Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of Putative UGTs in the Populus Genome
2.2. PCR Cloning and Sequencing of Selected Populus UGTs
2.3. Motif Alignment, Phylogenetic Analysis, and Comparison
2.4. Gene Duplication and Chromosomal Distribution
2.5. Gene Structure Analysis
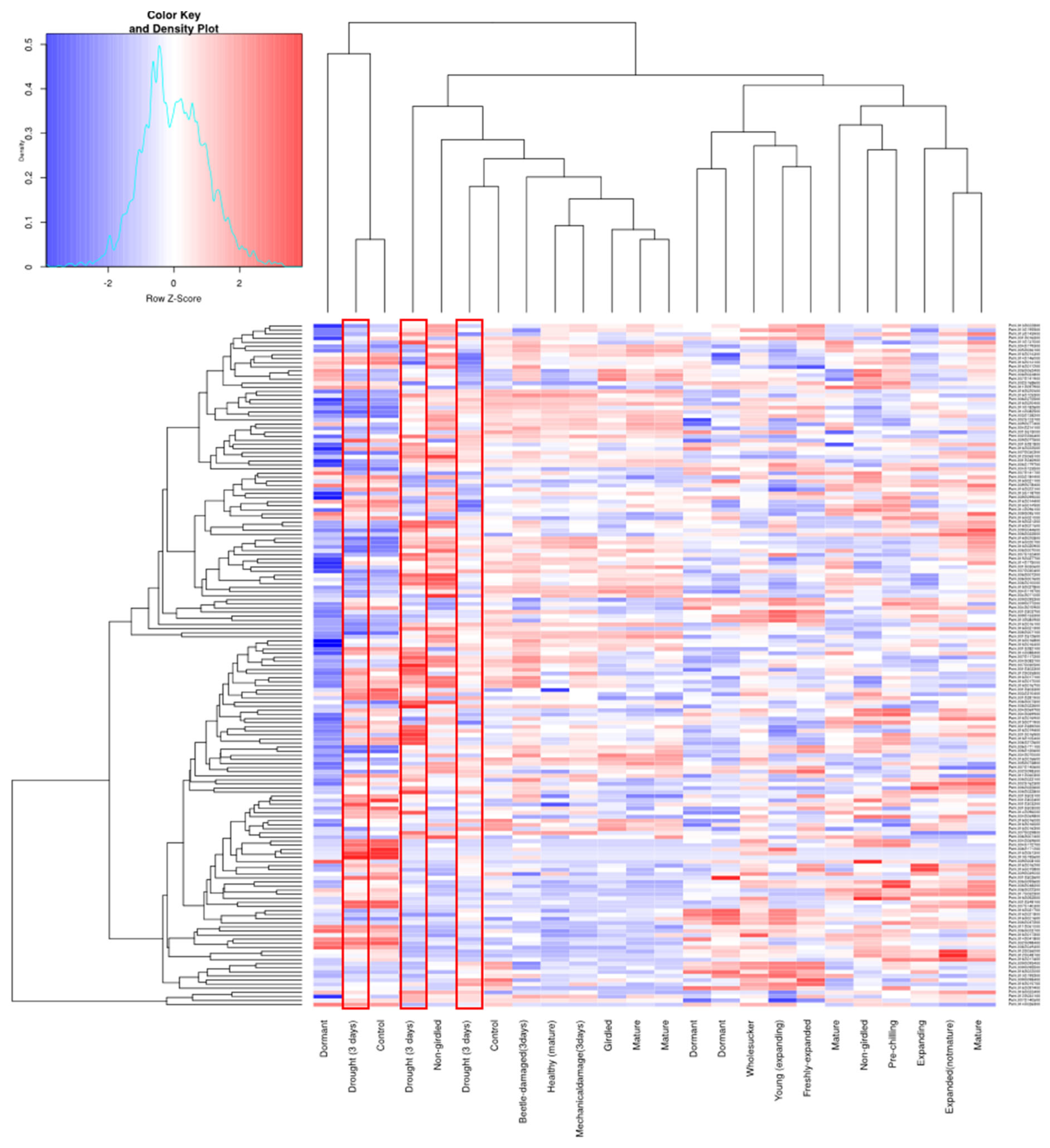
2.6. Digital Expression Analysis
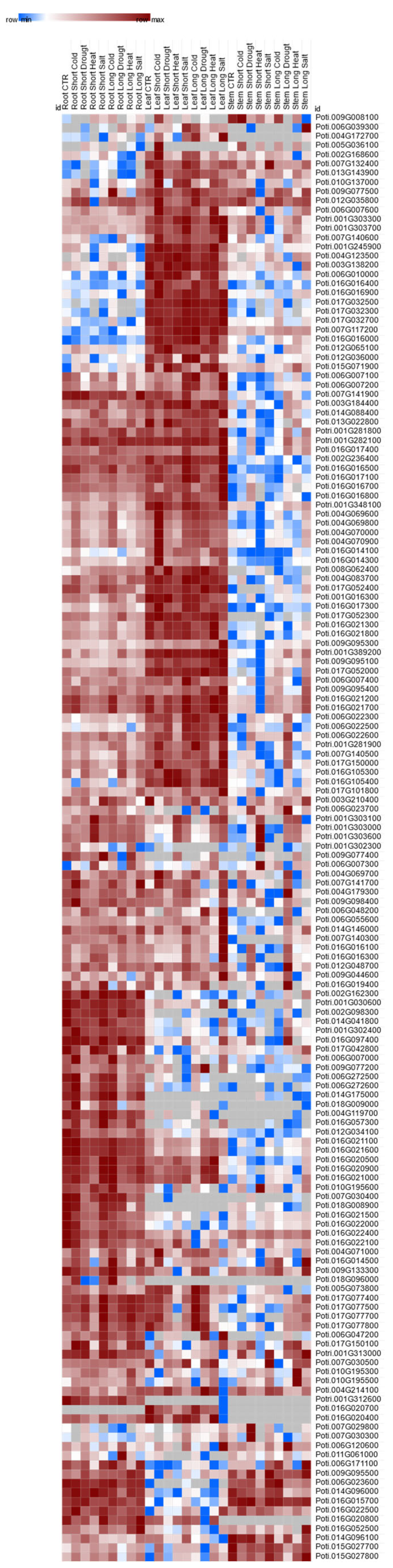
2.7. RNA Sequencing based UGTs Model in Tissues and under Drought Stress
2.8. Co-Expression Networking of Putative UGTs
3. Results
3.1. Identified Putative UGTs in the Populus Genome
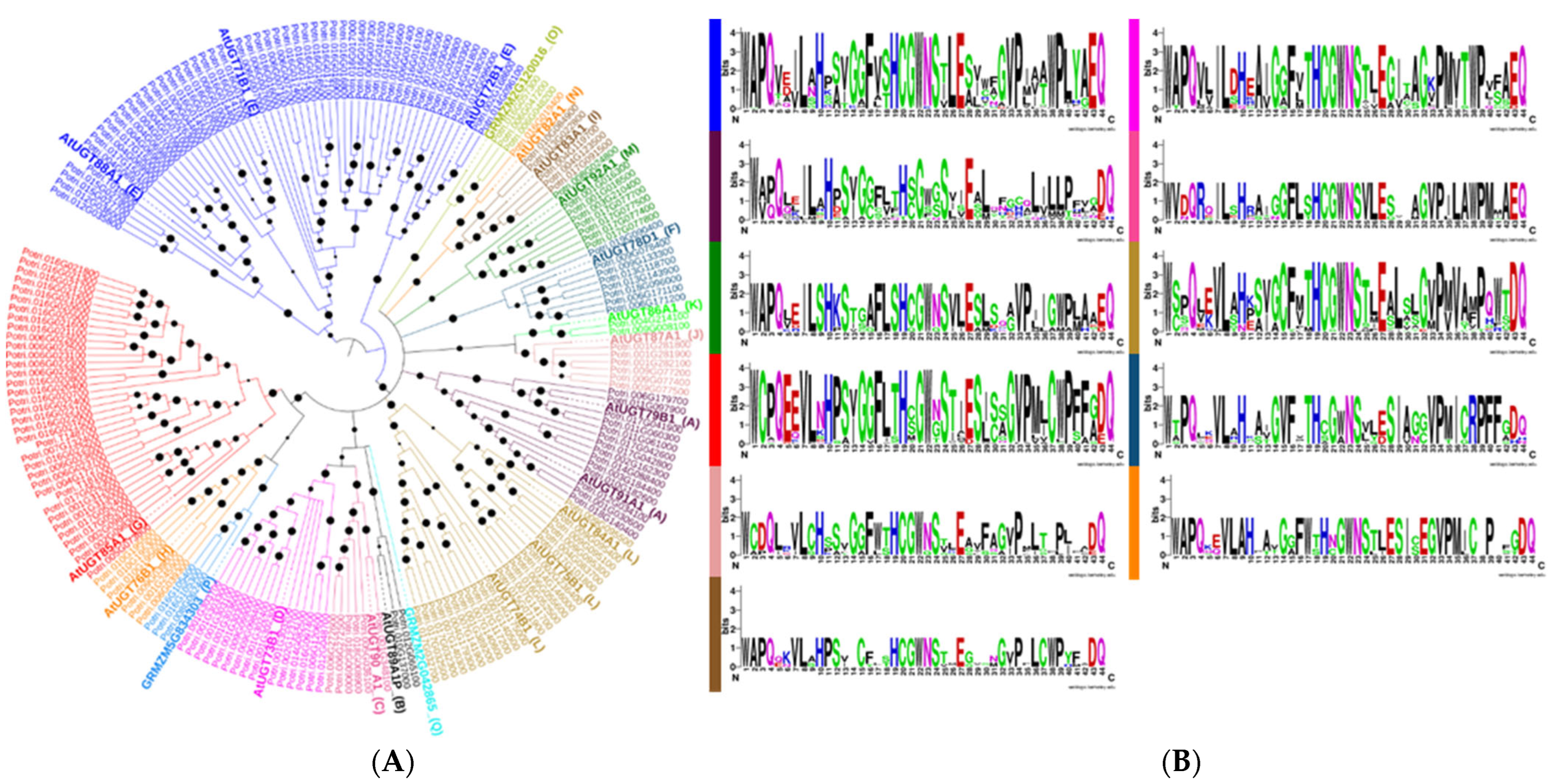
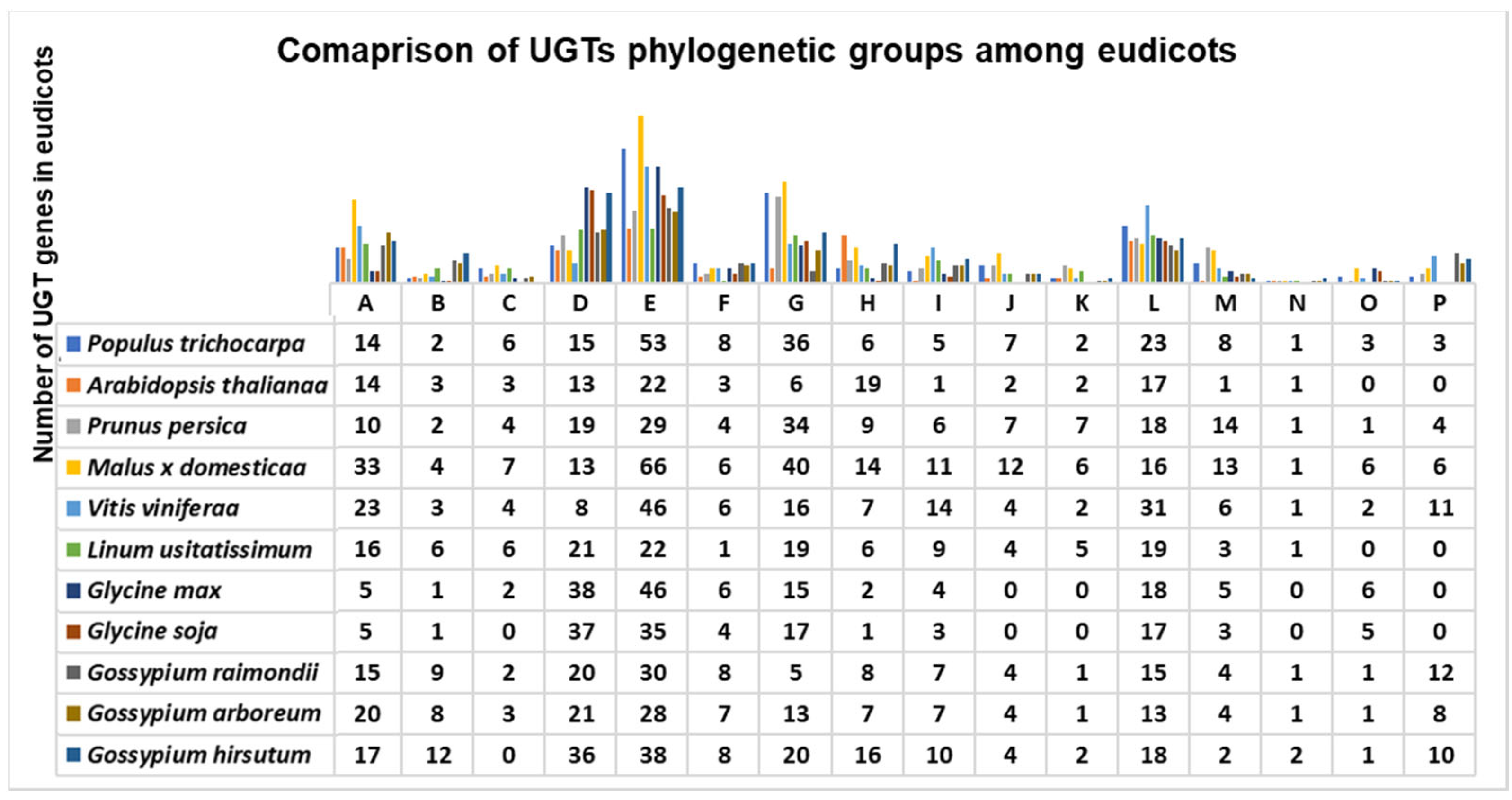
3.2. Motif Alignment, Phylogenetic Analysis, and Comparison
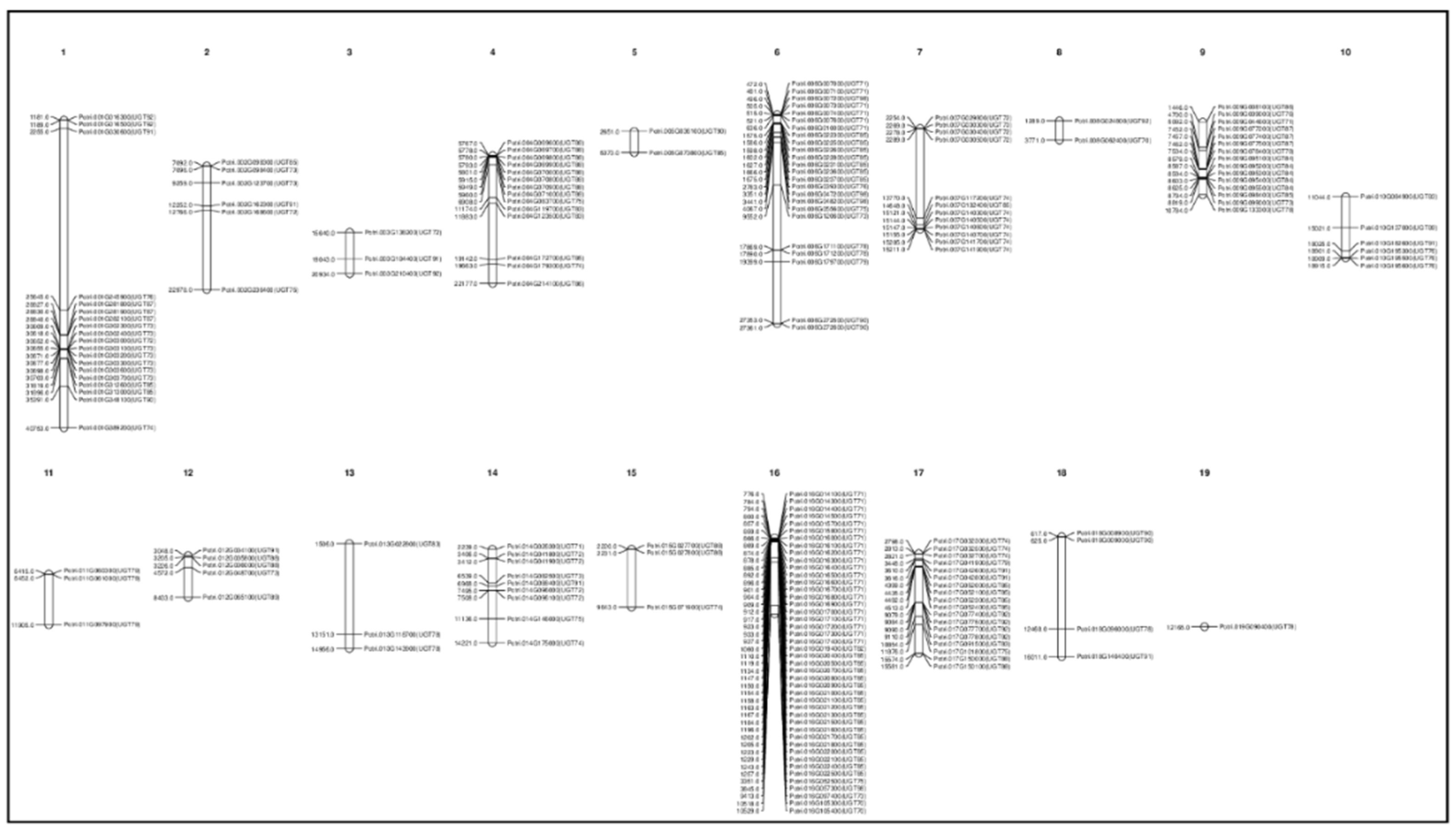
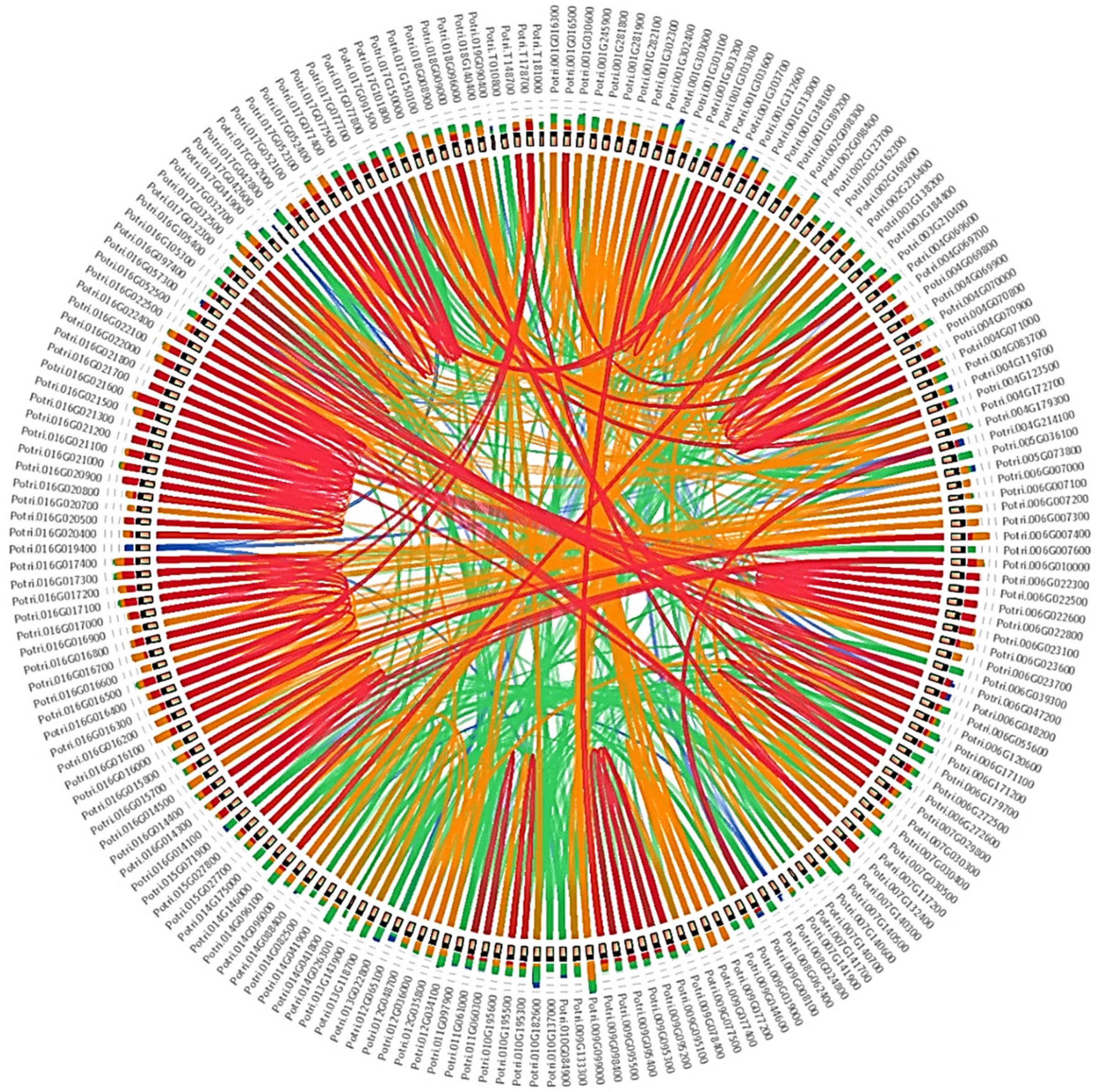
3.3. Gene Duplication and Chromosomal Distribution
3.4. Gene Structure Analysis
3.5. Digital Expression Analysis
3.6. RNA-Seq-based UGT Model under Drought
3.7. Co-Expression Networking of Putative UGTs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
Abbreviations
References
- Evans, L.M.; Slavov, G.T.; Rodgers-Melnick, E.; Martin, J.; Ranjan, P.; Muchero, W.; Brunner, A.M.; Schackwitz, W.; Gunter, L.; Chen, J.-G. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 2014, 46, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Zannini, P.; Frascaroli, F.; Nascimbene, J.; Halley, J.M.; Stara, K.; Cervellini, M.; Di Musciano, M.; De Vigili, F.; Rocchini, D.; Piovesan, G. Investigating sacred natural sites and protected areas for forest area changes in Italy. Conserv. Sci. Pract. 2022, 4, e12695. [Google Scholar] [CrossRef]
