The Promising Role of Non-Coding RNAs as Biomarkers and Therapeutic Targets for Leukemia
Abstract
:1. Introduction
2. Regulatory Non-Coding RNAs as Biomarkers and Therapeutic Targets in Leukemia
2.1. MicroRNAs
2.2. Long Non-Coding RNAs Role in Leukemia
2.3. Circular RNAs and Circ/Mir Axis
2.4. PiRNA Role in Leukemia
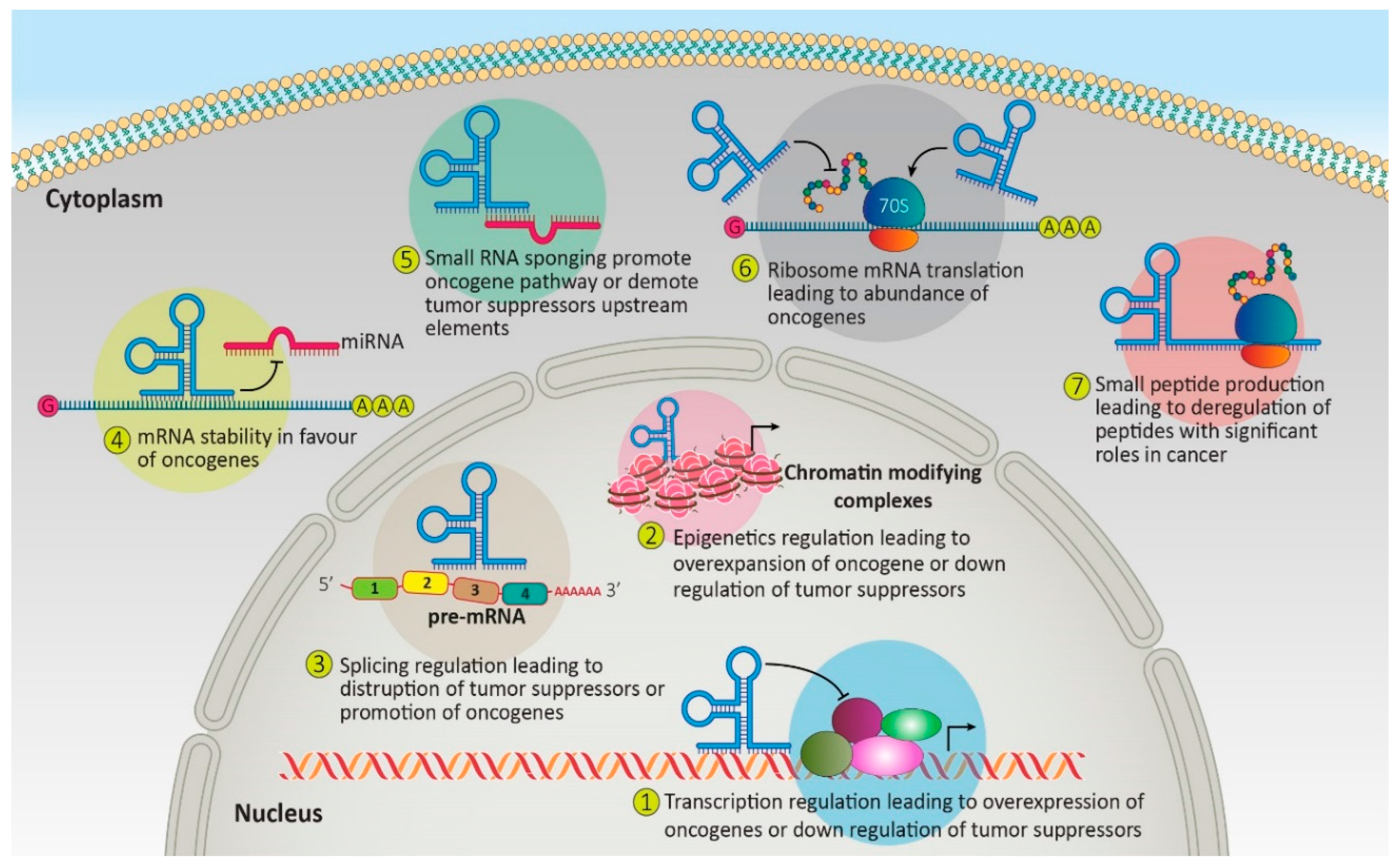
3. Non-Coding RNAs as Biomarkers and Therapeutic Targets in Leukemia
4. Non-Coding RNAs Role in Medicine and Their Limitation
5. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kantarjian, H.; Kadia, T.; DiNardo, C.; Daver, N.; Borthakur, G.; Jabbour, E.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F. Acute myeloid leukemia: Current progress and future directions. Blood Cancer J. 2021, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A. Treatment of chronic lymphocytic leukemia. N. Engl. J. Med. 2020, 383, 460–473. [Google Scholar] [CrossRef]
- Tebbi, C.K. Etiology of acute leukemia: A review. Cancers 2021, 13, 2256. [Google Scholar] [CrossRef] [PubMed]
- Nichol, J.N.; Kinal, M.; Miller, W.H. The etiology of acute leukemia. In Neoplastic Diseases of the Blood; Springer: Cham, Switzerland, 2018; pp. 161–177. [Google Scholar]
- DiNardo, C.D.; Wei, A.H. How I treat acute myeloid leukemia in the era of new drugs. Blood 2020, 135, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Shi, O.; Zeng, Q.; Lu, X.; Wang, W.; Li, Y.; Wang, Q. Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp. Hematol. 2020, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H. Precision medicine in acute lymphoblastic leukemia. Front. Med. 2020, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Hamamyh, T.; Yassin, M.A. Autoimmune hemolytic anemia in chronic myeloid leukemia. Pharmacology 2020, 105, 630–638. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-coding RNAs and their integrated networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef]
- Deogharia, M.; Gurha, P. The “guiding” principles of noncoding RNA function. Wiley Interdiscip. Rev. RNA 2022, 13, e1704. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and functions of long non-coding RNAs at multiple regulatory levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Non-Coding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Ghazimoradi, M.H.; Babashah, S. The role of CircRNA/miRNA/mRNA axis in breast cancer drug resistance. Front. Oncol. 2022, 5, 4714. [Google Scholar] [CrossRef]
- Rahnama, S.; Bakhshinejad, B.; Farzam, F.; Bitaraf, A.; Ghazimoradi, M.H.; Babashah, S. Identification of dysregulated competing endogenous RNA networks in glioblastoma: A way toward improved therapeutic opportunities. Life Sci. 2021, 277, 119488. [Google Scholar] [CrossRef]
- Fabbri, M.; Girnita, L.; Varani, G.; Calin, G.A. Decrypting noncoding RNA interactions, structures, and functional networks. Genome Res. 2019, 29, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Stav, S.; Atilho, R.M.; Mirihana Arachchilage, G.; Nguyen, G.; Higgs, G.; Breaker, R.R. Genome-wide discovery of structured noncoding RNAs in bacteria. BMC Microbiol. 2019, 19, 66. [Google Scholar] [CrossRef]
- Lambert, M.; Benmoussa, A.; Provost, P. Small non-coding RNAs derived from eukaryotic ribosomal RNA. Non-Coding RNA 2019, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Almouh, M.; Razmara, E.; Bitaraf, A.; Ghazimoradi, M.H.; Hassan, Z.M.; Babashah, S. Circular RNAs play roles in regulatory networks of cell signaling pathways in human cancers. Life Sci. 2022, 120975. [Google Scholar] [CrossRef]
- Dell’Aversana, C.; Giorgio, C.; D’Amato, L.; Lania, G.; Matarese, F.; Saeed, S.; Di Costanzo, A.; Petrizzi, V.B.; Ingenito, C.; A Martens, J.H.; et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia 2017, 31, 2315–2325. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, X.; Chen, H.; Han, Q.; Liu, C.; Sun, M. microRNA-628 inhibits the proliferation of acute myeloid leukemia cells by directly targeting IGF-1R. OncoTargets Ther. 2019, 12, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Kang, J.; Liu, L.; Chen, L.; Ren, S.; Tao, Y. MicroRNA-143 sensitizes acute myeloid leukemia cells to cytarabine via targeting ATG7-and ATG2B-dependent autophagy. Aging 2020, 12, 20111. [Google Scholar] [CrossRef] [PubMed]
- Liao, Q.; Wang, B.; Li, X.; Jiang, G. miRNAs in acute myeloid leukemia. Oncotarget 2017, 8, 3666. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, I.M.; Otero, D.; Kao, E.; Miletic, A.V.; Hother, C.; Ralfkiaer, E.; Rickert, R.C.; Gronbaek, K.; David, M. Onco-miR-155 targets SHIP1 to promote TNFα-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009, 1, 288–295. [Google Scholar] [CrossRef]
- Yamamoto, H.; Lu, J.; Oba, S.; Kawamata, T.; Yoshimi, A.; Kurosaki, N.; Yokoyama, K.; Matsushita, H.; Kurokawa, M.; Tojo, A.; et al. miR-133 regulates Evi1 expression in AML cells as a potential therapeutic target. Sci. Rep. 2016, 6, 19204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, Y.; Su, C.; Deng, T. miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol. Lett. 2016, 12, 3531–3536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Yao, Q.; Hou, Y.; Xu, M.; Liu, S.; Yang, L.; Zhang, L.; Xu, H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed. Pharmacother. 2013, 67, 381–386. [Google Scholar] [CrossRef]
- Zhou, X.; Jin, W.; Jia, H.; Yan, J.; Zhang, G. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J. Exp. Clin. Cancer Res. 2015, 34, 175–205. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Liao, W.; Peng, H.; Luo, X.; Luo, Z.; Jiang, H.; Xu, L. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J. Cancer Res. Clin. Oncol. 2016, 142, 77–87. [Google Scholar] [CrossRef]
