High Coverage Mitogenomes and Y-Chromosomal Typing Reveal Ancient Lineages in the Modern-Day Székely Population in Romania
Abstract
:1. Introduction
2. Material and Methods
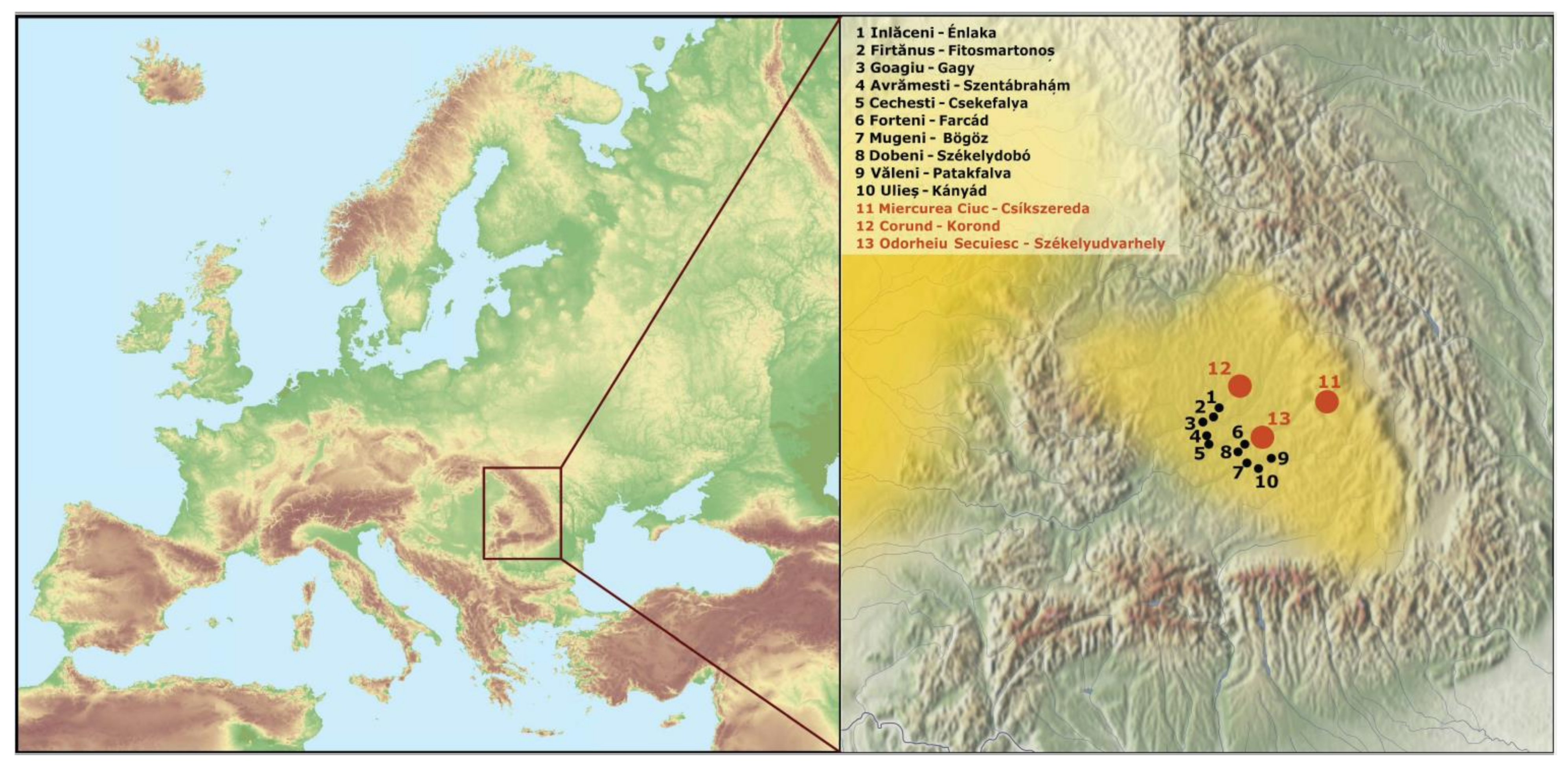
2.1. DNA Samples, Extraction, Amplification, and Sequencing
2.2. Pre-Processing of the Sequencing Data
2.3. Phylogenetic Analysis of the mtDNA
