Genetic Sequence Variation in the Plasmodium falciparum Histidine-Rich Protein 2 Gene from Field Isolates in Tanzania: Impact on Malaria Rapid Diagnosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Areas and Samples
2.2. Plasmodium Falciparum Detection
2.3. Pfhrp2 Exon 2 Amplification and Sequencing
2.4. Sequence Data Analysis
2.5. Statistical Analysis
3. Results
3.1. Distribution of PfHRP2 Amino Acid Repeats in Tanzania
3.2. HRP2-RDT Sensitivity Prediction in Detecting P. falciparum in Tanzania
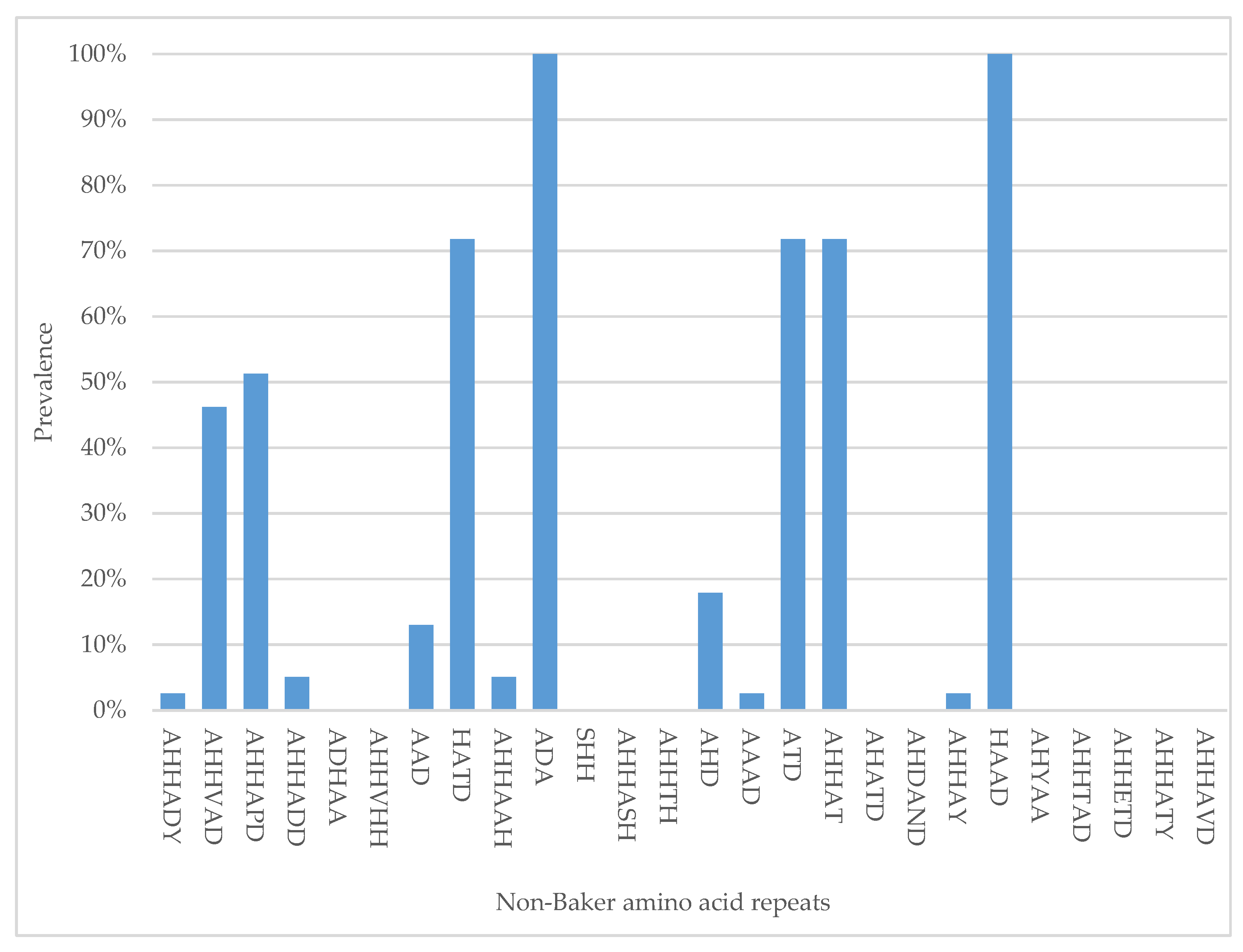
3.3. Distribution of “Non-Baker” Amino Acid Repeats
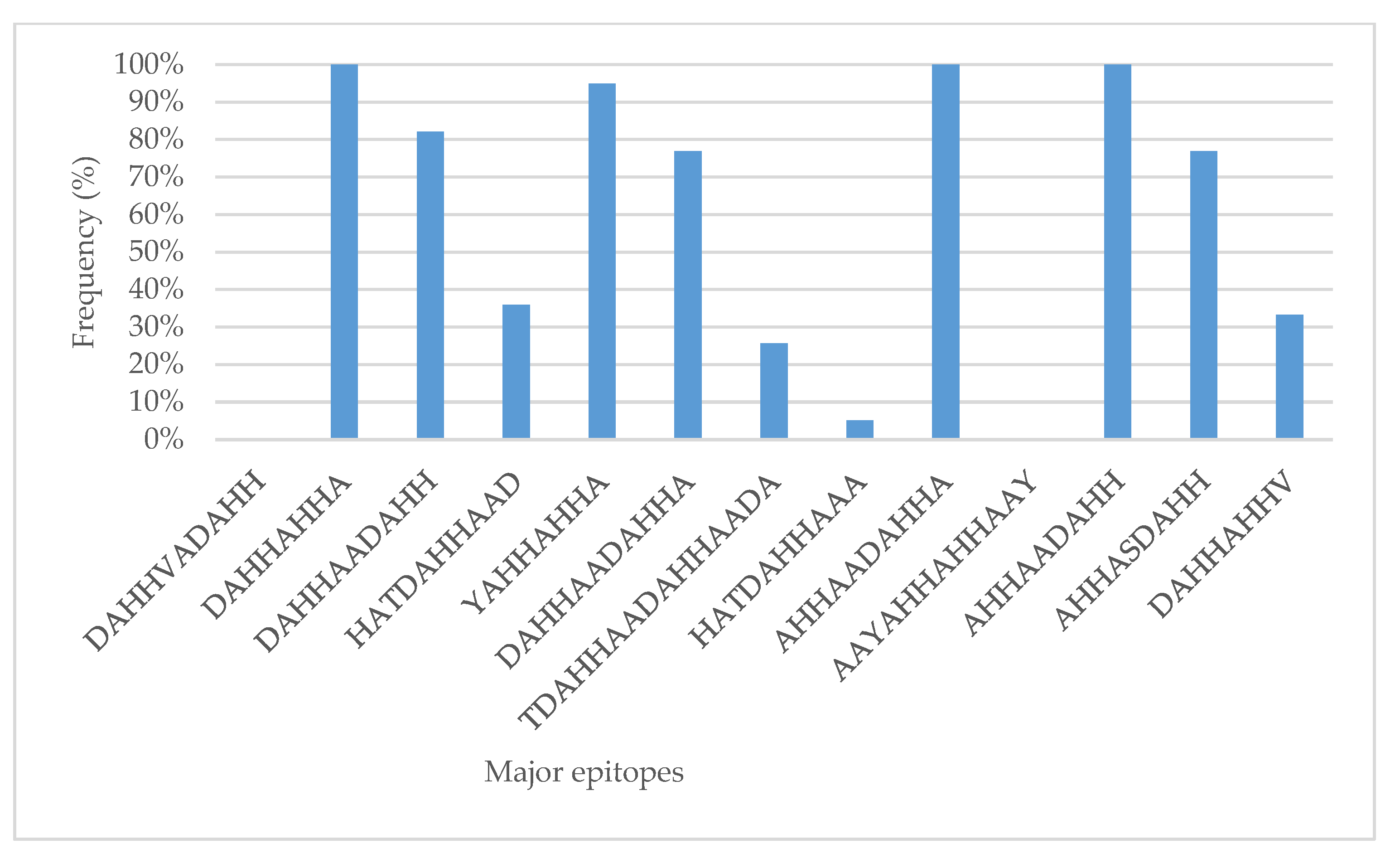
3.4. RDT Major Epitopes in Tanzania
4. Discussion
5. Conclusions
6. Study Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Guidelines for Malaria; WHO: Geneva, Switzerland, 2021; p. 225. [Google Scholar]
- Thepsamarn, P.; Prayoollawongsa, N.; Puksupa, P.; Puttoom, P.; Thaidumrong, P.; Wongchai, S.; Doddara, J.; Tantayarak, J.; Buchachart, K.; Wilairatana, P.; et al. The ICT Malaria Pf: A Simple, Rapid Dipstick Test for the Diagnosis of Plasmodium Falciparum Malaria at the Thai-Myanmar Border. Southeast Asian J. Trop. Med. Public Health 1997, 28, 723–726. [Google Scholar] [PubMed]
- Anthony, M. Rapid Diagnostic Tests for Malaria Parasites. Clin. Microbiol. Rev. 2002, 15, 66–78. [Google Scholar] [CrossRef]
- Zhao, J.; Lama, M.; Korenromp, E.; Aylward, P.; Shargie, E.; Filler, S.; Komatsu, R.; Atun, R. Adoption of Rapid Diagnostic Tests for the Diagnosis of Malaria, a Preliminary Analysis of the Global Fund Program Data, 2005 to 2010. PLoS ONE 2012, 7, e43549. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. World Malaria Report: 20 Years of Global Progress and Challenges; WHO: Geneva, Switzerland, 2020; Volume WHO/HTM/GM, ISBN 978-92-4-001579-1. [Google Scholar]
- Berhane, A.; Anderson, K.; Mihreteab, S.; Gresty, K.; Rogier, E.; Mohamed, S.; Hagos, F.; Embaye, G.; Chinorumba, A.; Zehaie, A.; et al. Major Threat to Malaria Control Programs by Plasmodium falciparum Lacking Histidine-Rich Protein 2, Eritrea. Emerg. Infect. Dis. 2018, 24, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Bakari, C.; Jones, S.; Subramaniam, G.; Mandara, C.I.; Chiduo, M.G.; Rumisha, S.; Chacky, F.; Molteni, F.; Mandike, R.; Mkude, S.; et al. Community-based surveys for Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions in selected regions of mainland Tanzania. Malar. J. 2020, 19, 391. [Google Scholar] [CrossRef]