- Gefen, N.; Binder, V.; Zaliova, M.; Linka, Y.; Morrow, M.; Novosel, A.; Edry, L.; Hertzberg, L.; Shomron, N.; Williams, O.; et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 2010, 24, 89–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coskun, E.; Neumann, M.; Schlee, C.; Liebertz, F.; Heesch, S.; Goekbuget, N.; Hoelzer, D.; Baldus, C.D. MicroRNA profiling reveals aberrant microRNA expression in adult ETP-ALL and functional studies implicate a role for miR-222 in acute leukemia. Leuk. Res. 2013, 37, 647–656. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, T.; Duan, J.; Liu, X.; Liu, L. MicroRNA 223 induced inhibition of the FBXW7 gene affects the proliferation and apoptosis of colorectal cancer cells via the Notch and Akt/mTOR pathways. Mol. Med. Rep. 2021, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Xu, X. Upregulation of miR-142-3p improves drug sensitivity of acute myelogenous leukemia through reducing P-glycoprotein and repressing autophagy by targeting HMGB1. Transl. Oncol. 2017, 10, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Yang, Y.; Chen, J.; Li, W.; Zhang, Q.; Mi, Y.; Goswami, R.S.; You, J.Q.; Lin, D.; Qian, M.D.; et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: A novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia 2017, 31, 2355–2364. [Google Scholar] [CrossRef] [PubMed]
- Kogut, S.; Paculova, H.; Rodriguez, P.; Boyd, J.; Richman, A.; Palaria, A.; Schjerven, H.; Frietze, S. Ikaros regulates microRNA networks in acute lymphoblastic leukemia. Epigenomes 2022, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.; Bhatnagar, H.; Lin, A.P.; Wang, L.; Aster, J.C.; Sill, H.; Aguiar, R.C. A microRNA-mediated regulatory loop modulates NOTCH and MYC oncogenic signals in B-and T-cell malignancies. Leukemia 2015, 29, 968–976. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Cheng, Y.; Hu, C.; Zhang, A.; Ren, Y.; Xu, X. MicroRNA-221 sensitizes chronic myeloid leukemia cells to imatinib by targeting STAT5. Leuk Lymphoma. 2018, 60, 1709–1720. [Google Scholar] [CrossRef]
- Boldrin, E.; Gaffo, E.; Niedermayer, A.; Boer, J.M.; Zimmermann, M.; Weichenhan, D.; Claus, R.; Münch, V.; Sun, Q.; Enzenmüller, S.; et al. MicroRNA-497/195 is tumor suppressive and cooperates with CDKN2A/B in pediatric acute lymphoblastic leukemia. Blood 2021, 138, 1953–1965. [Google Scholar] [CrossRef]
- Wen, F.; Cao, Y.X.; Luo, Z.Y.; Liao, P.; Lu, Z.W. LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem. Biophys. Res. Commun. 2018, 507, 1–8. [Google Scholar] [CrossRef]
- Jin, J.; Yao, J.; Yue, F.; Jin, Z.; Li, D.; Wang, S. Decreased expression of microRNA 214 contributes to imatinib mesylate resistance of chronic myeloid leukemia patients by upregulating ABCB1 gene expression. Exp. Ther. Med. 2018, 16, 1693–1700. [Google Scholar] [CrossRef] [Green Version]
- Vu, T.T.; Stölzel, F.; Wang, K.W.; Röllig, C.; Tursky, M.L.; Molloy, T.J.; Ma, D.D. miR-10a as a therapeutic target and predictive biomarker for MDM2 inhibition in acute myeloid leukemia. Leukemia 2021, 35, 1933–1948. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, S.Y.; Yan, H.Z.; Xu, D.D.; Chen, H.X.; Wang, X.Y.; Wang, X.; Liu, Y.T.; Zhang, L.; Wang, S.; et al. miR-203 inhibits proliferation and self-renewal of leukemia stem cells by targeting survivin and Bmi-1. Sci. Rep. 2016, 6, 19995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, H.; Tao, K.; Xiao, Q.; Huang, Z.; Zhong, L.; Cao, W.; Wen, J.; Feng, W. miR-29b suppresses CML cell proliferation and induces apoptosis via regulation of BCR/ABL1 protein. Exp. Cell Res. 2013, 319, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Xishan, Z.; Ziying, L.; Jing, D.; Gang, L. MicroRNA-320a acts as a tumor suppressor by targeting BCR/ABL oncogene in chronic myeloid leukemia. Sci. Rep. 2015, 5, srep12460. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.X.; Zhu, W.; Fang, C.; Fan, L.; Zou, Z.J.; Wang, Y.H.; Liu, P.; Hong, M.; Miao, K.R.; Liu, P.; et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 2012, 33, 1294–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, F.; Zhang, J.; Ji, M.; Li, P.; Du, Y.; Wang, H.; Zang, S.; Ma, D.; Sun, X.; Ji, C. miR 181b increases drug sensitivity in acute myeloid leukemia via targeting, HMGB1 and Mcl-1. Int. J. Oncol. 2014, 45, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Bai, H.; Wang, C.; Wei, D.; Qin, Y.; Xu, X. microRNA 125b promotes leukemia cell resistance to daunorubicin by inhibiting apoptosis. Mol. Med. Rep. 2014, 9, 1909–1916. [Google Scholar] [CrossRef] [Green Version]
- Ouimet, M.; Drouin, S.; Lajoie, M.; Caron, M.; St-Onge, P.; Gioia, R.; Richer, C.; Sinnett, D. A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget 2017, 8, 7477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhou, Q.; Ma, J.J. High expression of lnc-CRNDE presents as a biomarker for acute myeloid leukemia and promotes the malignant progression in acute myeloid leukemia cell line U937. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 763–770. [Google Scholar]
- Yazdi, N.; Houshmand, M.; Atashi, A.; Kazemi, A.; Najmedini, A.A.; Zarif, M.N. Long noncoding RNA PVT1: Potential oncogene in the development of acutelymphoblastic leukemia. Turk. J. Biol. 2018, 42, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zheng, P.; Wang, H.; Ai, Y.; Mao, X. Long non-coding RNA TUG1 modulates proliferation, migration, and invasion of acute myeloid leukemia cells via regulating miR-370-3p/MAPK1/ERK. OncoTargets Ther. 2019, 12, 10375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Wang, W.; Cao, L.; Li, Z.; Wang, X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol. Cell. 2016, 39, 330. [Google Scholar]