- Abraham, P.E.; Wang, X.; Ranjan, P.; Nookaew, I.; Zhang, B.; Tuskan, G.A.; Hettich, R.L. Integrating mRNA and protein sequencing enables the detection and quantitative profiling of natural protein sequence variants of Populus trichocarpa. J. Proteome Res. 2015, 14, 5318–5326. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Veljanovski, V. Functional characterization of flavonoid glycosyltransferases and an acid phosphatase from poplar (Populus spp.). Ph.D. Thesis, University of Victoria, Victoria, BC, Canada, 2012. [Google Scholar]
- Brunner, A.M.; Beers, E.P. Transcription factors in poplar growth and development. In Poplar; Genomics of Industrial Crop Plants, Series; Taylor & Francis Group: Abingdon-on-Thames, UK, 2010. [Google Scholar]
- Bowles, D.; Lim, E.-K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Sawai, S.; Saito, K. Triterpenoid biosynthesis and engineering in plants. Front. Plant Sci. 2011, 2, 25. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Jones, P.; Vogt, T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta 2001, 213, 164–174. [Google Scholar] [CrossRef]
- Speeckaert, N.; El Jaziri, M.; Baucher, M.; Behr, M. UGT72, a Major Glycosyltransferase Family for Flavonoid and Monolignol Homeostasis in Plants. Biology 2022, 11, 441. [Google Scholar] [CrossRef]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Hughes, J.; Hughes, M.A. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 1994, 5, 41–49. [Google Scholar] [CrossRef]
- Lorenc-Kukuła, K.; Korobczak, A.; Aksamit-Stachurska, A.; Kostyń, K.; Lukaszewicz, M.; Szopa, J. Glucosyltransferase: The gene arrangement and enzyme function. Cell. Mol. Biol. Lett. 2004, 9, 935–946. [Google Scholar]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G.; Tarling, C.A.; Ford, C.M.; Bowles, D.J.; Davies, G.J. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef]
