GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss)
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of Animals and Challenge to P. salmonis
2.2. Genotyping of Challenged Animals
2.3. Whole-Genome Sequence Data
2.4. Imputation to Whole Genome Sequence Level
2.5. Genome-Wide Association Study (GWAS)
2.6. Identification of Candidate Genes
3. Results
3.1. Summary Statistics
3.2. Quality Control of Genotypes and Imputation
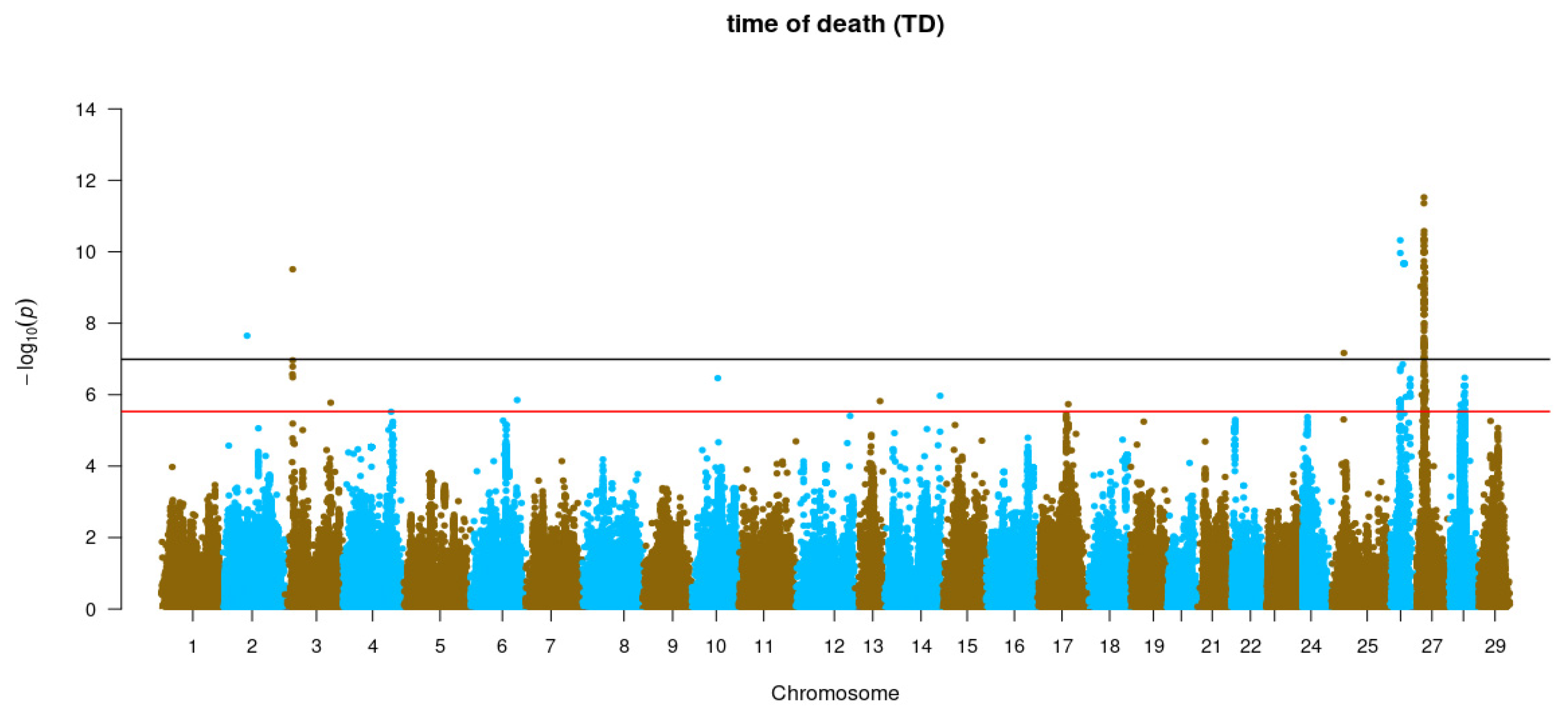
3.3. Genome-Wide Association Studies
3.4. Gene Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. El Estado Mundial de la Pesca y la Acuicultura 2022. Hacia la Transformación Azul; FAO: Rome, Italy, 2022. [Google Scholar]
- Subsecretaria de Pesca y Acuicultura (SUBPESCA). Informe Sectorial de Pesca y Acuicultura Noviembre 2021; Subsecretaria de Pesca y Acuicultura (SUBPESCA): Valparaíso, Chile, 2021. [Google Scholar]
- Sernapesca. Informe Sanitario Con Información Sanitaria De Agua Dulce Y Mar 1° semestre Año 2022; Sernapesca: Santiago, Chile, 2022; pp. 1–54.
- Pérez-Valenzuela, J.; Mejías, M.; Ortiz, D.; Salgado, P.; Montt, L.; Chávez-Báez, I.; Vera-Tamargo, F.; Mandakovic, D.; Wacyk, J.; Pulgar, P. Increased dietary availability of selenium in rainbow trout (Oncorhynchus mykiss) improves its plasma antioxidant capacity and resistance to infection with Piscirickettsia salmonis. Veterinaty Res. 2021, 52, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.; Torrealba, D.; Morales-Lange, B.; Mercado, L.; Dixon, B.; Conejeros, P.; Silva, G.; Soto, C.; Gallardo, J.A. Commercial Vaccines Do Not Confer Protection against Two Genogroups of Piscirickettsia salmonis, LF-89 and EM-90, in Atlantic Salmon. Biology 2022, 11, 993. [Google Scholar] [CrossRef] [PubMed]
- Sernapesca. Informe Sobre Uso De Antimicrobianos En La Salmonicultura Nacional Año 2021; Sernapesca: Santiago, Chile, 2022.