- Feng, Y.; Hu, S.; Li, L.; Zhang, S.; Liu, J.; Xu, X.; Zhang, M.; Du, T.; Du, Y.; Peng, X.; et al. LncRNA NR-104098 inhibits AML proliferation and induces differentiation through repressing EZH2 transcription by interacting with E2F1. Front. Cell Dev. Biol. 2020, 8, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Kuai, W.X.; Sun, X.Z.; Lu, X.C.; Yuan, Y.F. Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia patients and functions as a promoter by activating PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7867–7876. [Google Scholar]
- Zeng, C.; Xu, Y.; Xu, L.; Yu, X.; Cheng, J.; Yang, L.; Chen, S.; Li, Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 2014, 14, 693. [Google Scholar] [CrossRef] [Green Version]
- Eldin, R.A.; Osman, A.A.; Hassan, M.F.; Ibrahim, S.A.; El-Sakhawy, Y.N. HOTAIR expression and prognostic impact in acute myeloid leukemia patients. Egypt. J. Med. Hum. Genet. 2021, 22, 36. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.; Tang, S.; Li, M.; Feng, A.; Qin, L.; Liu, Z.; Wang, X. The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology 2017, 22, 208–216. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Bai, Y.; Kang, W.; Zhang, X.; Jiang, X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am. J. Cancer Res. 2017, 7, 1704. [Google Scholar]
- Choudhury, S.R.; Dutta, S.; Bhaduri, U.; Rao, M.R. LncRNA Hmrhl regulates expression of cancer related genes in Chronic Myelogenous Leukemia through chromatin association. NAR Cancer 2021, 3, zcab042. [Google Scholar] [CrossRef]
- Wu, D.M.; Wen, X.; Han, X.R.; Wang, S.; Wang, Y.J.; Shen, M.; Fan, S.H.; Zhang, Z.F.; Shan, Q.; Li, M.Q.; et al. Role of circular RNA DLEU2 in human acute myeloid leukemia. Mol. Cell. Biol. 2018, 38, e00259-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Chen, X.; Liu, J.; Jin, Y.; Wang, W. Circular RNA circ_0004277 Inhibits Acute Myeloid Leukemia Progression Through MicroRNA-134-5p/Single stranded DNA binding protein 2. Bioengineered 2022, 13, 9662–9673. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jin, P.; Zhang, Y.; Wang, K. CircSPI1 acts as an oncogene in acute myeloid leukemia through antagonizing SPI1 and interacting with microRNAs. Cell Death Dis. 2021, 12, 297. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Wang, W.T.; Zeng, Z.C.; Chen, T.Q.; Han, C.; Pan, Q.; Huang, W.; Fang, K.; Sun, L.Y.; Zhou, Y.F.; et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 2019, 134, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, G.; Miyamoto, T.; Jabbarzadeh-Tabrizi, S.; Iino, T.; Rocnik, J.L.; Kikushige, Y.; Mori, Y.; Shima, T.; Iwasaki, H.; Takenaka, K.; et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD–specific STAT5 activation. Blood Am. J. Hematol. 2009, 114, 5034–5043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheijen, B.; Ngo, H.T.; Kang, H.; Griffin, J.D. FLT3 receptors with internal tandem duplications promote cell viability and proliferation by signaling through Foxo proteins. Oncogene 2004, 23, 3338–3349. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Li, Y.; Wang, H.; Zhu, K.; Zhang, G. The regulation of circRNA RNF13/miRNA-1224-5p Axis promotes the malignant evolution in acute myeloid leukemia. BioMed Res. Int. 2020, 2020, 5654380. [Google Scholar] [CrossRef]
- Yuan, D.M.; Ma, J.; Fang, W.B. Identification of non-coding RNA regulatory networks in pediatric acute myeloid leukemia reveals circ-0004136 could promote cell proliferation by sponging miR-142. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9251–9258. [Google Scholar]
- Chen, J.J.; Lei, P.; Zhou, M. hsa_circ_0121582 inhibits leukemia growth by dampening Wnt/β-catenin signaling. Clin. Transl. Oncol. 2020, 22, 2293–2302. [Google Scholar] [CrossRef]
- Fan, H.; Li, Y.; Liu, C.; Liu, Y.; Bai, J.; Li, W. Circular RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem. Biophys. Res. Commun. 2018, 507, 178–184. [Google Scholar] [CrossRef]
- Hu, Q.; Gu, Y.; Chen, S.; Tian, Y.; Yang, S. Hsa_circ_0079480 promotes tumor progression in acute myeloid leukemia via miR-654-3p/HDGF axis. Aging 2021, 13, 1120. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ming, X.; Wu, J. Hsa_circ_0002483 regulates miR-758-3p/MYC axis to promote acute myeloid leukemia progression. Hematol. Oncol. 2021, 39, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and resistance to BCR-ABL1-targeted therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liang, Y.; Qin, Y.; Jiang, G.; Peng, Y.; Feng, W. circCRKL, a circRNA derived from CRKL, regulates BCR-ABL via sponging miR-877-5p to promote chronic myeloid leukemia cell proliferation. J. Transl. Med. 2022, 20, 395. [Google Scholar] [CrossRef]
- Zhong, A.N.; Yin, Y.; Tang, B.J.; Chen, L.; Shen, H.W.; Tan, Z.P.; Li, W.Q.; He, Q.; Sun, B.; Zhu, Y.; et al. CircRNA Microarray Profiling Reveals hsa_circ_0058493 as a Novel Biomarker for Imatinib-Resistant CML. Front. Pharmacol. 2021, 12, 2479. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wu, Z.; Xia, Y.; Qin, S.; Li, Y.; Wu, J.; Liang, J.; Wang, L.; Zhu, H.; Fan, L.; et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging 2019, 11, 3561. [Google Scholar] [CrossRef]