- Li, Y.; Baldauf, S.; Lim, E.-K.; Bowles, D.J. Phylogenetic Analysis of the UDP-glycosyltransferase Multigene Family of Arabidopsis thaliana* 210. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 175. [Google Scholar] [CrossRef]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 67. [Google Scholar] [CrossRef]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Ma, Q.; Li, L.; Zhang, C.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. 2015, 290, 1805–1818. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef]
- Ouyang, D.; Wang, L.-C.; Tang, T.; Feng, H. Genomic-wide identification and characterization of the uridine diphosphate glycosyltransferase family in eucommia ulmoides oliver. Plants 2021, 10, 1934. [Google Scholar] [CrossRef]
- Rehman, H.M.; Nawaz, M.A.; Bao, L.; Shah, Z.H.; Lee, J.-M.; Ahmad, M.Q.; Chung, G.; Yang, S.H. Genome-wide analysis of Family-1 UDP-glycosyltransferases in soybean confirms their abundance and varied expression during seed development. J. Plant Physiol. 2016, 206, 87–97. [Google Scholar] [CrossRef]
- Wei, Y.; Mu, H.; Xu, G.; Wang, Y.; Li, Y.; Li, S.; Wang, L. Genome-Wide Analysis and Functional Characterization of the UDP-Glycosyltransferase Family in Grapes. Horticulturae 2021, 7, 204. [Google Scholar] [CrossRef]
- Wu, B.; Gao, L.; Gao, J.; Xu, Y.; Liu, H.; Cao, X.; Zhang, B.; Chen, K. Genome-wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017, 8, 389. [Google Scholar] [CrossRef]
- Xiao, X.; Lu, Q.; Liu, R.; Gong, J.; Gong, W.; Liu, A.; Ge, Q.; Li, J.; Shang, H.; Li, P. Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. 3 Biotech 2019, 9, 453. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, L.-M.; Cheng, W.-W.; Song, X.; Long, Y.-H.; Xing, Z.-B. Genome-wide identification and expression analysis of glycosyltransferase gene family 1 in Quercus robur L. J. Appl. Genet. 2021, 62, 559–570. [Google Scholar] [CrossRef]