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Schar, D.; Klein, E.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T. Global Trends in antimicrobial use in aquaculture. Sci. Rep. 2020, 10, 21878. [Google Scholar] [CrossRef]
- Maisey, K.; Montero; Christodoulides, M. Vaccines for piscirickettsiosis (salmonid rickettsial septicaemia, SRS): The Chile perspective. Expert Rev. Vaccines 2016, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, C.; Veloso, P.; Espin, L.; Dixon, B.; Torrealba, D.; Said, I.; Afonso, J.; Soto, C.; Conejeros, P.; Gallardo, J. Host genetic variation explains reduced protection of commercial vaccines against Piscirickettsia salmonis in Atlantic salmon. Sci. Rep. 2020, 10, 18252. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P. Salmon aquaculture, Piscirickettsia salmonis virulence, and One Health: Dealing with harmful synergies between heavy antimicrobial use and piscine and human health comment on Avendaño-Herrera (2021). Aquaculture 2021, 537, 451–456. [Google Scholar] [CrossRef]
- Caruffo, M.; Vidal, S.; Santis, L.; Siel, D.; Pérez, O.; Huenchullan, P.; Sáenz, L. Effectiveness of a proteoliposome-based vaccine against salmonid rickettsial septicaemia in Oncorhynchus mykiss. Vet. Res. 2021, 52, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Ildefonso, R.; Peña, A.; Enríquez, R.; Barrientos, S.; Maldonado, L. Comparative pathogenesis of piscirickettsiosis in Atlantic salmon (Salmo salar L.) post-smolt experimentally challenged with LF-89-like and EM-90-like Piscirickettsia salmonis isolates. J. Fish Dis. 2017, 40, 1451–1472. [Google Scholar] [CrossRef]
- Rozas-Serri, M.; Peña, A.; Maldonado, L. Gene expression associated with immune response in Atlantic salmon head-kidney vaccinated with inactivated whole-cell bacterin of Piscirickettsia salmonis and pathogenic isolates. Fish Shellfish. Immunol. 2019, 93, 789–795. [Google Scholar] [CrossRef]
- Rozas-Serri, M. Why Does Piscirickettsia salmonis Break the Immunological Paradigm in Farmed Salmon? Biological Context to Understand the Relative Control of Piscirickettsiosis. Front. Immunol. 2022, 21, 856896. [Google Scholar] [CrossRef]
- Yáñez, J.; Martínez, V. Factores genéticos que inciden en la resistencia a enfermedades infecciosas en salmónidos y su aplicación en programas de mejoramiento. Arch. De Med. Vet. 2010, 42, 1–13. [Google Scholar] [CrossRef][Green Version]
- Yáñez, J.; Houston, R.; Newman, S. Genetics and genomics of disease resistance in salmonid species. Front. Genet. 2014, 5, 415. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.; Bangera, R.; Carvalheiro, R.; Correa, K.; Figueroa, R.; Lhorente, J.; Yáñez, J. Genomic prediction accuracy for resistance against Piscirickettsia salmonis in farmed rainbow trout. G3: Genes Genomes Genet. 2018, 8, 719–726. [Google Scholar] [CrossRef]
- Yáñez, J.; Peng, X.; Carvalheiro, R.; Hayes, B. Genomics applied to livestock and aquaculture breeding. Evol. Appl. 2022, 15, 517–522. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; and Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Bangera, R.; Correa, K.; Lhorente, J.; Figueroa, R.; Yáñez, J. Genomic predictions can accelerate selection for resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar). BMC Genom. 2017, 18, 121. [Google Scholar] [CrossRef]
- Barría, A.; Christensen, K.; Yoshida, G.; Correa, K.; Jedlicki, A.; Lhorente, J.; Davidson, W.; Yáñez, J. Genomic predictions and genome-wide association study of resistance against Piscirickettsia salmonis in coho salmon (Oncorhynchus kisutch) using ddRAD sequencing. G3: Genes Genomes Genet. 2018, 8, 1183–1194. [Google Scholar] [CrossRef]
- Barría, A.; Marín-Nahuelpi, R.; Cáceres, P.; López, M.; Bassini, L.; Lhorente, J.; Yáñez, J. Single-step genome-wide association study for resistance to Piscirickettsia salmonis in rainbow trout (Oncorhynchus mykiss). G3: Genes Genomes Genet. 2019, 9, 3833–3841. [Google Scholar] [CrossRef]
- McCarthy, M.; Abecasis, G.; Cardon, L.; Goldstein, D.; Little, J.; Ioannidis, J.; Hirschhorn, J. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat. Rev. Genet. 2008, 9, 356–369. [Google Scholar] [CrossRef]
- Goddard, M.; Hayes, B. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 2009, 10, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Christensen, O.F.; Janss, L.; Lund, M.S. Comparison of genomic predictions using genomic relationship matrices built with different weighting factors to account for locus-specific variances. J. Dairy Sci. 2014, 10, 6547–6559. [Google Scholar] [CrossRef] [PubMed]
- Van Binsbergen, R.; Bink, M.; Calus, M.; van Eeuwijk, F.; Hayes, B.; Hulsegge, I.; Veerkamp, R. Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle. Genet. Sel. Evol. 2014, 41, 41. [Google Scholar] [CrossRef]
- Tan, C.H.; Wu, Z.; Ren, J.; Huang, Z.; Liu, D.; He, X.; Prakapenka, D.; Zhang, R.; Li, N.; Yang, D.; et al. Genome-wide association study and accuracy of genomic prediction for teat number in Duroc pigs using genotyping-by-sequencing. Genet. Sel. Evol. 2017, 49, 35. [Google Scholar] [CrossRef]
- Zhang, H.; Yin, L.; Wang, M.; Yuan, Y.; Liu, X. Factors affecting the accuracy of genomic selection for agricultural economic traits in maize, cattle, and pig populations. Front. Genet. 2019, 10, 189. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.; Munung, N.; Vries, J.; Okada, Y.; Martin, A.; Martin, H.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. 2021, 1, 59. [Google Scholar] [CrossRef]