- Li, S.; Chen, J.; Fan, Y.; Xu, X.; Xiong, M.; Qi, Y.; Wu, W.; Zhao, Y. circZNF91 Promotes the Malignant Phenotype of Chronic Lymphocytic Leukemia Cells by Targeting the miR-1283/WEE1 Axis. Biomed. Res. Int. 2022, 2022, 2855394. [Google Scholar] [CrossRef]
- Papaioannou, D.; Petri, A.; Dovey, O.M.; Terreri, S.; Wang, E.; Collins, F.A.; Woodward, L.A.; Walker, A.E.; Nicolet, D.; Pepe, F.; et al. The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat. Commun. 2019, 10, 5351. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Fu, H.; Zhou, H.; Su, J.; Zhang, F.; Shen, J. Effects of novel ncRNA molecules, p15-piRNAs, on the methylation of DNA and histone H3 of the CDKN2B promoter region in U937 cells. J. Cell. Biochem. 2015, 116, 2744–2754. [Google Scholar] [CrossRef]
- Yan, H.; Wu, Q.L.; Sun, C.Y.; Ai, L.S.; Deng, J.; Zhang, L.; Chen, L.; Chu, Z.B.; Tang, B.; Wang, K.; et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 2015, 29, 196–206. [Google Scholar] [CrossRef]
- Pauli, C.; Liu, Y.; Rohde, C.; Cui, C.; Fijalkowska, D.; Gerloff, D.; Walter, C.; Krijgsveld, J.; Dugas, M.; Edemir, B.; et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood 2020, 135, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Berquet, L.; Valleron, W.; Grgurevic, S.; Quelen, C.; Zaki, O.; Quillet-Mary, A.; Davi, F.; Brousset, P.; Bousquet, M.; Ysebaert, L. Small nucleolar RNA expression profiles refine the prognostic impact of IGHV mutational status on treatment-free survival in chronic lymphocytic leukaemia. Br. J. Haematol. 2015, 172, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Babashah, S.; Soleimani, M. The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur. J. Cancer. 2011, 47, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Segal, M.; Slack, F.J. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin. Drug Discov. 2020, 15, 987–991. [Google Scholar] [CrossRef]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628. [Google Scholar] [CrossRef] [PubMed]
- Babashah, S.; Sadeghizadeh, M.; Rezaei Tavirani, M.; Farivar, S.; Soleimani, M. Aberrant microRNA expression and its implications in the pathogenesis of leukemias. Cell. Oncol. 2012, 35, 317–334. [Google Scholar] [CrossRef]
- Bi, L.; Sun, L.; Jin, Z.; Zhang, S.; Shen, Z. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol. Lett. 2018, 15, 5611–5619. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Lu, Q.; Zhu, J.; Fu, J.; Chen, Y.X. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn. Pathol. 2014, 9, 76. [Google Scholar] [CrossRef] [Green Version]
- Wallace, J.; Kagele, M.; Hu, R.; Runtsch, M.; Alexander, M.; Huffaker, T.; Mosbruger, T.; Rao, D.S.; Miles, R.R.; Round, J.; et al. MiR-155 Promotes FLT3-ITD-Induced Myeloproliferative Disease through Inhibition of Interferon Signaling. Blood 2016, 128, 2853. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood Am. J. Hematol. 2013, 122, 1891–1899. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Wang, Z.; Wang, Y.; Feng, W. Serum MicroRNA-370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int. J. Exp. Pathol. 2015, 8, 14658. [Google Scholar]
- Hornick, N.I.; Huan, J.; Doron, B.; Goloviznina, N.A.; Lapidus, J.; Chang, B.H.; Kurre, P. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci. Rep. 2015, 5, 11295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trino, S.; Lamorte, D.; Caivano, A.; Laurenzana, I.; Tagliaferri, D.; Falco, G.; Del Vecchio, L.; Musto, P.; De Luca, L. MicroRNAs as new biomarkers for diagnosis and prognosis, and as potential therapeutic targets in acute myeloid leukemia. Int. J. Mol. Sci. 2018, 19, 460. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.J.; Wu, D.H.; Zhou, J.D.; Li, X.X.; Zhang, W.; Guo, H.; Ma, J.C.; Deng, Z.Q.; Lin, J.; Qian, J. Overexpression of miR-216b: Prognostic and predictive value in acute myeloid leukemia. J. Cell. Physiol. 2018, 233, 3274–3281. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.L.; Wang, J.H.; Yang, M.; Wang, H.P.; Jin, J. MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J. Transl. Med. 2018, 16, 68. [Google Scholar] [CrossRef] [Green Version]
- Eisfeld, A.K.; Schwind, S.; Patel, R.; Huang, X.; Santhanam, R.; Walker, C.J.; Markowitz, J.; Hoag, K.W.; Jarvinen, T.M.; Leffel, B.; et al. Intronic miR-3151 within BAALC drives leukemogenesis by deregulating the TP53 pathway. Sci. Signal. 2014, 7, ra36. [Google Scholar] [CrossRef]
- Huang, Y.; Zou, Y.; Lin, L.; Ma, X.; Chen, H. Identification of serum miR-34a as a potential biomarker in acute myeloid leukemia. CBMJ 2018, 22, 799–805. [Google Scholar] [CrossRef]
- Cao, E.A.Z.; Qiu, J.; Yang, G.; Liu, Y.; Luo, W.; You, L.; Zheng, L.; Zhang, T. MiR-135a biogenesis and regulation in malignancy: A new hope for cancer research and therapy. Cancer Biol. Med. 2020, 17, 569. [Google Scholar] [CrossRef]
- Zhu, R.; Lin, W.; Zhao, W.; Fan, F.; Tang, L.; Hu, Y. A 4-microRNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. J. Cell. Biochem. 2019, 120, 3958–3968. [Google Scholar] [CrossRef]
- Seca, H.; Lima, R.; Almeida, G.; Sobrinho-Simoes, M.; Bergantim, R.; Guimaraes, J.; Helena Vasconcelos, M. Effect of miR-128 in DNA damage of HL-60 acute myeloid leukemia cells. Curr. Pharm. Biotechnol. 2014, 15, 492–502. [Google Scholar] [CrossRef] [Green Version]
- Fulci, V.; Colombo, T.; Chiaretti, S.; Messina, M.; Citarella, F.; Tavolaro, S.; Guarini, A.; Foà, R.; Macino, G. Characterization of B-and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer 2009, 48, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Luo, X.Q.; Feng, D.D.; Zhang, X.J.; Wu, J.; Zheng, Y.S.; Chen, X.; Xu, L.; Chen, Y.Q. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol. Cancer. 2011, 10, 108. [Google Scholar] [CrossRef] [Green Version]