- Zhao, X.; Dai, X.; Gao, L.; Guo, L.; Zhuang, J.; Liu, Y.; Ma, X.; Wang, R.; Xia, T.; Wang, Y. Functional analysis of an uridine diphosphate glycosyltransferase involved in the biosynthesis of polyphenolic glucoside in tea plants (Camellia sinensis). J. Agric. Food Chem. 2017, 65, 10993–11001. [Google Scholar] [CrossRef]
- Zhou, K.; Hu, L.; Li, P.; Gong, X.; Ma, F. Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci. 2017, 265, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.A.; Davies, G.J.; Bulone, V.; Henrissat, B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 1997, 326, 929. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.; Bowles, D. Higher plant glycosyltransferases. Genome Biol. 2001, 2, reviews3004.1. [Google Scholar]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Teze, D.; Coines, J.; Fredslund, F.; Dubey, K.D.; Bidart, G.N.; Adams, P.D.; Dueber, J.E.; Svensson, B.; Rovira, C.; Welner, D.H. O-/N-/S-Specificity in glycosyltransferase catalysis: From mechanistic understanding to engineering. ACS Catal. 2021, 11, 1810–1815. [Google Scholar] [CrossRef]
- Cantarel, B.; Coutinho, P.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Park, B.H.; Karpinets, T.V.; Syed, M.H.; Leuze, M.R.; Uberbacher, E.C. CAZymes Analysis Toolkit (CAT): Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 2010, 20, 1574–1584. [Google Scholar] [CrossRef] [Green Version]
- Rehman, H.M.; Nawaz, M.A.; Shah, Z.H.; Ludwig-Müller, J.; Chung, G.; Ahmad, M.Q.; Yang, S.H.; Lee, S.I. Comparative genomic and transcriptomic analyses of Family-1 UDP glycosyltransferase in three Brassica species and Arabidopsis indicates stress-responsive regulation. Sci. Rep. 2018, 8, 1875. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Trapero-Mozos, A.; Climent, M.F.L.; Gómez-Cadenas, A.; Gómez-Gómez, L. Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Sci. 2015, 234, 60–73. [Google Scholar] [CrossRef]