2.4. Population Genetic Analysis
2.5. Analysis of Y-STR Variations
3. Results and Discussion
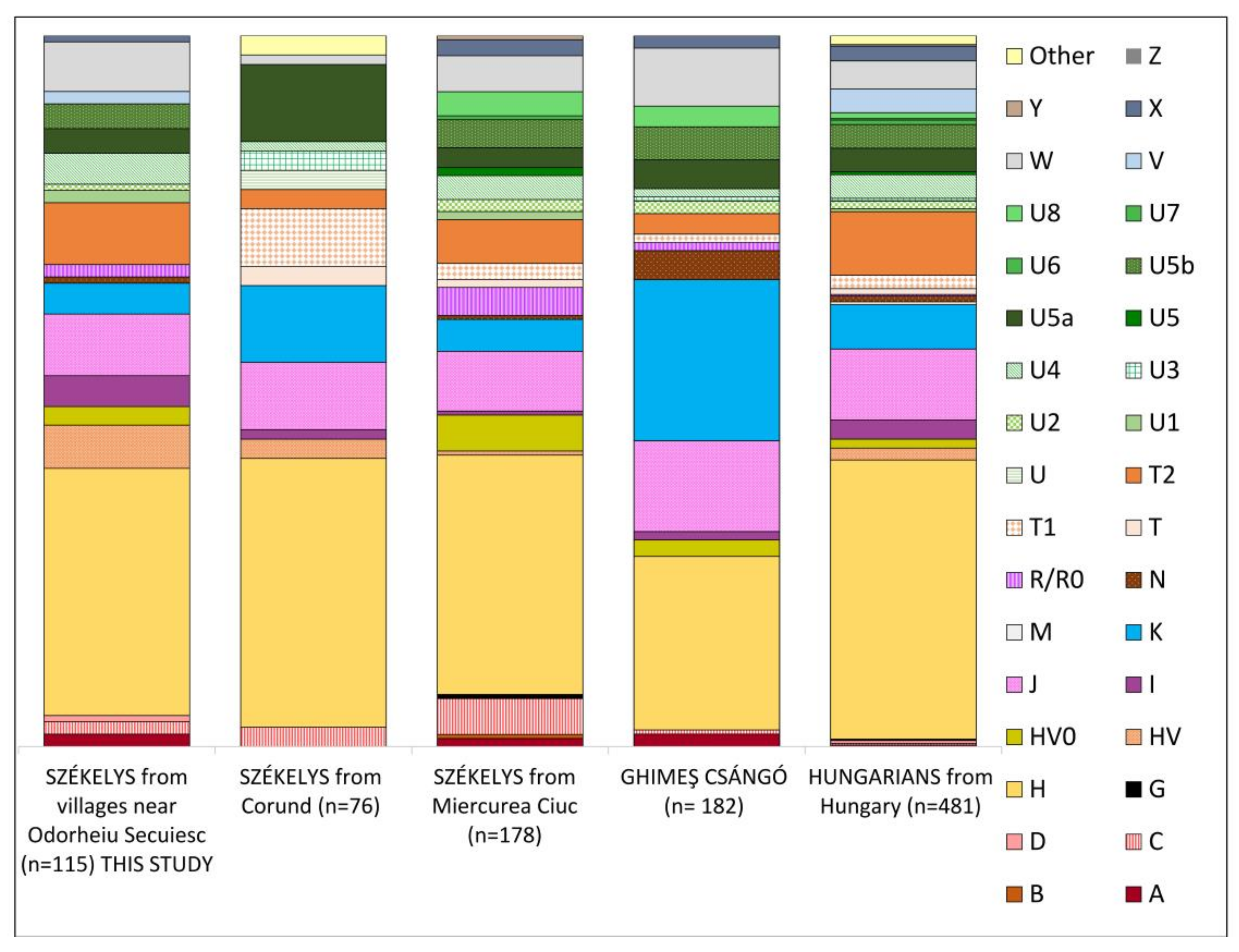
3.1. Maternal Lineages in the Dataset
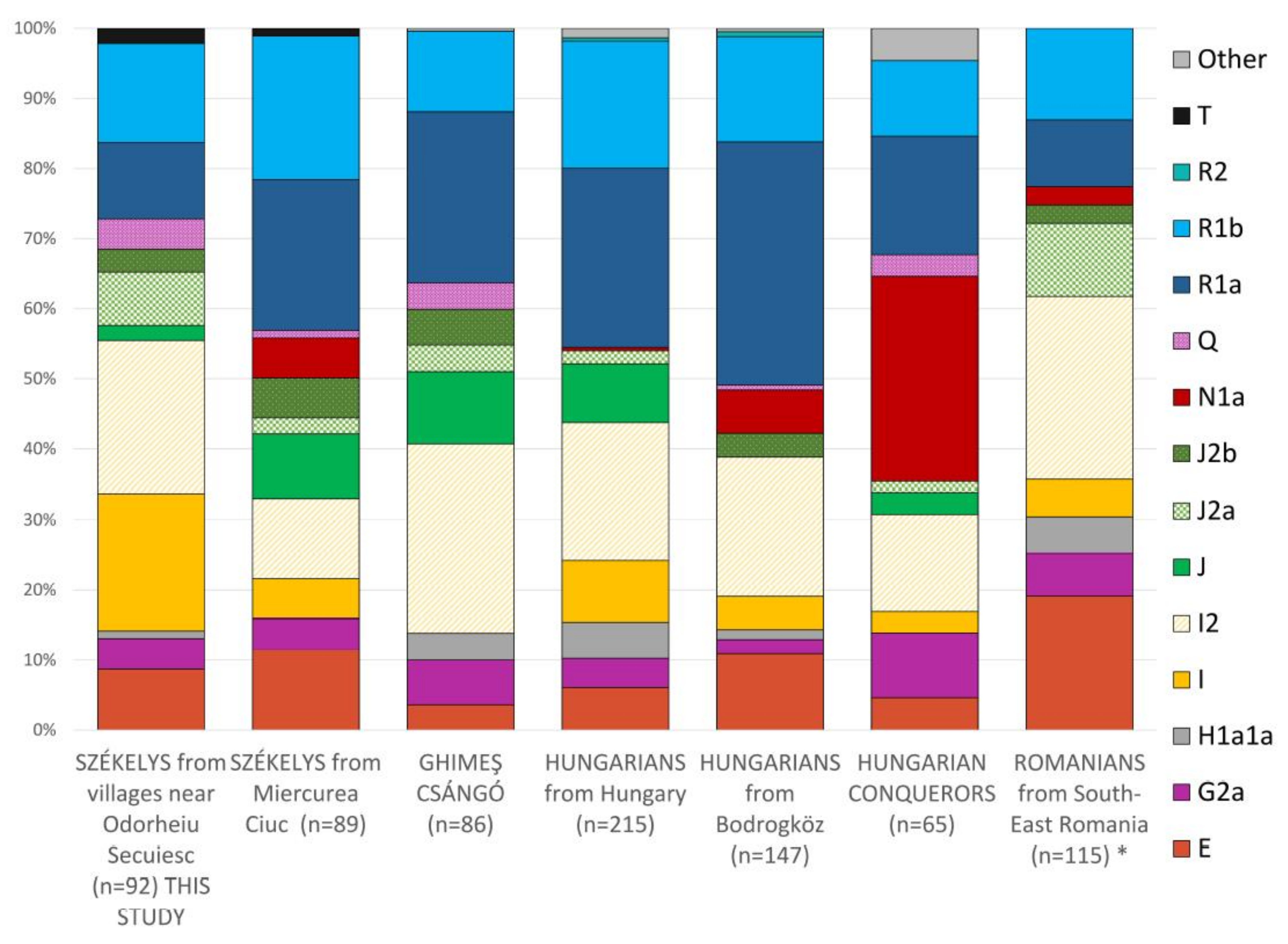
3.1.1. Haplogroup-Based Analyses
3.1.2. Whole Mitogenome Sequence-Based Evaluations
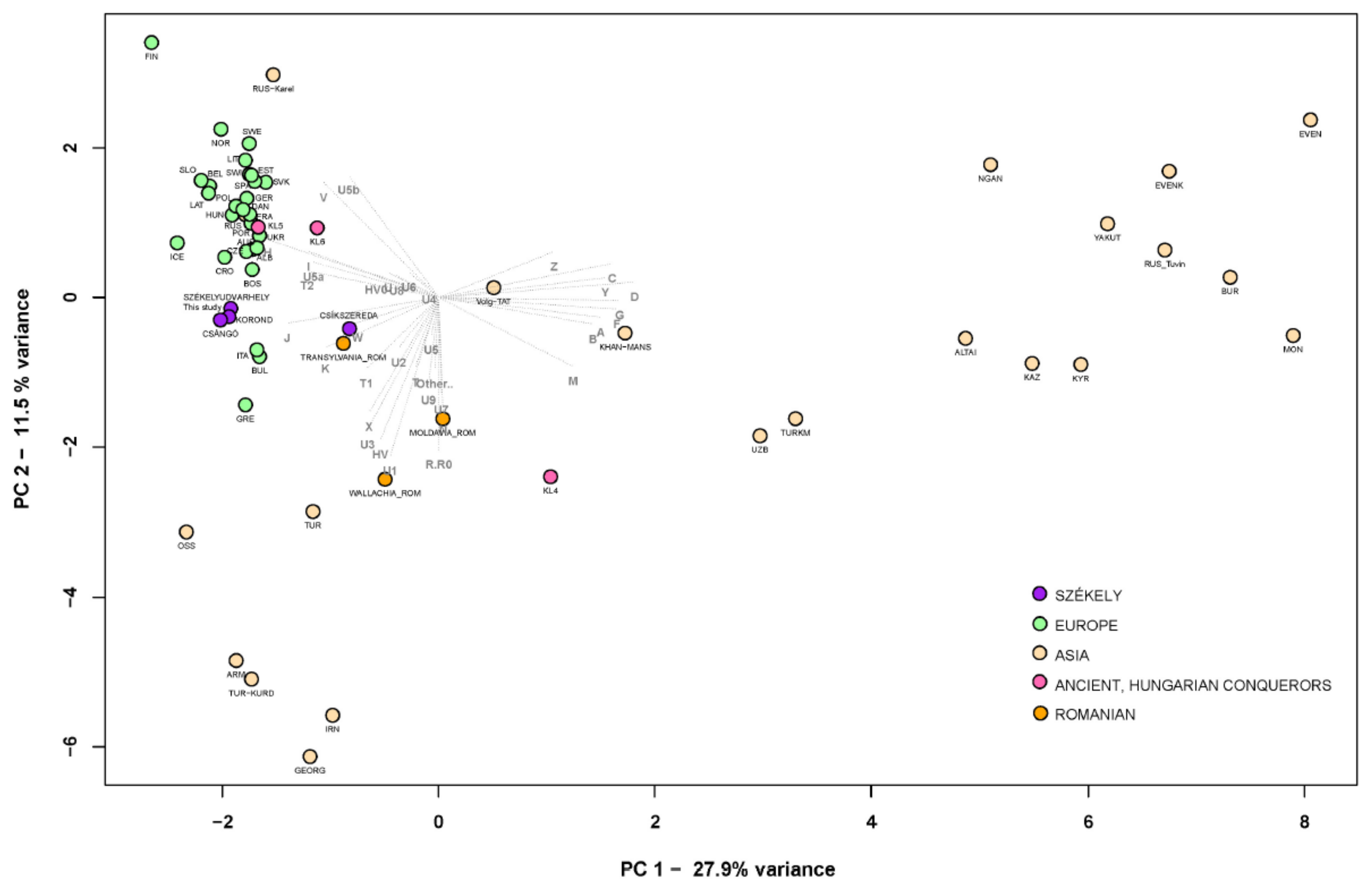
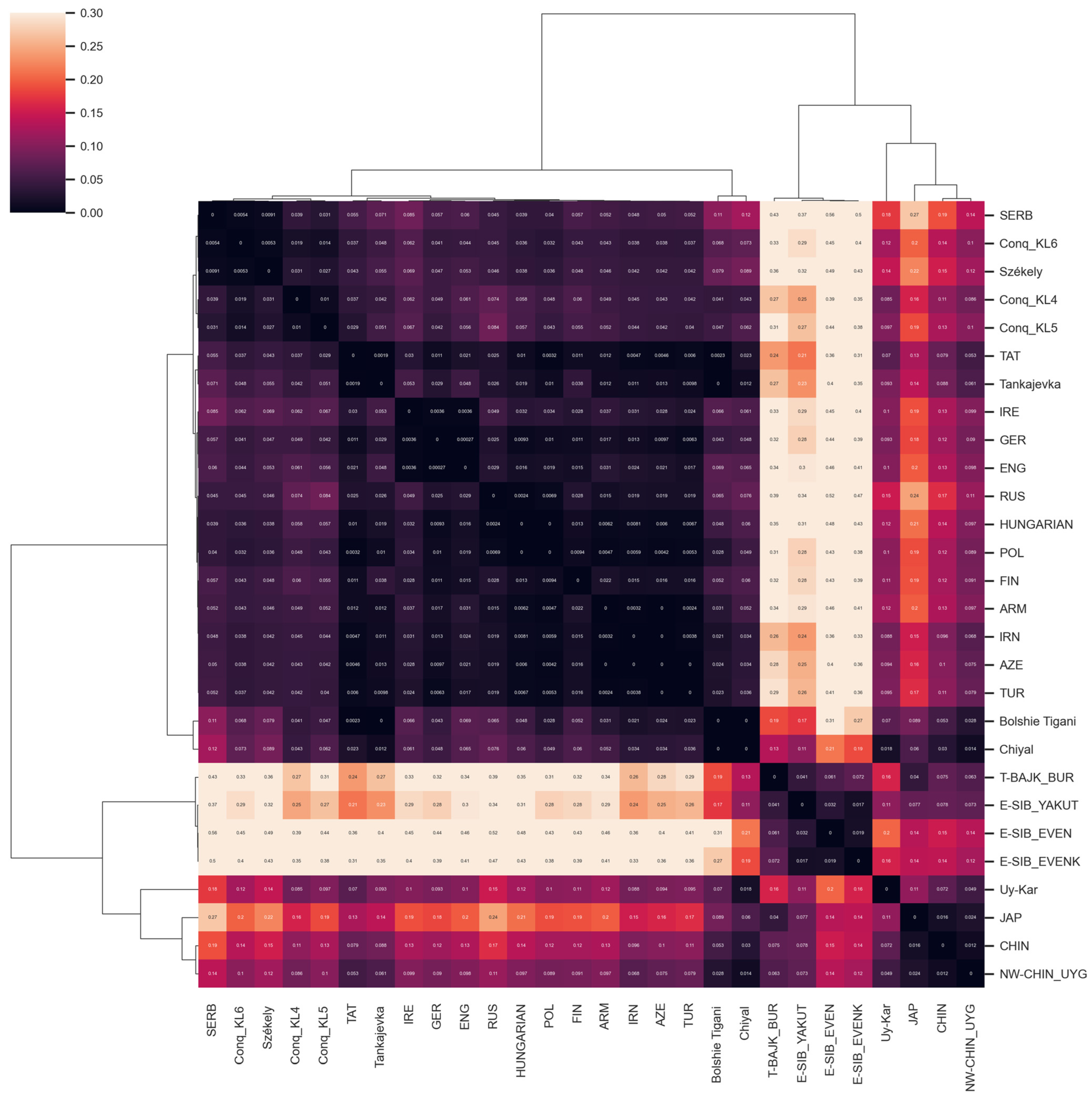