- Ghodousi, E.S.; Aberuyi, N.; Rahgozar, S. Simultaneous changes in expression levels of BAALC and miR-326: A novel prognostic biomarker for childhood ALL. Jpn. J. Clin. Oncol. 2020, 50, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, K.J.; Wolfe, A.L.; Oricchio, E.; Palomero, T.; de Keersmaecker, K.; McJunkin, K.; Zuber, J.; James, T.; Chang, K.; Khan, A.A.; et al. Genome-wide RNAi screen identifies miR-19 targets in Notch-induced acute T-cell leukaemia (T-ALL). Nat. Cell Biol. 2010, 12, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zhao, T.; Nie, L.; Zou, Y.; Zhang, Q. MicroRNA-223 decreases cell proliferation, migration, invasion, and enhances cell apoptosis in childhood acute lymphoblastic leukemia via targeting Forkhead box O 1. Biosci. Rep. 2020, 40, 10. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, X.; He, Q.; Gao, J.; Gao, Y.; Liu, B.; Liu, F. miR-142-3p regulates the formation and differentiation of hematopoietic stem cells in vertebrates. Cell Res. 2013, 23, 1356–1368. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhang, X.; Jia, H.; Li, D.; Zhang, B.; Zhang, H.; Hong, M.; Jiang, T.; Jiang, Q.; Lu, J.; et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia 2012, 26, 769–777. [Google Scholar] [CrossRef] [Green Version]
- Correia, N.C.; Barata, J.T. MicroRNAs and their involvement in T-ALL: A brief overview. Adv. Biol. Regul. 2019, 74, 100650. [Google Scholar] [CrossRef]
- Li, Z.; Huang, H.; Chen, P.; He, M.; Li, Y.; Arnovitz, S.; Jiang, X.; He, C.; Hyjek, E.; Zhang, J.; et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012, 3, 688. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Ye, Z.; Weyand, C.M.; Goronzy, J.J. miR-181a-regulated pathways in T-cell differentiation and aging. Immun. Ageing 2021, 18, 28. [Google Scholar] [CrossRef]
- Rawoof, A.; Swaminathan, G.; Tiwari, S.; Nair, R.A.; Dinesh Kumar, L. LeukmiR: A database for miRNAs and their targets in acute lymphoblastic leukemia. Database 2020, 2020, baz151. [Google Scholar] [CrossRef] [PubMed]
- Correia, N.C.; Fragoso, R.; Carvalho, T.; Enguita, F.J.; Barata, J.T. MiR-146b negatively regulates migration and delays progression of T-cell acute lymphoblastic leukemia. Sci. Rep. 2016, 6, 31894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.A.; Zhao, D.; Wang, H.; Liang, C.; Qiao, J.; Suo, S.; Elhajmoussa, Y.; Ghoda, L.; Frankhouser, D.; Rockne, R.; et al. Microrna-142 Deficiency Promotes Chronic Myeloid Leukemia (CML) Transformation from Chronic Phase (CP) to Blast Crisis (BC). Blood 2020, 136, 4. [Google Scholar] [CrossRef]
- Habib, E.M.; Nosiar, N.A.; Eid, M.A.; Taha, A.M.; Sherief, D.E.; Hassan, A.E.; Abdel Ghafar, M.T. MiR-150 expression in chronic myeloid leukemia: Relation to imatinib response. Lab. Med. 2022, 53, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Kotagama, K.; Chang, Y.; Mangone, M. miRNAs as biomarkers in chronic myelogenous leukemia. Drug Dev. Res. 2015, 76, 278–285. [Google Scholar] [CrossRef] [Green Version]
- Srutova, K.; Curik, N.; Burda, P.; Savvulidi, F.; Silvestri, G.; Trotta, R.; Klamova, H.; Pecherkova, P.; Sovova, Z.; Koblihova, J.; et al. BCR-ABL1 mediated miR-150 downregulation through MYC contributed to myeloid differentiation block and drug resistance in chronic myeloid leukemia. Haematologica 2018, 103, 2016. [Google Scholar] [CrossRef]
- Zhao, W.; Gupta, A.; Krawczyk, J.; Gupta, S. The miR-17-92 Cluster: Yin and Yang in Human Cancers. Cancer Treat Res. Commun. 2022, 33, 100647. [Google Scholar] [CrossRef]
- Venturini, L.; Battmer, K.; Castoldi, M.; Schultheis, B.; Hochhaus, A.; Muckenthaler, M.U.; Ganser, A.; Eder, M.; Scherr, M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood 2007, 109, 4399–4405. [Google Scholar] [CrossRef]
- Mahdloo, T.; Sahami, P.; Ramezani, R.; Jafarinia, M.; Goudarzi, H.; Babashah, S. Up-regulation of miR-155 potentiates CD34+ CML stem/progenitor cells to escape from the growth-inhibitory effects of TGF-ß1 and BMP signaling. EXCLI J. 2021, 20, 748. [Google Scholar]
- Agirre, X.; Jiménez-Velasco, A.; San José-Enériz, E.; Garate, L.; Bandrés, E.; Cordeu, L.; Aparicio, O.; Saez, B.; Navarro, G.; Vilas-Zornoza, A.; et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Cancer Res. 2008, 6, 1830–1840. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Xu, Y.; Jing, Z.; Wang, X.; Zha, X.; Zeng, C.; Chen, S.; Yang, L.; Luo, G.; Li, B.; et al. Altered expression pattern of miR-29a, miR-29b and the target genes in myeloid leukemia. Exp. Hematol. Oncol. 2014, 3, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollinerova, S.; Divoka, M.; Jarosova, M.; Zapletalova, J.; Modriansky, M. miR-29 expression during imatinib treatment of chronic myeloid leukemia patients. Biomed. Pap. Med. Fac. Palacky Univ. Olomouc 2012, 156, 165. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Kasar, S.; Underbayev, C.; Hassan, M.; Ilev, I.; Degheidy, H.; Bauer, S.; Marti, G.; Lutz, C.; Raveche, E.; Batish, M. Alterations in the mir-15a/16-1 loci impairs its processing and augments B-1 expansion in de novo mouse model of chronic lymphocytic leukemia (CLL). PLoS ONE 2016, 11, e0149331. [Google Scholar] [CrossRef] [PubMed]