- Sun, Y.-G.; Wang, B.; Jin, S.-H.; Qu, X.-X.; Li, Y.-J.; Hou, B.-K. Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS ONE 2013, 8, e59924. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; Van De Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Vanderauwera, S.; Zimmermann, P.; Rombauts, S.; Vandenabeele, S.; Langebartels, C.; Gruissem, W.; Inzé, D.; Van Breusegem, F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005, 139, 806–821. [Google Scholar] [CrossRef]
- Hajihashemi, S.; Geuns, J.M. Gene transcription and steviol glycoside accumulation in Stevia rebaudiana under polyethylene glycol-induced drought stress in greenhouse cultivation. FEBS Open Bio 2016, 6, 937–944. [Google Scholar] [CrossRef]
- Gilbert, M.K.; Bland, J.M.; Shockey, J.M.; Cao, H.; Hinchliffe, D.J.; Fang, D.D.; Naoumkina, M. A transcript profiling approach reveals an abscisic acid-specific glycosyltransferase (UGT73C14) induced in developing fiber of Ligon lintless-2 mutant of cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e75268. [Google Scholar]
- Liu, Z.; Yan, J.-P.; Li, D.-K.; Luo, Q.; Yan, Q.; Liu, Z.-B.; Ye, L.-M.; Wang, J.-M.; Li, X.-F.; Yang, Y. UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef]
- Priest, D.M.; Jackson, R.G.; Ashford, D.A.; Abrams, S.R.; Bowles, D.J. The use of abscisic acid analogues to analyse the substrate selectivity of UGT71B6, a UDP-glycosyltransferase of Arabidopsis thaliana. FEBS Lett. 2005, 579, 4454–4458. [Google Scholar] [CrossRef]
- Zhang, G.-Z.; Jin, S.-H.; Jiang, X.-Y.; Dong, R.-R.; Li, P.; Li, Y.-J.; Hou, B.-K. Ectopic expression of UGT75D1, a glycosyltransferase preferring indole-3-butyric acid, modulates cotyledon development and stress tolerance in seed germination of Arabidopsis thaliana. Plant Mol. Biol. 2016, 90, 77–93. [Google Scholar] [CrossRef]