- Daetwyler, H.; Capitan, A.; Pausch, H.; Stothard, P.; Van Binsbergen, R.; Brøndum, R.; Liao, X.; Djari, A.; Rodriguez, S.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Rincón, J.; Legarra, A. Sequence- vs. chip-assisted genomic selection: Accurate biological information is advised. Genet. Sel. Evol. 2015, 47, 43. [Google Scholar] [CrossRef]
- MacLeod, I.; Bowman, P.; Vander, J.; Haile-Mariam, M.; Kemper, K.; Chamberlain, A.; Schrooten, C.; Hayes, B.; Goddard, E. Exploiting biological priors and sequence variants enhances QTL discovery and genomic prediction of complex traits. BMC Genom. 2016, 17, 144. [Google Scholar] [CrossRef]
- Fernandes Júnior, F.; Carvalheiro, R.; de Oliveira, H.; Sargolzaei, M.; Costilla, R.; Ventura, R.; Fonseca, L.; Neves, H.; de Hayes, B.; de Albuquerque, L.G. Imputation accuracy to whole-genome sequence in Nellore cattle. Genet. Sel. Evol. 2021, 53, 27. [Google Scholar] [CrossRef]
- Yoshida, G.; Carvalheiro, R.; Lhorente, J.; Correa, K.; Figueroa, R.; Houston, R.; Yáñez, J.M. Accuracy of genotype imputation and genomic predictions in a two-generation farmed Atlantic salmon population using high-density and low-density SNP panels. Aquaculture 2018, 491, 147–154. [Google Scholar] [CrossRef]
- Van den Berg, S.; Vandenplas, J.; van Eeuwijk, F.; Bouwman, A.; Lopes, M.; Veerkamp, R. Imputation to whole-genome sequence using multiple pig populations and its use in genome-wide association studies. Genet. Sel. Evol. 2019, 51, 2. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Duan, D.; Xue, X.; Han, X.; Wang, K.; Qiao, R.; Li, X.L.; Li, X.J. An association study on imputed whole-genome resequencing from high-throughput sequencing data for body traits in crossbred pig. Anim. Genet. 2022, 53, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Yuan, X.; Lin, X.; Gao, N.; Luo, Y.; Chen, Z.; Li, J.; Zhang, X.; Zhang, Z. Imputation from SNP chip to sequence: A case study in a Chinese indigenous chicken population. J. Anim. Sci. Biotechnol. 2018, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Teng, J.; Ye, S.; Gao, N.; Chen, Z.; Diao, S.; Li, X.; Yuan, X.; Zhang, H.; Li, J.; Zhang, X.; et al. Incorporating genomic annotation into single-step genomic prediction with imputed whole-genome sequence data. J. Integr. Agric. 2022, 4, 1126–1136. [Google Scholar] [CrossRef]
- Lopez, B.I.M.; An, N.; Srikanth, K.; Lee, S.; Oh, J.-D.; Park, W.; Chai, H.-H.; Park, J.-E.; Lim, D. Genomic Prediction Based on SNP Functional Annotation Using Imputed Whole-Genome Sequence Data in Korean Hanwoo Cattle. Front. Genet. 2021, 11, 603822. [Google Scholar] [CrossRef]
- Li, H.; Zhu, B.; Xu, L.; Wang, Z.; Xu, L.; Zhou, P.; Gao, H.; Guo, P.; Chen, Y.; Gao, X.; et al. Genomic Prediction Using LD-Based Haplotypes Inferred From High-Density Chip and Imputed Sequence Variants in Chinese Simmental Beef Cattle. Front. Genet. 2021, 12, 665382. [Google Scholar] [CrossRef]
- Mancin, E.; Sosa-Madrid, B.; Blasco, A.; Ibáñez-Escriche, N. Genotype imputation to improve the cost-efficiency of genomic selection in rabbits. Animals 2021, 11, 803. [Google Scholar] [CrossRef]
- Yoshida, G.; Yáñez, J. Increased accuracy of genomic predictions for growth under chronic thermal stress in rainbow trout by prioritizing variants from GWAS using imputed sequence data. Evol. Appl. 2021, 15, 537–552. [Google Scholar] [CrossRef]
- Garcia, B.; Yoshida, G.; Carvalheiro, R.; Yáñez, J. Accuracy of genotype imputation to whole genome sequencing level using different populations of Nile tilapia. Aquaculture 2022, 551, 737947. [Google Scholar] [CrossRef]
- Yoshida, G.; Yáñez, J. Multi-trait GWAS using imputed high-density genotypes from whole-genome sequencing identifies genes associated with body traits in Nile tilapia. BMC Genom. 2021, 22, 57. [Google Scholar] [CrossRef]
- Liu, S.; Vallejo, R.L.; Palti, Y.; Gao, G.; Marancik, D.P.; Hernandez, A.G.; Wiens, G.D. Identification of single nucleotide polymorphism markers associated with bacterial cold water disease resistance and spleen size in rainbow trout. Front. Genet. 2015, 6, 298. [Google Scholar] [CrossRef][Green Version]
- Vallejo, R.; Liu, S.; Gao, G.; Fragomeni, B.; Hernandez, A.; Leeds, T.; Parsons, J.; Martin, K.; Evenhuis, J.; Welch, T.; et al. Similar genetic architecture with shared and unique quantitative trait loci for bacterial cold water disease resistance in two rainbow trout breeding populations. Front. Genet. 2017, 8, 156. [Google Scholar] [CrossRef]
- Liu, S.; Vallejo, R.L.; Evenhuis, J.P.; Martin, K.E.; Hamilton, A.; Gao, G.; Leeds, T.D.; Wiens, G.D.; Palti, Y. Retrospective evaluation of marker-assisted selection for resistance to bacterial cold water disease in three generations of a commercial rainbow trout breeding population. Front. Genet. 2018, 9, 286. [Google Scholar] [CrossRef]
- Marana, M.; Karami, A.; Ødegård, J.; Zuo, S.; Jaafar, R.; Mathiessen, H.; Jørgensen, V.L.; Kania, P.; Dalsgaard, I.; Nielsen, T.; et al. Whole-genome association study searching for QTL for Aeromonas salmonicida resistance in rainbow trout. Sci. Rep. 2021, 11, 17857. [Google Scholar] [CrossRef]
- Karami, A.M.; Ødegård, J.; Marana, M.H.; Zuo, S.; Jaafar, R.; Mathiessen, H.; von Gersdorff, J.; Kania, P.W.; Dalsgaard, I.; Nielsen, T.; et al. A Major QTL for Resistance to Vibrio anguillarum in Rainbow Trout. Front. Genet. 2020, 11, 607558. [Google Scholar] [CrossRef]