- Vera-Arias, C.A.; Holzschuh, A.; Oduma, C.O.; Badu, K.; Abdul-Hakim, M.; Yukich, J.; Hetzel, M.W.; Fakih, B.S.; Ali, A.; Ferreira, M.U.; et al. High-throughput Plasmodium falciparum hrp2 and hrp3 gene deletion typing by digital PCR to monitor malaria rapid diagnostic test efficacy. eLife 2022, 11, e72083. [Google Scholar] [CrossRef]
- Kaaya, R.D.; Kavishe, R.A.; Tenu, F.F.; Matowo, J.J.; Mosha, F.W.; Drakeley, C.; Sutherland, C.J.; Beshir, K.B. Deletions of the Plasmodium falciparum histidine-rich protein 2/3 genes are common in field isolates from north-eastern Tanzania. Sci. Rep. 2022, 12, 5802. [Google Scholar] [CrossRef]
- Grignard, L.; Nolder, D.; Sepúlveda, N.; Berhane, A.; Mihreteab, S.; Kaaya, R.; Phelan, J.; Moser, K.; van Schalkwyk, D.A.; Campino, S.; et al. A novel multiplex qPCR assay for detection of Plasmodium falciparum with histidine-rich protein 2 and 3 (pfhrp2 and pfhrp3) deletions in polyclonal infections. EBioMedicine 2020, 55, 102757. [Google Scholar] [CrossRef]
- USAID-PMI. President’ S Malaria Initiative Tanzania Malaria Operational Plan FY 2016. Pres. Malar. Initiat. 2016, 1–79. [Google Scholar]
- Kalinga, A.K.; Kavishe, R.A.; Ishengoma, D.S.; Kagaruki, G.B.; Mweya, C.N.; Temu, L.; Chiduo, S.; Mswanya, C.; Mwanziva, C.; Mgata, S.; et al. Prevalence of asymptomatic Malaria infections in selected military camps in Tanzania. Tanzan. J. Health Res. 2019, 21, 1–11. [Google Scholar] [CrossRef]
- Yman, V.; Wandell, G.; Mutemi, D.D.; Miglar, A.; Asghar, M.; Hammar, U.; Karlsson, M.; Lind, I.; Nordfjell, C.; Rooth, I.; et al. Persistent transmission of Plasmodium malariae and Plasmodium ovale species in an area of declining Plasmodium falciparum transmission in eastern Tanzania. PLoS Negl. Trop. Dis. 2019, 13, e0007414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kavanaugh, M.J.; Azzam, S.E.; Rockabrand, D.M. Malaria Rapid Diagnostic Tests: Literary Review and Recommendation for a Quality Assurance, Quality Control Algorithm. Diagnostics 2021, 11, 768. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.D. Genomic organization, structure and possible function of histidine-rich proteins of malaria parasites. Int. J. Biochem. 1988, 20, 471–477. [Google Scholar] [CrossRef]
