2022, 13, 1611. https://doi.org/10.3390/genes13091611
Kulinczak M, Sromek M, Panek G, Zakrzewska K, Lotocka R, Szafron LM, Chechlinska M, Siwicki JK. Endometrial Cancer-Adjacent Tissues Express Higher Levels of Cancer-Promoting Genes than the Matched Tumors. Genes. 2022; 13(9):1611. https://doi.org/10.3390/genes13091611
Chicago/Turabian StyleKulinczak, Mariusz, Maria Sromek, Grzegorz Panek, Klara Zakrzewska, Renata Lotocka, Lukasz Michal Szafron, Magdalena Chechlinska, and Jan Konrad Siwicki. 2022. "Endometrial Cancer-Adjacent Tissues Express Higher Levels of Cancer-Promoting Genes than the Matched Tumors" Genes 13, no. 9: 1611. https://doi.org/10.3390/genes13091611