- Jaafar, R.; Ødegård, J.; Mathiessen, H.; Karami, A.; Marana, A.; von Gersdorff, L.J.; Zuo, S.; Nielsen, T.; Kania, P.; Buchmann, K. Quantitative trait loci (QTL) associated with resistance of rainbow trout Oncorhynchus mykiss against the parasitic ciliate Ichthyophthirius multifiliis. J. Fish Dis. 2020, 43, 1591–1602. [Google Scholar] [CrossRef]
- Rodríguez, F.; Flores-Mara, R.; Yoshida, G.; Barría, A.; Jedlicki, A.; Lhorente, J.; Reyes-López, F.; Yáñez, J. Genome-Wide Association Analysis for Resistance to Infectious Pancreatic Necrosis Virus Identifies Candidate Genes Involved in Viral Replication and Immune Response in Rainbow Trout (Oncorhynchus mykiss). G3: Genes Genomes Genet. 2019, 9, 2897–2904. [Google Scholar] [CrossRef]
- Silva, R.; Evenhuis, J.; Vallejo, R.; Gao, G.; Martin, K.; Leeds, T.; Palti, Y.; Lourenco, D.R. Whole-genome mapping of quantitative trait loci and accuracy of genomic predictions for resistance to columnaris disease in two rainbow trout breeding populations. Genet. Sel. Evol. 2019, 51, 42. [Google Scholar] [CrossRef]
- Zuo, S.; Karami, A.M.; Ødegård, J.; Mathiessen, H.; Marana, M.H.; Jaafar, R.M.; von Gersdorff, J.L.; Abdu, M.; Kania, P.W.; Dalsgaard, I.; et al. Immune gene expression and genome-wide association analysis in rainbow trout with different resistance to Yersinia ruckeri infection. Fish Shellfish. Immunol. 2020, 106, 441–450. [Google Scholar] [CrossRef]
- Correa, K.; Lhorente, J.; López, L.; Bassini, L.; Naswa, S.; Deeb, N.; Di Genova, A.; Maass, A.; Davidson, W.; Yáñez, J. Genome-wide association analysis reveals loci associated with resistance against Piscirickettsia salmonis in two Atlantic salmon (Salmo salar L.) chromosomes. BMC Genom. 2015, 16, 854. [Google Scholar] [CrossRef]
- Yáñez, J.; Yoshida, G.; Parra, A.; Correa, K.; Barría, A.; Bassini, L.; Christensen, K.; López, L.; Carvalheiro, R.; Lhorente, J.; et al. Comparative genomic analysis of three salmonid species identifies functional candidate genes involved in resistance to the intracellular bacterium Piscirickettsia salmonis. Front. Genet. 2019, 10, 00665. [Google Scholar] [CrossRef]
- Palti, Y.; Vallejo, R.; Gao, G.; Liu, S.; Hernandez, A.; Rexroad III, C.; Wiens, G. Detection and Validation of QTL AffectingBacterial Cold Water Disease Resistance inRainbow Trout Using Restriction-SiteAssociated DNA Sequencing. PLoS ONE 2015, 10, e0138435. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 5, 559–575. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A new approach for efficient genotype imputation using information from relatives. BMC Genom. 2014, 15, 478. [Google Scholar] [CrossRef]
- Yang., J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- de Leeuw, C.; Mooij, J.; Heskes, T.; Posthuma, D. MAGMA:Generalized Gene-Set Analysis of GWAS Data. PLoS Computational. Biol. 2015, 11, 11–19. [Google Scholar] [CrossRef]
- Kaplan, E.; Meier, L. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1985, 53, 457–481. [Google Scholar] [CrossRef]
- Martinez, V. Genomic Selection Applied to Piscirickettsia salmonis Resistance in Chilean Atlantic Salmon. In Proceedings of the International Plant & Animal Genome XXII, San Diego, CA, USA, 11–15 January 2014. [Google Scholar]
- Dettleff, P.; Bravo, C.; Patel, A.; Martinez, V. Patterns of Piscirickettsia salmonis load in susceptible and resistant families of Salmo salar. Fish Shellfish. Immunol. 2015, 45, 67–71. [Google Scholar] [CrossRef]
- Barría, A.; Trịnh, Q.; Mahmuddin, M.; Peñaloza, C.; Papadopoulou, A.; Gervais, O.; Chadag, V.; Benzie, J.; Houston, R. A major quantitative trait locus affecting resistance to Tilapia lake virus in farmed Nile tilapia (Oreochromis niloticus). Heredity 2021, 127, 334–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.Y.; Yuan, W.J.; Xu, J.D.; Wang, J.X. Cation-dependent mannose-6-phosphate receptor functions as a pattern recognition receptor in anti-bacterial immunity of Marsupenaeus japonicus. Dev. Comp. Immunol. 2018, 89, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Nadimpalli, S.; Yerramalla, U.; Hille-Rehfeld, A.; Figura, K. Mannose 6-phosphate receptors (MPR 300 and MPR 46) from a teleostean fish (trout). Physiol.—B Biochem. Mol. Biol. 1999, 23, 261–265. [Google Scholar] [CrossRef]
- Dahms, N.M.; Hancock, M.K. P-type lectins. Biochim. Et Biophys. Acta—Gen. Subjects 2002, 1572, 317–340. [Google Scholar] [CrossRef]
- Nolan, C.; McCarthy, M.; Eivers, E.; Jirtle, R.; Byrnes, L. Mannose 6-phosphate receptors in an ancient vertebrate, zebrafish. Dev. Genes Evol. 2006, 216, 144–151. [Google Scholar] [CrossRef]
- Pérez-Stuardo, D.; Morales-Reyes, J.; Tapia, S.; Ahumada, D.; Espinoza, A.; Soto-Herrera, V.; Brianson, B.; Ibaceta, V.; Sandino, A.; Spencer, E.; et al. Non-lysosomal activation in macrophages of atlantic salmon (Salmo salar) after infection with Piscirickettsia salmonis. Front. Immunol. 2019, 10, 434. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Asiedu, M.; Adelstein, R.; Wei, Q. A novel guanine nucleotide exchange factor MyoGEF is required for cytokinesis. Cell Cycle 2006, 5, 1234–1239. [Google Scholar] [CrossRef]
- Jiao, M.; Wu, D.; Wei, Q. Myosin II-interacting guanine nucleotide exchange factor promotes bleb retraction via stimulating cortex reassembly at the bleb membran. Mol. Biol. Cell 2018, 29, 643–656. [Google Scholar] [CrossRef]