- Panton, L.J.; McPhie, P.; Lee Maloy, W.; Wellems, T.E.; Taylor, D.W.; Howard, R.J. Purification and partial characterization of an unusual protein of Plasmodium falciparum: Histidine-rich protein II. Mol. Biochem. Parasitol. 1989, 35, 149–160. [Google Scholar] [CrossRef]
- Ghimire, P.; Samantaray, J.C.; Mirdha, B.R.; Patra, A.K.; Panda, A.K. Purification and partial characterization of PfHRP-II protein of Plasmodium falciparum. Southeast Asian J. Trop. Med. Public Health 2003, 34, 739–743. [Google Scholar]
- Papalexis, V.; Siomos, M.A.; Campanale, N.; Guo, X.; Kocak, G.; Foley, M.; Tilley, L. Histidine-rich protein 2 of the malaria parasite, Plasmodium falciparum, is involved in detoxification of the by-products of haemoglobin degradation. Mol. Biochem. Parasitol. 2001, 115, 77–86. [Google Scholar] [CrossRef]
- Akinyi, S.; Hayden, T.; Gamboa, D.; Torres, K.; Bendezu, J.; Abdallah, J.F.; Griffing, S.M.; Quezada, W.M.; Arrospide, N.; De Oliveira, A.M.E.; et al. Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru. Sci. Rep. 2013, 3, 2797. [Google Scholar] [CrossRef]
- Wellems, T.E.; Howard, R.J. Homologous genes encode two distinct histidine-rich proteins in a cloned isolate of Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 1986, 83, 6065–6069. [Google Scholar] [CrossRef]
- Rock, E.P.; Marsh, K.; Saul, A.J.; Wellems, T.E.; Taylor, D.W.; Maloy, W.L.; Howard, R.J. Comparative analysis of the Plasmodium falciparum histidine-rich proteins HRP-I, HRP-II and HRP-III in malaria parasites of diverse origin. Parasitology 1987, 95, 209–227. [Google Scholar] [CrossRef]
- Scherf, A.; Mattei, D. Cloning and characterization of chromosome breakpoints of Plasmodium falciparum: Breakage and new telomere formation occurs frequently and randomly in subtelomeric genes. Nucleic Acids Res. 1992, 20, 1491–1496. [Google Scholar] [CrossRef]
- Lee, N.; Gatton, M.L.; Pelecanos, A.; Bubb, M.; Gonzalez, I.; Bell, D.; Cheng, Q.; McCarthy, J.S. Identification of optimal epitopes for Plasmodium falciparum rapid diagnostic tests that target histidine-rich proteins 2 and 3. J. Clin. Microbiol. 2012, 50, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Baker, J.; Andrews, K.T.; Gatton, M.L.; Bell, D.; Cheng, Q.; McCarthy, J. Effect of sequence variation in Plasmodium falciparum histidine- rich protein 2 on binding of specific monoclonal antibodies: Implications for rapid diagnostic tests for malaria. J. Clin. Microbiol. 