- Underbayev, C.; Kasar, S.; Ruezinsky, W.; Degheidy, H.; Schneider, J.S.; Marti, G.; Bauer, S.R.; Fraidenraich, D.; Lightfoote, M.M.; Parashar, V.; et al. Role of mir-15a/16-1 in early B cell development in a mouse model of chronic lymphocytic leukemia. Oncotarget 2016, 7, 60986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balatti, V.; Pekarky, Y.; Croce, C.M. Role of microRNA in chronic lymphocytic leukemia onset and progression. J. Hematol. Oncol. 2015, 8, 12. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta-Gene Regul. Mech. 2014, 1839, 1097–1109. [Google Scholar] [CrossRef]
- Graf, J.; Kretz, M. From structure to function: Route to understanding lncRNA mechanism. BioEssays 2020, 42, 2000027. [Google Scholar] [CrossRef]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Pan, J.Q.; Zhang, Y.Q.; Wang, J.H.; Xu, P.; Wang, W. lncRNA co-expression network model for the prognostic analysis of acute myeloid leukemia. Int. J. Mol. Med. 2017, 39, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, Y.; Song, H.Q.; Sun, G.W. Long non-coding RNA LINC00899 as a novel serum biomarker for diagnosis and prognosis prediction of acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7364–7370. [Google Scholar] [PubMed]
- He, C.; Wang, X.; Luo, J.; Ma, Y.; Yang, Z. Long Noncoding RNA maternally expressed Gene 3 is downregulated, and its insufficiency correlates with poor-risk stratification, worse treatment response, as well as unfavorable survival data in patients with acute myeloid leukemia. Technol. Cancer Res. Treat. 2020, 19, 1533033820945815. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Kou, D.; Liu, B.; Huang, Y.; Li, S.; Qi, Y.; Guo, Y.; Huang, T.; Qi, X.; Jia, L. LncRNA MEG3 contributes to drug resistance in acute myeloid leukemia by positively regulating ALG9 through sponging miR-155. Int. J. Lab. Hematol. 2020, 42, 464–472. [Google Scholar] [CrossRef]
- Sellers, Z.P.; Bolkun, L.; Kloczko, J.; Wojtaszewska, M.L.; Lewandowski, K.; Moniuszko, M.; Ratajczak, M.Z.; Schneider, G. Increased methylation upstream of the MEG3 promotor is observed in acute myeloid leukemia patients with better overall survival. Clin. Epigenetics 2019, 11, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghazimoradi, M.H.; Farivar, S. The role of DNA demethylation in induction of stem cells. Prog. Biophys. Mol. Biol. 2020, 153, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Ghazimoradi, M.H.; Khalafizadeh, A.; Babashah, S. A critical review on induced totipotent stem cells: Types and methods. Stem Cell Res. 2022, 63, 102857. [Google Scholar] [CrossRef]
- Lajoie, M.; Drouin, S.; Caron, M.; St-Onge, P.; Ouimet, M.; Gioia, R.; Lafond, M.H.; Vidal, R.; Richer, C.; Oualkacha, K.; et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0174124. [Google Scholar] [CrossRef] [Green Version]
- Garitano-Trojaola, A.; San José-Enériz, E.; Ezponda, T.; Unfried, J.P.; Carrasco-León, A.; Razquin, N.; Barriocanal, M.; Vilas-Zornoza, A.; Sangro, B.; Segura, V.; et al. Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget 2018, 9, 12842. [Google Scholar] [CrossRef]
- Durinck, K.; Wallaert, A.; Van de Walle, I.; Van Loocke, W.; Volders, P.J.; Vanhauwaert, S.; Geerdens, E.; Benoit, Y.; Van Roy, N.; Poppe, B.; et al. The Notch driven long non-coding RNA repertoire in T-cell acute lymphoblastic leukemia. Haematologica 2014, 99, 1808. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, A.; Kaviani, S.; Yaghmaie, M.; Pashaiefar, H.; Ahmadvand, M.; Jalili, M.; Alimoghaddam, K.; Eslamijouybari, M.; Ghavamzadeh, A. Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings. Blood Res. 2018, 53, 320. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.L.; Liu, W.; Tian, L.H.; Chai, T.T.; Liu, Y.; Zhang, F.; Fu, H.Y.; Zhou, H.R.; Shen, J.Z. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol. Rep. 2017, 38, 1353–1362. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Ju, J.K.; Han, W.N.; Shi, C.L. Long non-coding RNA (lncRNA) plasmacytoma variant translocation 1 gene (PVT1) modulates the proliferation and apoptosis of acute lymphoblastic leukemia cells by sponging miR-486-5p. Bioengineered 2022, 13, 4587–4597. [Google Scholar] [CrossRef]
- Hirano, T.; Yoshikawa, R.; Harada, H.; Harada, Y.; Ishida, A.; Yamazaki, T. Long noncoding RNA, CCDC26, controls myeloid leukemia cell growth through regulation of KIT expression. Mol. Cancer 2015, 14, 90. [Google Scholar] [CrossRef] [Green Version]
- Zeng, C.; Liu, S.; Lu, S.; Yu, X.; Lai, J.; Wu, Y.; Chen, S.; Wang, L.; Yu, Z.; Luo, G.; et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer 2018, 17, 130. [Google Scholar] [CrossRef]
- Guo, G.; Kang, Q.; Zhu, X.; Chen, Q.; Wang, X.; Chen, Y.; Ouyang, J.; Zhang, L.; Tan, H.; Chen, R.; et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene 2015, 34, 1768–1779. [Google Scholar] [CrossRef]
- Shehata, A.M.; Gohar, S.F.; Muharram, N.M.; Eldin, S.M. LncRNA CCAT2 expression at diagnosis predicts imatinib response in chronic phase chronic myeloid leukemia patients. Leuk. Res. 2022, 116, 106838. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, C.K. Long noncoding RNA SNHG5 is up-regulated and serves as a potential prognostic biomarker in acute myeloid leukemia. Eur. Rev. Med.Pharmacol. Sci. 2018, 22, 3342–3347. [Google Scholar]
- Huang, L.; Rong, Y.; Tang, X.; Yi, K.; Wu, J.; Wang, F. Circular RNAs are promising biomarkers in liquid biopsy for the diagnosis of non-small cell lung cancer. Front. Mol. Biosci. 2021, 8, 625722. [Google Scholar] [CrossRef]
- Guo, S.; Li, B.; Chen, Y.; Zou, D.; Yang, S.; Zhang, Y.; Wu, N.; Sheng, L.; Huang, H.; Ouyang, G.; et al. Hsa_circ_0012152 and Hsa_circ_0001857 accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Front. Oncol. 2020, 10, 1655. [Google Scholar] [CrossRef]