FST Analyses
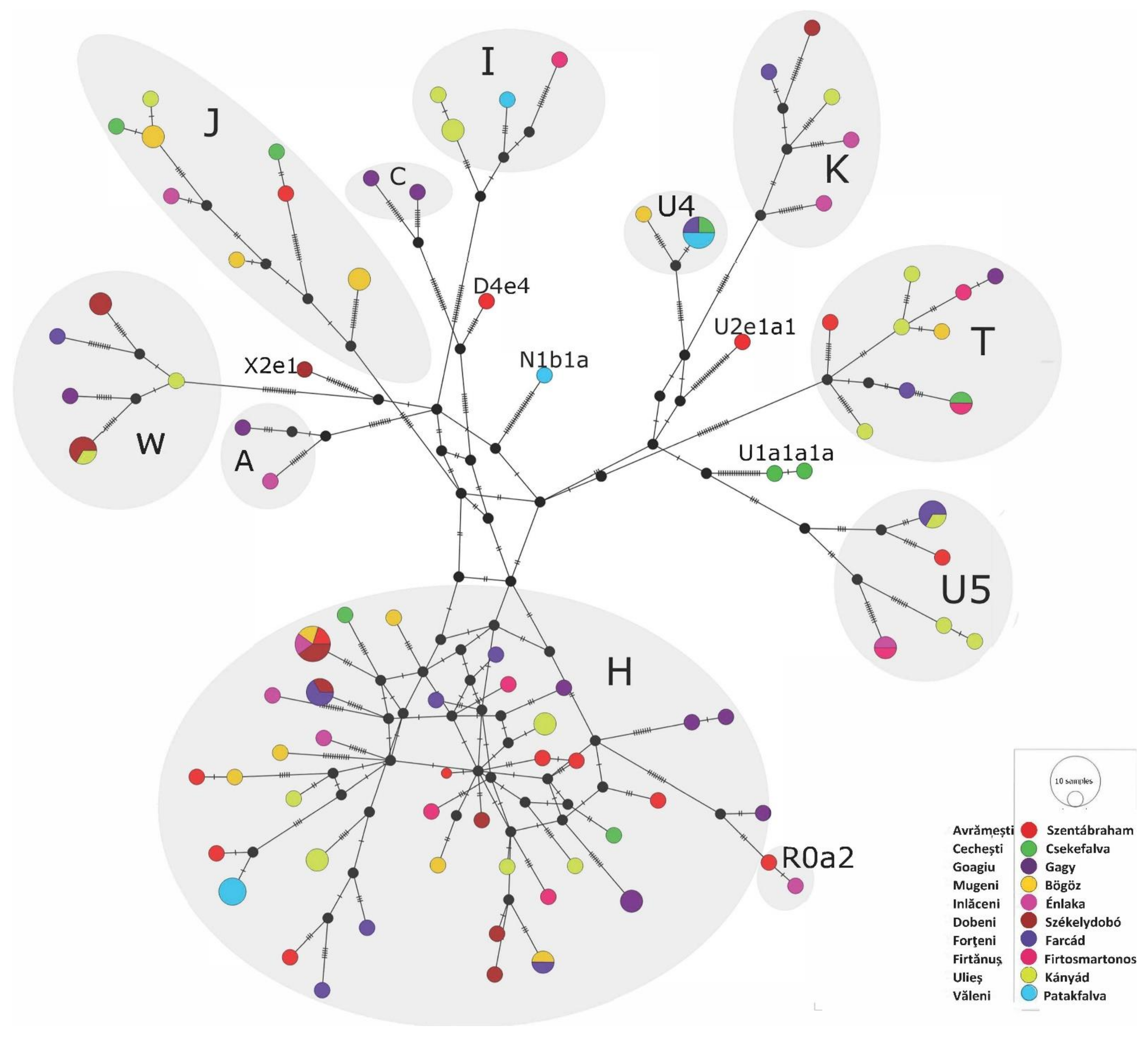
Phylogenetic Analyses of the Székely Maternal Lineages
3.2. Paternal Lineages in the New Székely Dataset
3.2.1. Haplogroup-Based Analyses
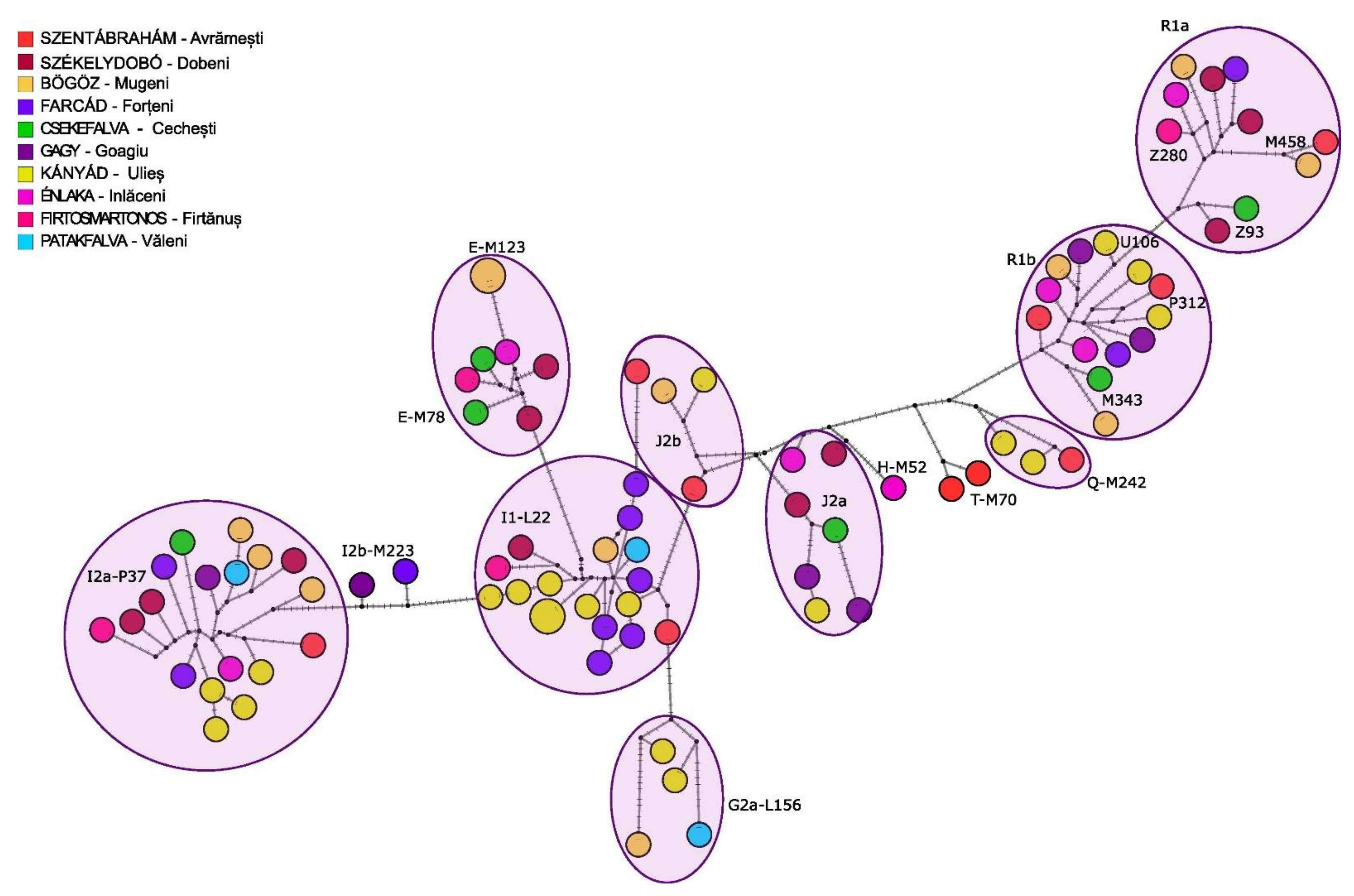
3.2.2. Y Chromosome Phylogenetic Analyses
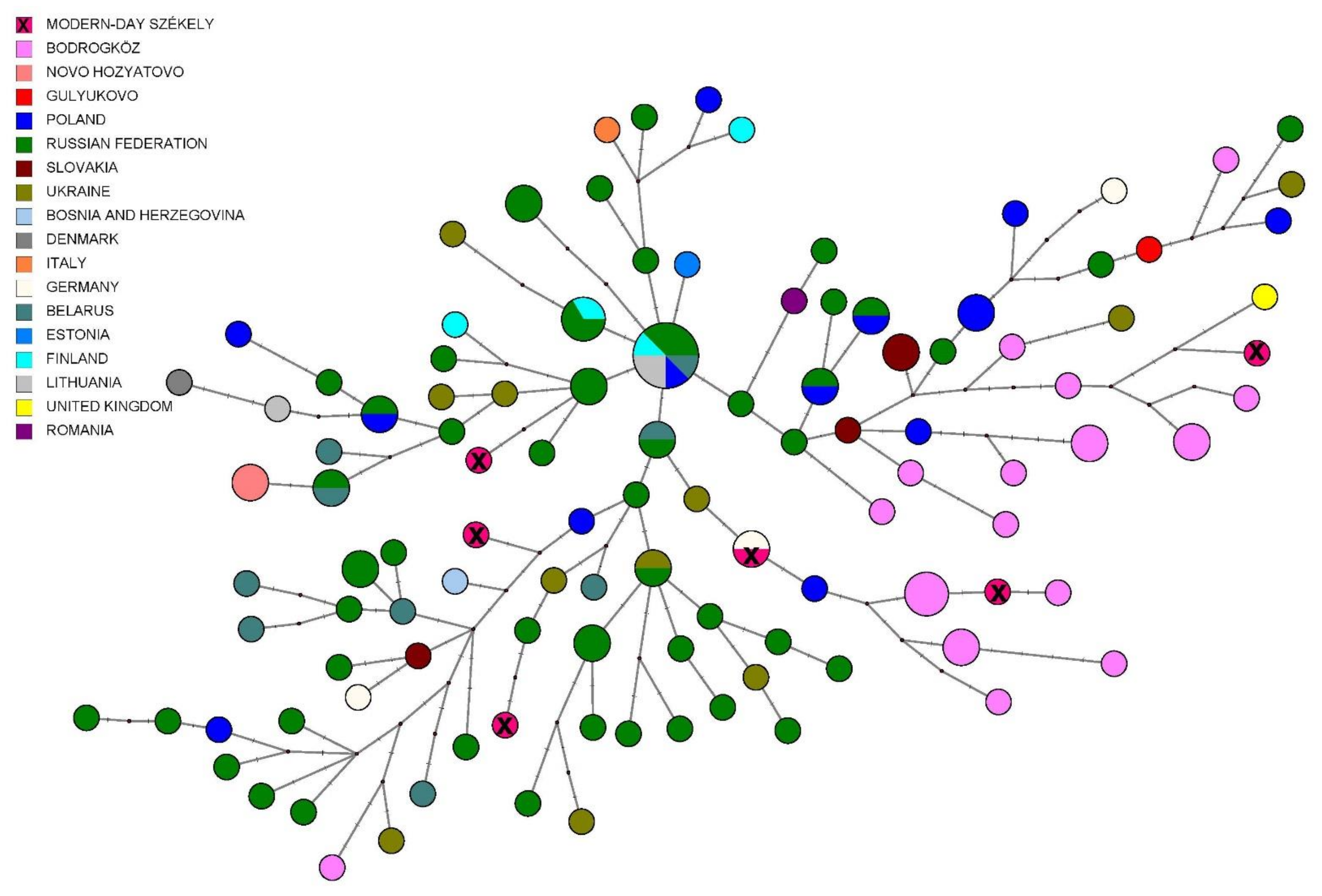