2006, 44, 2773–2778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Addai-Mensah, O.; Dinko, B.; Noagbe, M.; Ameke, S.L.; Annani-Akollor, M.E.; Owiredu, E.-W.; Mensah, K.; Tackie, R.; Togbe, E.; Agyare-Kwabi, C.; et al. Plasmodium falciparum histidine-rich protein 2 diversity in Ghana. Malar. J. 2020, 19, 256. [Google Scholar] [CrossRef]
- Willie, N.; Zimmerman, P.A.; Mehlotra, R.K. Plasmodium falciparum Histidine-Rich Protein 2 Gene Variation in a Malaria-Endemic Area of Papua New Guinea. Am. J. Trop. Med. Hyg. 2018, 99, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.; Ho, M.-F.; Pelecanos, A.; Gatton, M.; Chen, N.; Abdullah, S.; Albertini, A.; Ariey, F.; Barnwell, J.; Bell, D.; et al. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: Implications for the performance of malaria rapid diagnostic tests. Malar. J. 2010, 9, 129. [Google Scholar] [CrossRef]
- Baker, J.; McCarthy, J.; Gatton, M.; Kyle, D.E.; Belizario, V.; Luchavez, J.; Bell, D.; Cheng, Q. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. J. Infect. Dis. 2005, 192, 870–877. [Google Scholar] [CrossRef]
- Beshir, K.B.; Sepúlveda, N.; Bharmal, J.; Robinson, A.; Mwanguzi, J.; Busula, A.O.; de Boer, J.G.; Sutherland, C.; Cunningham, J.; Hopkins, H. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci. Rep. 2017, 7, 14718. [Google Scholar] [CrossRef]
- World Health Organization. Response Plan to Pfhrp2 Gene Deletions; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Kaaya, R.D.; Kajeguka, D.C.; Matowo, J.J.; Ndaro, A.J.; Mosha, F.W.; Chilongola, J.O.; Kavishe, R.A. Predictive markers of transmission in areas with different malaria endemicity in north-eastern Tanzania based on seroprevalence of antibodies against Plasmodium falciparum. BMC Res. Notes 2021, 14, 404. [Google Scholar] [CrossRef]
- Kassam, N.A.; Kaaya, R.D.; Damian, D.J.; Schmiegelow, C.; Kavishe, R.A.; Alifrangis, M.; Wang, C.W. Ten years of monitoring malaria trend and factors associated with malaria test positivity rates in Lower Moshi. Malar. J. 2021, 20, 193. [Google Scholar] [CrossRef]
- Kruhøffer, M.; Voss, T.; Beller, K.; Scherer, M.; Cramer, J.; Deutschmann, T.; Homberg, C.; Schlumpberger, M.; Lenz, C. Evaluation of the QIAsymphony SP Workstation for Magnetic Particle—Based Nucleic Acid Purification from Different Sample Types for Demanding Downstream Applications. JALA J. Assoc. Lab. Autom. 2010, 15, 41–51. [Google Scholar] [CrossRef]