- Min, Q.H.; Wang, X.Z.; Zhang, J.; Chen, Q.G.; Li, S.Q.; Liu, X.Q.; Li, J.; Liu, J.; Yang, W.M.; Jiang, Y.H.; et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Experimental. Cell Res. 2018, 362, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol. Ther. Nucleic Acids. 2020, 20, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ben, S.; Xin, J.; Li, S.; Zheng, R.; Wang, H.; Fan, L.; Du, M.; Zhang, Z.; Wang, M. The biogenesis and biological function of PIWI-interacting RNA in cancer. J. Hematol. Oncol. 2021, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Weng, W.; Li, H.; Goel, A. Piwi-interacting RNAs (piRNAs) and cancer: Emerging biological concepts and potential clinical implications. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2019, 1871, 160–169. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Y.; Ma, N.; Sang, B.; Hu, X.; Cong, X.; Liu, Z. Overexpression of Hiwi inhibits the growth and migration of chronic myeloid leukemia cells. Cell Biochem. Biophys. 2015, 73, 117–124. [Google Scholar] [CrossRef]
- Ghaseminezhad, Z.; Sharifi, M.; Bahreini, A.; Mehrzad, V. Investigation of the expression of P-element-induced wimpy testis-interacting RNAs in human acute myeloid leukemia. Meta Gene 2022, 31, 100998. [Google Scholar] [CrossRef]
- Wilkes, M.C.; Repellin, C.E.; Sakamoto, K.M. Beyond mRNA: The role of non-coding RNAs in normal and aberrant hematopoiesis. Mol. Genet. Metab. 2017, 122, 28–38. [Google Scholar] [CrossRef]
- Goudarzi, K.M.; Lindström, M.S. Role of ribosomal protein mutations in tumor development. Int. J. Oncol. 2016, 48, 1313–1324. [Google Scholar] [CrossRef]
- Orellana, E.A.; Siegal, E.; Gregory, R.I. tRNA dysregulation and disease. Nat. Rev. Genet. 2022, 23, 651–664. [Google Scholar] [CrossRef]
- Thandapani, P.; Kloetgen, A.; Witkowski, M.T.; Glytsou, C.; Lee, A.K.; Wang, E.; Wang, J.; LeBoeuf, S.E.; Avrampou, K.; Papagiannakopoulos, T.; et al. Valine tRNA levels and availability regulate complex I assembly in leukaemia. Nature 2022, 601, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Veneziano, D.; Tomasello, L.; Balatti, V.; Palamarchuk, A.; Rassenti, L.Z.; Kipps, T.J.; Pekarsky, Y.; Croce, C.M. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2019, 116, 24252–24258. [Google Scholar] [CrossRef] [PubMed]
- Hershberger, C.E.; Daniels, N.J.; Padgett, R.A. Spliceosomal factor mutations and mis-splicing in MDS. Best Pract. Res. Clin. Haematol. 2020, 33, 101199. [Google Scholar] [CrossRef] [PubMed]
- Makishima, H.; Visconte, V.; Sakaguchi, H.; Jankowska, A.M.; Abu Kar, S.; Jerez, A.; Przychodzen, B.; Bupathi, M.; Guinta, K.; Afable, M.G.; et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood Am. J. Hematol. 2012, 119, 3203–3210. [Google Scholar] [CrossRef] [Green Version]
- Quesada, V.; Conde, L.; Villamor, N.; Ordóñez, G.R.; Jares, P.; Bassaganyas, L.; Ramsay, A.J.; Beà, S.; Pinyol, M.; Martínez-Trillos, A.; et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012, 44, 47–52. [Google Scholar] [CrossRef]
- Shuai, S.; Suzuki, H.; Diaz-Navarro, A.; Nadeu, F.; Kumar, S.A.; Gutierrez-Fernandez, A.; Delgado, J.; Pinyol, M.; López-Otín, C.; Puente, X.S.; et al. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature 2019, 574, 712–716. [Google Scholar] [CrossRef]
- Visconte, V.; O Nakashima, M.J.; Rogers, H. Mutations in splicing factor genes in myeloid malignancies: Significance and impact on clinical features. Cancers 2019, 11, 1844. [Google Scholar] [CrossRef] [Green Version]
- Kurtovic-Kozaric, A.; Przychodzen, B.; Singh, J.; Konarska, M.M.; Clemente, M.J.; Otrock, Z.K.; Nakashima, M.; His, E.D.; Yoshida, K.; Shiraishi, Y.; et al. PRPF8 defects cause missplicing in myeloid malignancies. Leukemia 2015, 29, 126–136. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, K.; Nimura, K. Regulation of RNA splicing: Aberrant splicing regulation and therapeutic targets in cancer. Cells 2021, 10, 923. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small nucleolar RNAs: Insight into their function in cancer. Front Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Köhn, M.; Misiak, D.; Bäumer, N.; Cui, C.; Göllner, S.; et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef]
- Ronchetti, D.; Mosca, L.; Cutrona, G.; Tuana, G.; Gentile, M.; Fabris, S.; Agnelli, L.; Ciceri, G.; Matis, S.; Massucco, C.; et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. BMC Med. Genet. 2013, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Werf, J.; Chin, C.V.; Fleming, N.I. SnoRNA in cancer progression, metastasis and immunotherapy response. Biology 2021, 10, 809. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Shotorbani, S.S.; Baradaran, B. RNA interference and its role in cancer therapy. Adv. Pharm. Bull. 2014, 4, 313. [Google Scholar]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Henry, S.P.; Grillone, L.R. Fomivirsen. Clin. Pharmacokinet. 2002, 41, 255–260. [Google Scholar] [CrossRef]
- Migliorati, J.M.; Liu, S.; Liu, A.; Gogate, A.; Nair, S.; Bahal, R.; Rasmussen, T.P.; Manautou, J.E.; Zhong, X.B. Absorption, Distribution, Metabolism, and Excretion of US Food and Drug Administration–Approved Antisense Oligonucleotide Drugs. Drug Metab. Dispos. 2022, 50, 888–897. [Google Scholar] [CrossRef]
- Raal, F.J.; Kallend, D.; Ray, K.K.; Turner, T.; Koenig, W.; Wright, R.S.; Wijngaard, P.L.; Curcio, D.; Jaros, M.J.; Leiter, L.A.; et al. Inclisiran for the treatment of heterozygous familial hypercholesterolemia. N. Engl. J. Med. 2020, 382, 1520–1530. [Google Scholar] [CrossRef]