- Ke, J.; Tian, J.; Mei, S.; Ying, P.; Yang, N.; Wang, X.; Zou, D.; Peng, X.; Yang, Y.; Zhu, Y.; et al. Genetic Predisposition to Colon and Rectal Adenocarcinoma Is Mediated by a Super-enhancer Polymorphism Coactivating CD9 and PLEKHG6. Cancer Epidemiol. Biomark. Prev. 2020, 29, 850–859. [Google Scholar] [CrossRef]
- Botwright, N.A.; Mohamed, A.R.; Slinger, J.; Lima, P.C.; Wynne, J.W. Host-Parasite Interaction of Atlantic salmon (Salmo salar) and the Ectoparasite Neoparamoeba perurans in Amoebic Gill Disease. Front. Immunol. 2021, 12, 672700. [Google Scholar] [CrossRef]
- Katakura, F.; Katzenback, B.A.; Belosevic, M. Molecular and functional characterization of erythropoietin receptor of the goldfish (Carassius auratus L.). Dev. Comp. Immunol. 2014, 45, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Malik, M.S.; Bjørgen, H.; Dhamotharan, K.; Wessel, Ø.; Koppang, E.O.; Di Cicco, E.; Hansen, E.F.; Dahle, M.K.; Rimstad, E. Erythroid Progenitor Cells in Atlantic Salmon (Salmo salar) May Be Persistently and Productively Infected with Piscine Orthoreovirus (PRV). Viruses 2019, 11, 824. [Google Scholar] [CrossRef]
- Chu, C.-Y.; Cheng, C.-H.; Yang, C.-H.; Huang, C.-J. Erythropoietins from teleosts. Cell. Mol. Life Sci. 2008, 65, 3545–3552. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, D.; Heinrich, R. Alternative Erythropoietin Receptors in the Nervous System. J. Clin. Med. 2018, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, X.; Gao, R.; Wang, K.; Zheng, W.; Liu, H. A cytokine receptor domeless promotes white spot syndrome virus infection via JAK/STAT signaling pathway in red claw crayfish Cherax quadricarinatus. Dev. Comp. Immunol. 2020, 111, 103749. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Caballero-Solares, A.; Hall, J.R.; Umasuthan, N.; Kumar, S.; Jakob, E.; Skugor, S.; Hawes, C.; Santander, J.; Taylor, R.G.; et al. Transcriptome Profiling of Atlantic Salmon (Salmo salar) Parr With Higher and Lower Pathogen Loads Following Piscirickettsia salmonis Infection. Front. Immunol. 2021, 12, 789465. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, X.; Lu, Z.; Huang, R.; Tuan, N.; Wu, J.; Yang, F.; Ge, H.; Zhong, C.; Sun, Q.; et al. Transcriptome and Metabolome Analyses of Sea Cucumbers Apostichopus japonicus in Southern China During the Summer Aestivation Period. J. Ocean. Univ. China 2021, 20, 198–212. [Google Scholar] [CrossRef]
- Kumar, R.; Cheney, K.; McKirdy, R.; Neilsen, P.; Schulz, R.; Lee, J.; Cohen, J.; Booker, G.; Callen, D. CBFA2T3-ZNF652 corepressor complex regulates transcription of the E-box gene HEB. J. Biol. Chem. 2008, 283, 19026–19038. [Google Scholar] [CrossRef]
- Brown, R.; Jacobse, J.; Anant, S.; Blunt, K.; Chen, B.; Vega, P.; Jones, C.; Pilat, J.; Revetta, R.; Gorby, A.; et al. MTG16 (CBFA2T3) regulates colonic epithelial differentiation, colitis, and tumorigenesis by repressing E protein transcription factors. bioRXiv 2022, 11, 1–41. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-wide scan of selection signatures in Dehong humped cattle for heat tolerance and disease resistance. Anim. Genet. 2019, 51, 292–299. [Google Scholar] [CrossRef]
- Alshawi, A.; Essa, A.; Al-Bayatti, S.; Hanotte, O. Genome Analysis Reveals Genetic Admixture and Signature of Selection for Productivity and Environmental Traits in Iraqi Cattle. Front. Genet. 2019, 10, 609. [Google Scholar] [CrossRef]
- Steinauer, N.; Guo, C.; Zhang, J. The transcriptional corepressor CBFA2T3 inhibits all- trans-retinoic acid-induced myeloid gene expression and differentiation in acute myeloid leukemia. J. Biol. Chem. 2020, 295, 8887–8900. [Google Scholar] [CrossRef]
- Kasuya, Y.; Kim, J.-D.; Hatano, M.; Tatsumi, K.; Matsuda, S. Pathophysiological Roles of Stress-Activated Protein Kinases in Pulmonary Fibrosis. Int. J. Mol. Sci. 2021, 11, 6041. [Google Scholar] [CrossRef]
- Gordon, E.A.; Whisenan, T.C.; Zeller, M.; Kaake, R.M.; Gordon, W.M.; Krotee, P.; Patel, V.; Huang, L.; Baldi, P.; Bardwell, L. Combining docking site and phosphosite predictions to find new substrates: Identification of smoothelin-like-2 (SMTNL2) as a c-Jun N-terminal kinase (JNK) substrate. Cell. Signal. 2013, 25, 2518–2529. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Zhang, R.; Li, D.; Gao, M.Q. LRRC75A antisense lncRNA1 knockout attenuates inflammatory responses of bovine mammary epithelial cells. Int. J. Biol. Sci. 2020, 16, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, J.; Huang, Z.; Jing, H.; Yin, B.; Guo, S.; Deng, G.; Guo, M. Exosomal lnc-AFTR as a novel translation regulator of FAS ameliorates Staphylococcus aureus-induced mastitis. BioFactors 2021, 48, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.K.; Wen, H.; Ting, J.P.Y. The Inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef]
- Lin, S.; Ke, C.; Liu, L.; Gao, Y.; Xu, L.; Han, B.; Zhao, Y.; Zhang, S.; Sun, D. Genome-wide association studies for immunoglobulin concentrations in colostrum and serum in Chinese Holstein. BMC Genom. 2022, 23, 41. [Google Scholar] [CrossRef]
- Brandstaetter, H.; Kendrick-Jones, J.; Buss, F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J. Cell Sci. 2012, 125, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Nissa, M.U.; Pinto, N.; Ghosh, B.; Singh, U.; Goswami, M.; Srivastava, S. Proteomic analysis of liver tissue reveals Aeromonas hydrophila infection mediated modulation of host metabolic pathways in Labeo rohita. bioRXiv 2021. [Google Scholar] [CrossRef]