- Snounou, G.; Singh, B. Nested PCR analysis of Plasmodium parasites. Methods Mol. Med. 2002, 72, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Muwanguzi, J.; Henriques, G.; Sawa, P.; Bousema, T.; Sutherland, C.J.; Beshir, K.B. Lack of K13 mutations in Plasmodium falciparum persisting after artemisinin combination therapy treatment of Kenyan children. Malar. J. 2016, 15, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariette, N.; Barnadas, C.; Bouchier, C.; Tichit, M.; Ménard, D. Country-wide assessment of the genetic polymorphism in Plasmodium falciparum and Plasmodium vivax antigens detected with rapid diagnostic tests for malaria. Malar. J. 2008, 7, 219. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Gatton, M.L.; Barnwell, J.; Chiodini, P.; McCarthy, J.; Bell, D.; Cunningham, J. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: A review and recommendations for accurate reporting. Malar. J. 2014, 13, 283. [Google Scholar] [CrossRef] [PubMed]
- Nolder, D.; Stewart, L.; Tucker, J.; Ibrahim, A.; Gray, A.; Corrah, T.; Gallagher, C.; John, L.; O’Brien, E.; Aggarwal, D.; et al. Failure of rapid diagnostic tests in Plasmodium falciparum malaria cases among travelers to the UK and Ireland: Identification and characterisation of the parasites. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2021, 108, 137–144. [Google Scholar] [CrossRef]
- Sepúlveda, N.; Phelan, J.; Diez-Benavente, E.; Campino, S.; Clark, T.G.; Hopkins, H.; Sutherland, C.; Drakeley, C.J.; Beshir, K.B. Global analysis of Plasmodium falciparum histidine-rich protein-2 (pfhrp2) and pfhrp3 gene deletions using whole-genome sequencing data and meta-analysis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2018, 62, 211–219. [Google Scholar] [CrossRef]
- Muralidharan, V.; Oksman, A.; Iwamoto, M.; Wandless, T.J.; Goldberg, D.E. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc. Natl. Acad. Sci. USA 2011, 108, 4411–4416. [Google Scholar] [CrossRef]
- Zilversmit, M.M.; Volkman, S.K.; DePristo, M.A.; Wirth, D.F.; Awadalla, P.; Hartl, D.L. Low-complexity regions in Plasmodium falciparum: Missing links in the evolution of an extreme genome. Mol. Biol. Evol. 2010, 27, 2198–2209. [Google Scholar] [CrossRef]