- Ohanian, M.; Lin, T.L.; Ritchie, E.; Craig, M.; Agrawal, A.; Halka, K.G.; Pemmaraju, N.; Borthakur, G.; Lim, M.; Pierce, S.A.; et al. Safety and Efficacy of Lower Intensity Induction Therapy with Intravenous Prexigebersen (BP1001) in Patients with High-Risk and Relapsed/Refractory Acute Myeloid Leukemia (AML). Blood 2021, 138, 1281. [Google Scholar] [CrossRef]
- Elena, M.; Eleftheria, G.; Yiannis, S.; Lefteris, Z.C.; Michael, P.; Georgios, A.; Christos, P.C. Clinical trials of nanovesicles for drug delivery applications. In Applications of Nanovesicular Drug Delivery; Academic Press: Cambridge, MA, USA, 2022; pp. 467–486. [Google Scholar]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Qiu, S.; Adema, C.M.; Lane, T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005, 33, 1834–1847. [Google Scholar] [CrossRef] [Green Version]
- Kaczmarek, J.C.; Kowalski, P.S.; Anderson, D.G. Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Ning, B.; Yu, D.; Yu, A.M. Advances and challenges in studying noncoding RNA regulation of drug metabolism and development of RNA therapeutics. Biochem. Pharmacol. 2019, 169, 113638. [Google Scholar] [CrossRef]
- Sun, Y.M.; Chen, Y.Q. Principles and innovative technologies for decrypting noncoding RNAs: From discovery and functional prediction to clinical application. J. Hematol. Oncol. 2020, 13, 109. [Google Scholar] [CrossRef]
- Gardner, P.P.; Daub, J.; Tate, J.G.; Nawrocki, E.P.; Kolbe, D.L.; Lindgreen, S.; Wilkinson, A.C.; Finn, R.D.; Griffiths-Jones, S.; Eddy, S.R.; et al. Rfam: Updates to the RNA families database. Nucleic Acids Res. 2009, 37 (Suppl. 1), D136–D140. [Google Scholar] [CrossRef] [Green Version]
- Zeka, F.; Mestdagh, P. and Vandesompele, J. RT-qPCR-based quantification of small non-coding RNAs. In Small Non-Coding RNAs; Humana Press: New York, NY, USA, 2015; pp. 85–102. [Google Scholar]
- Grillone, K.; Riillo, C.; Scionti, F.; Rocca, R.; Tradigo, G.; Guzzi, P.H.; Alcaro, S.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020, 39, 117. [Google Scholar] [CrossRef]
- Cheng, C.; Moore, J.; Greene, C. Applications of Bioinformatics to Non-Coding RNAs in the Era of Next-Generation Sequencing. In Biocomputing; Springer: Berlin/Heidelberg, Germany, 2014; pp. 412–416. [Google Scholar]
| Type | Effector | Target | Reference(s) |
|---|---|---|---|
| miRNA | miR-194-5p | BCLAF1 | [21] |
| miR-628 | IGF-1R | [22] | |
| miR-143 | Autophagy | [23] | |
| miR-126 | SHIP1 and CEBPB | [24,25] | |
| miR-133 | Evi1 | [26] | |
| miR-223 | G1/S cell cycle | [27,28,29] | |
| miR-181a | Cell proliferation and enhanced cell cycling | [30] | |
| miR-128 | Apoptosis, genome instability, and drug sensitivity | [31] | |
| miR-125b | Cell survival and lowered apoptosis | [32] | |
| miR-222 | ETS1 | [33] | |
| miR-223 | FOXO1 | [34] | |
| miR-142-3p | Drug resistance | [35] | |
| miR-26b | PIK3CD | [36] | |
| miR-26 | Ikaros | [37] | |
| miR-30a | NOTCH1 and NOTCH2 | [38] | |
| miR-221 | STAT5 | [39] | |
| miR-497 | CDKN2A/B | [40] | |
| miR-328 | MALAT1 | [41] | |
| miR-214 | ABCB1 | [42] | |
| miR-10a | USF1 | [43] | |
| miR-203 | Proliferation | [44] | |
| miR-29b | Abl-1 | [45] | |
| miR-320 | ABL | [46] | |
| miR -181a and miR-181b | Drug resistance | [47,48] | |
| miR-125b | GRK2 and PUMA | [49] | |
| lncRNA | RP11-137H2.4 | NR3C1 | [50] |
| CRNDE | Proliferation | [51] | |
| PVT1 | Myc | [52] | |
| TUG1 | Proliferation | [53] | |
| CCAT1 | Proliferation and myeloid cell differentiation | [54] | |
| NR-104098 | EZH2 | [55] | |
| LINC00265 | PI3K-Akt | [56] | |
| NEAT1 | Proliferation and drug resistance | [57] | |
| HOTAIR | MRP1 | [58,59] | |
| SGNH5 | Drug resistance | [60] | |
| Hmrhl | Tumor development | [61] | |
| circRNA | Circ-DLEU2 | proliferation | [62] |
| Circ_0004277 | miR-134-5p and SSBP2 | [63] | |
| CircSPI1 | SPI1 | [64] | |
| CircMYBL2 | FLT3 | [65,66,67] | |
| Circ _004136 | miR-142 and miR-29a | [68] | |
| CircRNF13 | miR-1224-5p | [69] | |
| Circ_0121582 | miR-224 | [70] | |
| Circ_100290 | miR-203 | [71] | |
| Circ_0079480 | miR-655-3p | [72] | |
| Circ_0002483 | Cell proliferation and cell cycle arrest | [73] | |
| CircBA1 | Cell growth and medication resistance | [74] | |
| CircCRKL | miR-877-5p | [75] | |
| Circ-0058493 | miR-548b-3p | [76] | |
| Circ-0132266 | miR-337-3p | [77] | |
| CircZNF91 | miR-1283 | [78] | |
| HOXB-AS3 | Ribosomal RNA transcription | [79] | |
| piRNA and snoRNA | 11186 | CDKN2B | [80] |
| 823 | DNA methylation and angiogenesis | [81] | |
| SNORD42A | Proliferation | [82] | |
| SNHG12 | miR-195 | [83] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghazimoradi, M.H.; Karimpour-Fard, N.; Babashah, S. The Promising Role of Non-Coding RNAs as Biomarkers and Therapeutic Targets for Leukemia. Genes 2023, 14, 131. https://doi.org/10.3390/genes14010131
Ghazimoradi MH, Karimpour-Fard N, Babashah S. The Promising Role of Non-Coding RNAs as Biomarkers and Therapeutic Targets for Leukemia. Genes. 2023; 14(1):131. https://doi.org/10.3390/genes14010131
Chicago/Turabian StyleGhazimoradi, Mohammad H., Naeim Karimpour-Fard, and Sadegh Babashah. 2023. "The Promising Role of Non-Coding RNAs as Biomarkers and Therapeutic Targets for Leukemia" Genes 14, no. 1: 131. https://doi.org/10.3390/genes14010131
APA StyleGhazimoradi, M. H., Karimpour-Fard, N., & Babashah, S. (2023). The Promising Role of Non-Coding RNAs as Biomarkers and Therapeutic Targets for Leukemia. Genes, 14(1), 131. https://doi.org/10.3390/genes14010131