- Chen, T.-T.; Liu, F.-F.; Xiao, D.-W.; Jiang, X.-Y.; Li, P.; Zhao, S.-M.; Hou, B.-K.; Li, Y.-J. The Arabidopsis UDP-glycosyltransferase75B1, conjugates abscisic acid and affects plant response to abiotic stresses. Plant Mol. Biol. 2020, 102, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Gordon, H.; Fellenberg, C.; Lackus, N.D.; Archinuk, F.; Sproule, A.; Nakamura, Y.; Köllner, T.G.; Gershenzon, J.; Overy, D.P.; Constabel, C.P. CRISPR/Cas9 disruption of UGT71L1 in poplar connects salicinoid and salicylic acid metabolism and alters growth and morphology. Plant Cell 2022, 34, 2925–2947. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.j.; Wang, B.; Yu, H.m.; Li, Q.; Hou, B.k. The Arabidopsis UGT87A2, a stress-inducible family 1 glycosyltransferase, is involved in the plant adaptation to abiotic stresses. Physiol. Plant. 2017, 159, 416–432. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Cao, J.; Chen, Q.; He, J.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. Abscisic acid receptors maintain abscisic acid homeostasis by modulating UGT71C5 glycosylation activity. J. Integr. Plant Biol. 2021, 63, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Langlois-Meurinne, M.; Gachon, C.M.; Saindrenan, P. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005, 139, 1890–1901. [Google Scholar] [CrossRef]
- Mohnike, L.; Rekhter, D.; Huang, W.; Feussner, K.; Tian, H.; Herrfurth, C.; Zhang, Y.; Feussner, I. The glycosyltransferase UGT76B1 modulates N-hydroxy-pipecolic acid homeostasis and plant immunity. Plant Cell 2021, 33, 735–749. [Google Scholar] [CrossRef]
- Song, J.T.; Koo, Y.J.; Seo, H.S.; Kim, M.C.; Do Choi, Y.; Kim, J.H. Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase, leads to increased susceptibility to Pseudomonas syringae. Phytochemistry 2008, 69, 1128–1134. [Google Scholar] [CrossRef] [PubMed]