- Kumar, N.; Singh, J.P.N.; Pande, V.; Mishra, N.; Srivastava, B.; Kapoor, R.; Valecha, N.; Anvikar, A.R. Genetic variation in histidine rich proteins among Indian Plasmodium falciparum population: Possible cause of variable sensitivity of malaria rapid diagnostic tests. Malar. J. 2012, 11, 298. [Google Scholar] [CrossRef]
- Wurtz, N.; Briolant, S.; Lemarié, D.; Pommier de Santi, V.; Pascual, A.; Roodt, T.; Benoit, N.; Hupin, C.; Pradines, B. Delayed diagnosis of Plasmodium falciparum in a soldier in Uganda: False-positive rapid diagnostic test associated with reduced repeats in pfhrp2. Med. Sante Trop. 2013, 23, 181–184. [Google Scholar] [CrossRef]
- Willie, N.; Mehlotra, R.K.; Howes, R.E.; Rakotomanga, T.A.; Ramboarina, S.; Ratsimbasoa, A.C.; Zimmerman, P.A. Insights into the Performance of SD Bioline Malaria Ag P.f/Pan Rapid Diagnostic Test and Plasmodium falciparum Histidine-Rich Protein 2 Gene Variation in Madagascar. Am. J. Trop. Med. Hyg. 2018, 98, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.-F.; Baker, J.; Lee, N.; Luchavez, J.; Ariey, F.; Nhem, S.; Oyibo, W.; Bell, D.; González, I.; Chiodini, P.; et al. Circulating antibodies against Plasmodium falciparum histidine-rich proteins 2 interfere with antigen detection by rapid diagnostic tests. Malar. J. 2014, 13, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arndt, L.; Koleala, T.; Orbán, Á.; Ibam, C.; Lufele, E.; Timinao, L.; Lorry, L.; Butykai, Á.; Kaman, P.; Molnár, A.P.; et al. Magneto-optical diagnosis of symptomatic malaria in Papua New Guinea. Nat. Commun. 2021, 12, 969. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.K. Clustering Nuclear Magnetic Resonance: Machine learning assistive rapid two-dimensional relaxometry mapping. Eng. Rep. 2021, 3, e12383. [Google Scholar] [CrossRef]

| AA Code | AA Type | Occurrence | Frequency |
|---|---|---|---|
| TYPE 1 | AHHAHHVAD | 29 | 38.5% |
| TYPE 2 | AHHAHHAAD | 335 | 100% |
| TYPE 3 | AHHAHHAAY | 36 | 71.8% |
| TYPE 4 | AHH | 228 | 94.9% |
| TYPE 5 | AHHAHHASD | 35 | 76.9% |
| TYPE 6 | AHHATD | 50 | 69.2% |
| TYPE 7 | AHHAAD | 122 | 89.7% |
| TYPE 8 | AHHAAY | 32 | 66.7% |
| TYPE 9 | AAY | 2 | 5.1% |
| TYPE 10 | AHHAAAHHATD | 1 | 2.6% |
| TYPE 11 | AHN | 0 | 0% |
| TYPE 12 | AHHAAAHHEAATH | 1 | 2.6% |
| TYPE 13 | AHHASD | 2 | 5.1% |
| TYPE 14 | AHHAHHATD | 5 | 10.3% |
| TYPE 15 | AHHAHHAAN | 1 | 2.6% |