- Gomes, F.; Watanabe, L.; Vianez, J.; Nunes, M.; Cardoso, J.; Lima, C.; Schneider, H.; Sampaio, I. Comparative analysis of the transcriptome of the Amazonian fish species Colossoma macropomum (tambaqui) and hybrid tambacu by next generation sequencing. PLoS Genet. 2019, 14, e0212755. [Google Scholar] [CrossRef]
- To, V.P.T.H.; Masagounder, K.; Loewen, M.E. SLC transporters ASCT2, B0AT1-like, y+LAT1, and LAT4-like associate with methionine electrogenic and radio-isotope flux kinetics in rainbow trout intestine. Physiol. Rep. 2019, 7, e14274. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Ding, M.; Liang, Q.; Yang, Y.; Chen, M.; Wei, X.; Wang, A.; Liao, S.; Ye, J. The key differentially expressed genes and proteins related to immune response in the spleen of pufferfish (Takifugu obscurus) infected by Aeromonas hydrophila. Fish Shellfish. Immunol. 2019, 91, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, E.M.; Mason, M.W.; Camus, A.C.; Rhodes, O.E.; Parrott, B.B. Chronic low dose irradiation alters hepatic transcriptional profiles, but not global DNA methylation in medaka (Oryzias latipes). Sci. Total Environ. 2020, 729, 138680. [Google Scholar] [CrossRef] [PubMed]
- Cockcroft, S.; Garner, K. Function of the phosphatidylinositol transfer protein gene family: Is phosphatidylinositol transfer the mechanism of action? Crit. Rev. Biochem. Mol. Biol. 2011, 46, 89–117. [Google Scholar] [CrossRef] [PubMed]
- Vieira, N.M.; Spinazzola, J.M.; Alexander, M.S.; Moreira, Y.B.; Kawahara, G.; Gibbs, D.E.; Mead, L.C.; Verjovski-Almeida, S.; Zatz, M.; Kunkel, L.M. Repression of phosphatidylinositol transfer protein α ameliorates the pathology of Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA 2017, 114, 6080–6085. [Google Scholar] [CrossRef] [PubMed]
- Schurmans, S.; Vande Catsyne, C.A.; Desmet, C.; Moës, B. The phosphoinositide 5-phosphatase INPP5K: From gene structure to in vivo functions. Adv. Biol. Regul. 2021, 79, 100760. [Google Scholar] [CrossRef]
- McGrath, M.; Eramo, E.; Gurung, R.; Sriratana, A.; Gehrig, S.; Lynch, G.; Lourdes, S.; Koentgen, F.; Feeney, S.; Lazarou, L.; et al. Defective lysosome reformation during autophagy causes skeletal muscle disease. J. Clin. Investig. 2021, 4, 131. [Google Scholar] [CrossRef]
- Xiong, F.; Xiong, J.; Wu, Y.F.; Cao, L.; Huang, W.S.; Chang, M.X. Time-resolved RNA-seq provided a new understanding of intestinal immune response of European eel (Anguilla anguilla) following infection with Aeromonas hydrophila. Fish Shellfish. Immunol. 2020, 105, 297–309. [Google Scholar] [CrossRef]
- Zheng, J.; You, W.; Zheng, C.; Wan, P.; Chen, J.; Jiang, X.; Zhu, Z.; Zhang, Z.; Gong, A.; Li, W.; et al. Knockdown of fbxo39 inhibits proliferation and promotes apoptosis of human osteosarcoma u-2os cells. Oncol. Lett. 2018, 16, 1849–1854. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Zebunke, M.; Murani, E.; Trakooljul, N.; Krieter, J.; Puppe, B.; Schwerin, M.; Wimmers, K. Integrated Genome-wide association and hypothalamus eQTL studies indicate a link between the circadian rhythm-related gene PER1 and coping behavior. Sci. Rep. 2015, 5, 16264. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Chim, C.; Pang, R.; Zeng, H.; Dai, Y.; Zhang, R.; Lam, C.S.C.; Tan, V.; Hung, I.F.N.; Lan, H.; et al. XIAP-associated factor 1 (XAF1), a novel target of p53, enhances p53-mediated apoptosis via post-translational modification. Mol. Carcinog. 2012, 51, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Kim, J.; Ko, K.; Ryu, B.; Lee, M.; Kim, H.; Chi, S. XAF1 forms a positive feedback loop with IRF-1 to drive apoptotic stress response and suppress tumorigenesis. Cell Death Dis. 2018, 9, 806. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Timmerhaus, G.; Schiøtz, B.L.; Torgersen, J.; Afanasyev, S.; Iliev, D.; Jørgensen, J.; Takle, H.; Jørgensen, S.M. Genomic survey of early responses to viruses in Atlantic salmon, Salmo salar L. Mol. Immunol. 2011, 49, 163–174. [Google Scholar] [CrossRef]
- Xu, C.; Evensen, Ø.; Munang’andu, H.M. De novo assembly and transcriptome analysis of Atlantic salmon macrophage/dendritic-like TO cells following type I IFN treatment and Salmonid alphavirus subtype-3 infection. BMC Genom. 2015, 16, 96. [Google Scholar] [CrossRef]
- Riise, R.; Odqvist, L.; Mattsson, J.; Monkley, S.; Abdillahi, S.; Tyrchan, C.; Muthas, D.; Yrlid, L. Bleomycin hydrolase regulates the release of chemokines important for inflammation and wound healing by keratinocytes. Sci. Rep. 2019, 9, 20407. [Google Scholar] [CrossRef]
- Schwartz, D.R.; Homanics, G.E.; Hoyt, D.G.; Klein, E.; Abernethy, J.; Lazo, J.S. The neutral cysteine protease bleomycin hydrolase is essential for epidermal integrity and bleomycin resistance. Proc. Natl. Acad. Sci. USA 1999, 96, 4680–4685. [Google Scholar] [CrossRef]
- Fernández-Montero, Á.; Torrecillas, S.; Montero, D.; Acosta, F.; Prieto-Álamo, M.; Abril, N.; Jurado, J. Proteomic profile and protease activity in the skin mucus of greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girellae—An immunological approach. Fish Shellfish. Immunol. 2021, 110, 100–115. [Google Scholar] [CrossRef]
- Misk, E.; Huber, P.; MacInnes, J.I.; Sherif, S.M.; Abo-Ismail, M.; Lumsden, J.S. Innate response of rainbow trout gill epithelial (RTgill-W1) cell line to ultraviolet-inactivated VHSV and FliC and rhabdovirus infection. Fish Shellfish. Immunol. Rep. 2022, 3, 100043. [Google Scholar] [CrossRef]
- Dean, J.M.; He, A.; Tan, M.; Wang, J.; Lu, D.; Razani, B.; Lodhi, I. MED19 Regulates Adipogenesis and Maintenance of White Adipose Tissue Mass by Mediating PPARγ-Dependent Gene Expression. Cell Rep. 2020, 33, 108228. [Google Scholar] [CrossRef]