- von Saint Paul, V.; Zhang, W.; Kanawati, B.; Geist, B.; Faus-Keßler, T.; Schmitt-Kopplin, P.; Schäffner, A.R. The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 2011, 23, 4124–4145. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Yin, G.; Xu, H.; Xiao, S.; Qin, Y.; Li, Y.; Yan, Y.; Hu, Y. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013, 13, 148. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.; Li, W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Tang, S.; Liang, H.; Yan, D.; Zhao, Y.; Han, X.; Carlson, J.E.; Xia, X.; Yin, W. Populus euphratica: The transcriptomic response to drought stress. Plant Mol. Biol. 2013, 83, 539–557. [Google Scholar] [CrossRef]
- Ruprecht, C.; Proost, S.; Hernandez-Coronado, M.; Ortiz-Ramirez, C.; Lang, D.; Rensing, S.A.; Becker, J.D.; Vandepoele, K.; Mutwil, M. Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules. Plant J. 2017, 90, 447–465. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 5773–5778. [Google Scholar] [CrossRef] [PubMed]
- Sterck, L.; Rombauts, S.; Jansson, S.; Sterky, F.; Rouzé, P.; Van de Peer, Y. EST data suggest that poplar is an ancient polyploid. New Phytol. 2005, 167, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, H.M.; Meng, X.F.; Lin, J.S.; Li, Y.J.; Hou, B.K. Ectopic expression of glycosyltransferase UGT 76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. 2018, 20, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Husar, S.; Berthiller, F.; Fujioka, S.; Rozhon, W.; Khan, M.; Kalaivanan, F.; Elias, L.; Higgins, G.S.; Li, Y.; Schuhmacher, R. Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 51. [Google Scholar] [CrossRef] [Green Version]