| TYPE 16 | AHHAAN | 0 | 0% |
| TYPE 17 | AHHDG | 0 | 0% |
| TYPE 18 | AHHDD | 0 | 0% |
| TYPE 19 | AHHAA | 18 | 41% |
| TYPE 20 | SHHDD | 0 | 0% |
| TYPE 21 | AHHAHHATY | 0 | 0% |
| TYPE 22 | AHHAHHAGD | 0 | 0% |
| TYPE 23 | ARHAAD | 0 | 0% |
| TYPE 24 | AHHTHHAAD | 0 | 0% |
| Surveys | n | Length (aa) | Number of Individual Repeats | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 * | 2 * | 3 | 4 * | 5 | 6 * | 7 | 8 | 9 * | 10 * | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 * | 20 | 21 | 22 | 23 | 24 | |||
| Global # | 458 | 187–306 | 0–7 | 5–19 | 0–3 | 0–4 | 0–3 | 0–7 | 0–13 | 0–3 | 0–1 | 0–4 | 0–1 | 1 | 0–2 | 0–1 | - | - | - | - | 0–1 | 0–1 | 0–1 | 0–1 | 0–1 | 0–1 |
| Previous study # | 39 | 207–287 | 0–7 | 8–17 | 0–2 | 0–2 | 0–2 | 2–6 | 2–9 | 0–3 | 0 | 0–3 | 0 | 1 | 0–1 | 0–1 | - | - | - | - | 0 | 0 | 0 | 0 | 0–1 | 0 |
| Current study | 39 | 173–260 | 0–5 | 3–12 | 0–2 | 0–20 | 0–2 | 0–3 | 0–9 | 0–2 | 0–1 | 0–1 | 0 | 0–1 | 0–1 | 0–2 | 0–1 | 0 | 0 | 0 | 0–3 | 0 | 0 | 0 | 0 | 0 |
| Mean | 232 | 0.7 | 8.6 | 0.9 | 5.8 | 0.9 | 1.3 | 3.1 | 0.8 | 0.05 | 0.02 | 0 | 0.03 | 0.05 | 0.1 | 0.03 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 | 0 | |
| Median | 237 | 0 | 9 | 1 | 4 | 1 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| No | Sample | Type 2 (AHHAHHAAD) | Type 7 (AHHAAD) | Score (Type 2 × Type 7) | Sensitivity |
|---|---|---|---|---|---|
| 1 | B01_TZHRPR.ab1 | 11 | 1 | 11 | Non-sensitive |
| 2 | B02_TZHRPR.ab1 | 9 | 1 | 9 | Non-sensitive |
| 3 | B04_TZHRPR.ab1 | 9 | 1 | 9 | Non-sensitive |
| 4 | B05_TZHRPR.ab1 | 4 | 3 | 12 | Non-sensitive |
| 5 | B06_TZHRPR.ab1 | 10 | 6 | 60 | Sensitive |
| 6 | B07_TZHRPR.ab1 | 9 | 1 | 9 | Non-sensitive |
| 7 | B08_TZHRPR.ab1 | 9 | 5 | 45 | Borderline |
| 8 | B11_TZHRPR.ab1 | 8 | 2 | 16 | Non-sensitive |
| 9 | C01_TZHRPR.ab1 | 12 | 7 | 84 | Sensitive |
| 10 | C02_TZHRPR.ab1 | 10 | 6 | 60 | Sensitive |
| 11 | C03_TZHRPR.ab1 | 10 | 5 | 50 | Sensitive |
| 12 | C04_TZHRPR.ab1 | 5 | 2 | 10 | Non-sensitive |
| 13 | C06_TZHRPR.ab1 | 9 | 2 | 18 | Non-sensitive |
| 14 | C07_TZHRPR.ab1 | 10 | 2 | 20 | Non-sensitive |
| 15 | C08_TZHRPR.ab1 | 6 | 3 | 18 | Non-sensitive |
| 16 | D01_TZHRPR.ab1 | 7 | 0 | 0 | Non-sensitive |
| 17 | D03_TZHRPR.ab1 | 11 | 5 | 55 | Sensitive |
| 18 | D07_TZHRPR.ab1 | 8 | 0 | 0 | Non-sensitive |
| 19 | D11_TZHRPR.ab1 | 7 | 3 | 21 | Non-sensitive |