- Zelechower, H.; Elbert, A.E. PPARs—Receptores activados por proliferadores peroxisomales. Rev. Nefrol. Dial. Traspl. 2009, 29, 74–83. [Google Scholar]
- Chen, J.J.; Xia, X.H.; Wang, L.F.; Jia, Y.F.; Nan, P.; Li, L.; Chang, Z.J. Identification and comparison of gonadal transcripts of testis and ovary of adult common carp Cyprinus carpio using suppression subtractive hybridizatio. Theriogenology 2015, 83, 1416–1427. [Google Scholar] [CrossRef] [PubMed]
- Judycka, S.; Nynca, J.; Hliwa, P.; Ciereszko, A. Characteristics and Cryopreservation of Semen of Sex-Reversed Females of Salmonid Fish. Int. J. Mol. Sci. 2021, 22, 964. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.L.; Fox, M.A.; Timpano, K.R.; Moya, P.R.; Ren-Patterson, R.; Andrews, A.M.; Holmes, A.; Lesch, K.P.; Wendland, J.R. How the serotonin story is being rewritten by new gene-based discoveries principally related to SLC6A4, the serotonin transporter gene, which functions to influence all cellular serotonin systems. Neuropharmacology 2008, 55, 932–960. [Google Scholar] [CrossRef]
- Salem, M.; Kenney, P.B.; Rexroad, C.E.; Yao, J. Development of a 37 k high-density oligonucleotide microarray: A new tool for functional genome research in rainbow trout. J. Fish Biol. 2008, 72, 2187–2206. [Google Scholar] [CrossRef]
- Wu, X.; Yamada-Mabuchi, M.; Morris, E.J.; Tanwar, P.S.; Dobens, L.; Gluderer, S.; Khan, S.; Cao, J.; Stocker, H.; Hafen, E.; et al. The Drosophila homolog of human tumor suppressor TSC-22 promotes cellular growth, proliferation, and survival. Proc. Natl. Acad. Sci. USA 2008, 14, 5414–5419. [Google Scholar] [CrossRef]
- Dragotto, J.; Canterini, S.; Del Porto, P.; Bevilacqua, A.; Fiorenza, M.T. The interplay between TGF-β-stimulated TSC22 domain family proteins regulates cell-cycle dynamics in medulloblastoma cells. J. Cell. Physiol. 2019, 234, 18349–18360. [Google Scholar] [CrossRef]
- Kamimura, R.; Uchida, D.; Kanno, S.-I.; Shiraishi, R.; Hyodo, T.; Sawatani, Y.; Shimura, M.; Hasegawa, T.; Tsubura-Okubo, M.; Yaguchi, E.; et al. Identification of Binding Proteins for TSC22D1 Family Proteins Using Mass Spectrometry. Int. J. Mol. Sci. 2021, 22, 10913. [Google Scholar] [CrossRef]
- Vogel, P.; Hans-Jfirgen, M.; Cieslak, A.; Adermann, K.; Forssmann, W. hDIP—A potential transcriptional regulator related to murine TSC-22 and Drosophila shortsighted (shs)—Is expressed in a large number of human tissues. Biochim. Et Biophys. Acta (BBA)-Gene Struct. Expr. 1996, 1309, 200–204. [Google Scholar] [CrossRef]
- Tacchi, L.; Bron, J.E.; Taggart, J.B.; Secombes, C.J.; Bickerdike, R.; Adler, M.A.; Takle, H.; Martin, S.A. Multiple tissue transcriptomic responses to Piscirickettsia salmonis in Atlantic salmon (Salmo salar). Physiol. Genom. 2011, 43, 1241–1254. [Google Scholar] [CrossRef]
- Rodriguez-Cupello, C.; Dam, M.; Serini, L.; Wang, S.; Lindgren, D.; Englund, E.; Kjellman, P.; Axelson, H.; García-Mariscal, A.; Madsen, C.D. The STRIPAK Complex Regulates Response to Chemotherapy Through p21 and p27. Front. Cell Dev. Biol. 2020, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Dong, Y.; Qiang, R.; Zhang, Y.; Zhang, X.; Chen, Y.; Jiang, P.; Ma, X.; Wu, L.; Ai, J.; et al. Characterization of Strip1 Expression in Mouse Cochlear Hair Cells. Front. Genet. 2021, 12, 625867. [Google Scholar] [CrossRef] [PubMed]
- Kück, U.; Radchenko, D.; Teichert, I. STRIPAK, a highly conserved signaling complex, controls multiple eukaryotic cellular and developmental processes and is linked with human diseases. Biol. Chem. 2019, 400, 1005–1022. [Google Scholar] [CrossRef] [PubMed]
- Puente-Marin, S.; Nombela, I.; Ciordia, S.; Mena, M.C.; Chico, V.; Coll, J.; Ortega-Villaizan, M.D.M. In Silico Functional Networks Identified in Fish Nucleated Red Blood Cells by Means of Transcriptomic and Proteomic Profiling. Genes 2018, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Rowland, K.; Saelao, P.; Wang, Y.; Fulton, J.E.; Liebe, G.N.; McCarron, A.M.; Wolc, A.; Gallardo, R.A.; Kelly, T.; Zhou, H.; et al. Association of Candidate Genes with Response to Heat and Newcastle Disease Virus. Genes 2018, 9, 560. [Google Scholar] [CrossRef]
- Kaur, S.; Sang, Y.; Aballay, A. Myotubularin-related protein protects against neuronal degeneration mediated by oxidative stress or infection. J. Biol. Chem. 2022, 298, 101614. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Lin, Y.L.; Qin, H.; Xiong, Y.Y.; Jiang, D.L.; Lin, H.R.; Yu, Z.L.; Xia, J.L. Identifying a genome-wide QTL interval controlling for ammonia-nitrogen tolerance on chrLG1 of Nile tilapia. Aquaculture 2021, 543, 736946. [Google Scholar] [CrossRef]
- Stefanini, L.; Boulaftali, Y.; Ouellette, T.; Holinstat, M.; Désiré, L.; Leblond, B.; Andre, P.; Conley, P.; Bergmeier, W. Rap1-Rac1 Circuits Potentiate Platelet Activation. Arterioscler. Thromb. Vasc. Biol. 2011, 32, 434–441. [Google Scholar] [CrossRef]
- Nagy, Z.; Smolenski, A. Cyclic nucleotide-dependent inhibitory signaling interweaves with activating pathways to determine platelet responses. Res. Pract. Trombos. Haemost. 2018, 2, 558–571. [Google Scholar] [CrossRef]
- Neumüller, O.; Hoffmeister, M.; Babica, J.; Prelle, C.; Gegenbauer, K.; Smolenski, A.P. Synaptotagmin-like protein 1 interacts with the GTPase-activating protein Rap1GAP2 and regulates dense granule secretion in platelets. Blood. 2009, 114, 1396–1404. [Google Scholar] [CrossRef]