- Mazel, A.; Levine, A. Induction of glucosyltransferase transcription and activity during superoxide-dependent cell death in Arabidopsis plants. Plant Physiol. Biochem. 2002, 40, 133–140. [Google Scholar] [CrossRef]
- Poppenberger, B.; Fujioka, S.; Soeno, K.; George, G.L.; Vaistij, F.E.; Hiranuma, S.; Seto, H.; Takatsuto, S.; Adam, G.; Yoshida, S. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 2005, 102, 15253–15258. [Google Scholar] [CrossRef]
- Tanaka, K.; Hayashi, K.-i.; Natsume, M.; Kamiya, Y.; Sakakibara, H.; Kawaide, H.; Kasahara, H. UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 2014, 55, 218–228. [Google Scholar] [CrossRef]
- Meißner, D.; Albert, A.; Böttcher, C.; Strack, D.; Milkowski, C. The role of UDP-glucose: Hydroxycinnamate glucosyltransferases in phenylpropanoid metabolism and the response to UV-B radiation in Arabidopsis thaliana. Planta 2008, 228, 663–674. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, H.M.; Khan, U.M.; Nawaz, S.; Saleem, F.; Ahmed, N.; Rana, I.A.; Atif, R.M.; Shaheen, N.; Seo, H. Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms. Genes 2022, 13, 1640. https://doi.org/10.3390/genes13091640
Rehman HM, Khan UM, Nawaz S, Saleem F, Ahmed N, Rana IA, Atif RM, Shaheen N, Seo H. Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms. Genes. 2022; 13(9):1640. https://doi.org/10.3390/genes13091640
Chicago/Turabian StyleRehman, Hafiz Mamoon, Uzair Muhammad Khan, Sehar Nawaz, Fozia Saleem, Nisar Ahmed, Iqrar Ahmad Rana, Rana Muhammad Atif, Nabeel Shaheen, and Hyojin Seo. 2022. "Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms" Genes 13, no. 9: 1640. https://doi.org/10.3390/genes13091640
APA StyleRehman, H. M., Khan, U. M., Nawaz, S., Saleem, F., Ahmed, N., Rana, I. A., Atif, R. M., Shaheen, N., & Seo, H. (2022). Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms. Genes, 13(9), 1640. https://doi.org/10.3390/genes13091640