| 20 | E01_TZHRPR.ab1 | 9 | 2 | 18 | Non-sensitive |
| 21 | E02_TZHRPR.ab1 | 12 | 2 | 24 | Non-sensitive |
| 22 | E03_TZHRPR.ab1 | 7 | 1 | 7 | Non-sensitive |
| 23 | E04_TZHRPR.ab1 | 8 | 0 | 0 | Non-sensitive |
| 24 | E05_TZHRPR.ab1 | 10 | 2 | 20 | Non-sensitive |
| 25 | E06_TZHRPR.ab1 | 10 | 7 | 70 | Sensitive |
| 26 | E07_TZHRPR.ab1 | 6 | 9 | 54 | Sensitive |
| 27 | E08_TZHRPR.ab1 | 9 | 2 | 18 | Non-sensitive |
| 28 | E11_TZHRPR.ab1 | 4 | 2 | 8 | Non-sensitive |
| 29 | E12_TZHRPR.ab1 | 9 | 7 | 63 | Sensitive |
| 30 | G02_TZHRPR.ab1 | 11 | 3 | 33 | Non-sensitive |
| 31 | G03_TZHRPR.ab1 | 10 | 6 | 60 | Sensitive |
| 32 | G06_TZHRPR.ab1 | 11 | 5 | 55 | Sensitive |
| 33 | G07_TZHRPR.ab1 | 10 | 4 | 40 | Non-sensitive |
| 34 | G11_TZHRPR.ab1 | 9 | 1 | 9 | Non-sensitive |
| 35 | G12_TZHRPR.ab1 | 3 | 2 | 6 | Non-sensitive |
| 36 | H02_TZHRPR.ab1 | 10 | 0 | 0 | Non-sensitive |
| 37 | H03_TZHRPR.ab1 | 11 | 7 | 77 | Sensitive |
| 38 | H06_TZHRPR.ab1 | 5 | 3 | 15 | Non-sensitive |
| 39 | H07_TZHRPR.ab1 | 7 | 2 | 14 | Non-sensitive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaaya, R.D.; Amour, C.; Matowo, J.J.; Mosha, F.W.; Kavishe, R.A.; Beshir, K.B. Genetic Sequence Variation in the Plasmodium falciparum Histidine-Rich Protein 2 Gene from Field Isolates in Tanzania: Impact on Malaria Rapid Diagnosis. Genes 2022, 13, 1642. https://doi.org/10.3390/genes13091642
Kaaya RD, Amour C, Matowo JJ, Mosha FW, Kavishe RA, Beshir KB. Genetic Sequence Variation in the Plasmodium falciparum Histidine-Rich Protein 2 Gene from Field Isolates in Tanzania: Impact on Malaria Rapid Diagnosis. Genes. 2022; 13(9):1642. https://doi.org/10.3390/genes13091642
Chicago/Turabian StyleKaaya, Robert D., Caroline Amour, Johnson J. Matowo, Franklin W. Mosha, Reginald A. Kavishe, and Khalid B. Beshir. 2022. "Genetic Sequence Variation in the Plasmodium falciparum Histidine-Rich Protein 2 Gene from Field Isolates in Tanzania: Impact on Malaria Rapid Diagnosis" Genes 13, no. 9: 1642. https://doi.org/10.3390/genes13091642
APA StyleKaaya, R. D., Amour, C., Matowo, J. J., Mosha, F. W., Kavishe, R. A., & Beshir, K. B. (2022). Genetic Sequence Variation in the Plasmodium falciparum Histidine-Rich Protein 2 Gene from Field Isolates in Tanzania: Impact on Malaria Rapid Diagnosis. Genes, 13(9), 1642. https://doi.org/10.3390/genes13091642