- Tan, S.; Wang, W.; Tian, C.; Niu, D.; Zhou, T.; Yang, Y.; Gao, D.; Liu, Z. Post-transcriptional regulation through alternative splicing after infection with Flavobacterium columnare in channel catfish (Ictalurus punctatus). Fish Shellfish. Immunol. 2019, 91, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Zhou, Q.; Li, K.; Xu, W.; Wang, L.; Hu, G.; Chen, S. Transcriptome analysis of Giant grouper (Epinephelus lanceolatus) kidney and spleen in response to spotted knifejaw iridovirus (SKIV) infection. Aquac. Res. 2020, 52, 1954–1964. [Google Scholar] [CrossRef]
- Niwa, V.; Nagata-Ohashi, K.; Takeichi, M.; Mizuno, K.; Uemura, T. Control of actin reorganization by slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 2002, 108, 233–246. [Google Scholar] [CrossRef]
- Sun, B.; Fang, Y.T.; Jin, D.J.; Chen, Z.Y.; Li, Z.Y.; Gu, X.D.; Xiang, J.B. miR-194 Inhibits the Proliferation of SW620 Colon Cancer Stem Cells Through Downregulation of SSH2 Expression. Cancer Manag. Res. 2019, 11, 10229–10238. [Google Scholar] [CrossRef]
- Zheng, C.; Li, R.; Zheng, S.; Fang, H.; Xu, M.; Zhong, L. LINC00174 Facilitates Cell Proliferation, Cell Migration and Tumor Growth of Osteosarcoma via Regulating the TGF-β/SMAD Signaling Pathway and Upregulating SSH2 Expression. Front. Mol. Biosci. 2021, 8, 697773. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Kim, K.T.; Kim, C.H.; Lee, E.Y.; Lee, S.G.; Seo, M.E.; Kim, J.H.; Chung, C.K. Unveiling the genetic variation of severe continuous/mixed-type ossification of the posterior longitudinal ligament by whole-exome sequencing and bioinformatic analysis. Spine J. 2021, 21, 1847–1856. [Google Scholar] [CrossRef] [PubMed]



| Number of samples | 2130 | 102 | 2130 |
| Initial SNPs | 46,482 | 22,649,022 | 579,960 |
| Call-rate | 38,744 | 3,787,940 | 579,960 |
| Minor allele frequency | 37,672 | 2,599,032 | 537,784 |
| Hardy–Weinberg equilibrium | 31,215 | 2,382,000 | 488,979 |
| Trait/Parameter | |||
|---|---|---|---|
| TD a | 13.59 | 36.18 | 0.27 (0.02) |
| BS b | 0.045 | 0.134 | 0.25 (0.02) |
| CHR a | START b | STOP c | p d | Name e | Trait f | Function g |
|---|---|---|---|---|---|---|
| 3 | 15,087,418 | 15,094,391 | Cation-dependent mannose-6-phosphate receptor | TD | Lysosomal | |
| 3 | 15,408,170 | 15,722,916 | Pleckstrin homology domain containing, family G (with RhoGef domain) member 6 | TD | Apoptosis | |
| 3 | 66,685,266 | 66,751,795 | TSC22 domain family protein 1 | BS | Inflammation/Apoptosis | |
| 13 | 40,702,364 | 40,707,470 | Erythropoietin receptor | TD | Inflammation | |
| 17 | 48,001,343 | 48,015,360 | Striatin-interacting protein 1 homolog | BS | Organization of the cytoskeleton | |
| 26 | 12,843,404 | 12,851,300 | N-acetylgalactosamine-6-sulfatase-like | TD | Phagocytosis | |
| 26 | 12,858,495 | 12,910,113 | protein CBFA2T3 | TD | Immune response | |
| 26 | 26,219,318 | 26,284,525 | TOX high mobility group box family member 3 | TD | Immune response | |
| 26 | 18,051,513 | 18,080,240 | Myotubularin related protein 10 | TD/BS | Lipid metabolism | |
| 26 | 21,925,171 | 21,942,499 | AP-2 complex subunit alpha-2 | BS | Immune response | |
| 27 | 9,997,966 | 10,017,806 | Smoothelin protein 2 | TD | Immune response | |
| 27 | 10,055,887 | 10,086,375 | Leucine-rich repeat-containing protein 75A | TD | Immune response | |
| 27 | 10,112,119 | 10,185,117 | Unconventional myosin-Ic | TD | Immune response | |
| 27 | 10,192,105 | 10,217,956 | Large neutral amino acids transporter small subunit 4 | TD | Organization of the cytoskeleton | |
| 27 | 10,221,995 | 10,236,446 | Phosphatidylinositol transfer protein alpha | TD | Organization of the cytoskeleton | |
| 27 | 10,238,047 | 10,250,315 | Inositol polyphosphate 5-phosphatase K | TD | Organization of the cytoskeleton | |
| 27 | 10,252,627 | 10,257,723 | Tektin-1 | TD | Organization of the cytoskeleton | |
| 27 | 10,257,875 | 10,260,754 | F-box only protein 39-like | TD | immune response | |
| 27 | 10,260,862 | 10,263,488 | XIAP-associated factor 1 | TD | Apoptosis | |
| 27 | 10,384,815 | 10,405,300 | Bleomycin Hydrolase | TD | Immune response | |
| 27 | 10,409,313 | 10,439,943 | Sodium-dependent serotonin transporter | TD | Posttraumatic stress | |
| 27 | 10,606,714 | 10,608,776 | Mediator of RNA polymerase II transcription subunit 19a | TD | Inflammation | |
| 27 | 10,300,075 | 10,379,486 | RAP1 GTPase activating protein 2a | TD/BS | immune response | |
| 27 | 10,493,554 | 10,513,502 | Protein phosphatase Slingshot homolog 2 | TD/BS | Inflammation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez-Roncancio, C.; García, B.; Gallardo-Hidalgo, J.; Yáñez, J.M. GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss). Genes 2023, 14, 114. https://doi.org/10.3390/genes14010114
Sánchez-Roncancio C, García B, Gallardo-Hidalgo J, Yáñez JM. GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss). Genes. 2023; 14(1):114. https://doi.org/10.3390/genes14010114
Chicago/Turabian StyleSánchez-Roncancio, Charles, Baltasar García, Jousepth Gallardo-Hidalgo, and José M. Yáñez. 2023. "GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss)" Genes 14, no. 1: 114. https://doi.org/10.3390/genes14010114
APA StyleSánchez-Roncancio, C., García, B., Gallardo-Hidalgo, J., & Yáñez, J. M. (2023). GWAS on Imputed Whole-Genome Sequence Variants Reveal Genes Associated with Resistance to Piscirickettsia salmonis in Rainbow Trout (Oncorhynchus mykiss). Genes, 14(1), 114. https://doi.org/10.3390/genes14010114