3.2.3. Comparison of Paternal Lineages with Ancient Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMOVA | Analysis of Molecular Variance |
| HG | Haplogroup |
| MDS | Multidimensional Scaling |
| MJ | Median-joining |
| NGS | Next-generation Sequencing |
| NJ | Neighbor-joining |
| PCA | Principal Component Analysis |
| SNP | Single Nucleotide Polymorphism |
| STR | Short Tandem Repeat |
| Y-STR | Y chromosome Short Tandem Repeat |
| YHRD | Y-STR Haplotype Reference Database |
| ybp | years before present |
References
- Benkő, E.; Oborni, T. Székelyföld Története I. [The History of Szeklerland I.]; MTA Bölcsészettudományi Kutatóközpont; Erdély Múzeum-Egyesület; Haáz Rezső Múzeum: Székelyudvarhely, Romania, 2016; pp. 94–103, 107–118. [Google Scholar]
- Benkő, E. A Középkori Székelyföld I. [The Medieval Székelyland I.]; MTA BTK Régészeti Intézet: Budapest, Hungary, 2012; pp. 12–13, 70–71. [Google Scholar]
- Kristó, G. A Székelyek Eredete. In A Székelyek Eredete; Balassi Kiadó: Budapest, Hungary, 2005; p. 176. [Google Scholar]
- Zimonyi, I. Muslim Sources on the Magyars in the Second Half of the 9th Century. In East Central and Eastern Europe in the Middle Ages, 450–1450; Brill Academic Publishers: Leiden, The Netherlands; Boston, MA, USA, 2015; pp. 75–76. [Google Scholar]
- Benkő, L.; Szabó, T.; Die Szekler, Á. Zur Siedlungsgeschichte Einer Ungarischen Volksgruppe. In Ungarn Jahrbuch; Stadtmüller, Georg: Mainz, Germany, 1986; pp. 207–224. [Google Scholar]
- Pardi, G. From Peace Negotiations to War? About the Battle of Olšava in 1116. In Magister Historiae; Belucz, M., Ed.; Eötvös Loránd University: Budapest, Hungary, 2014; p. 152. [Google Scholar]
- Szentpétery, E. Scriptores Rerum Hungaricarum Tempore Ducum Regumque Stirpis Arpadianae Gestarum; Nap Kiadó: Budapest, Hungary, 1937. [Google Scholar]
- Göckenjan, H. Hilfsvölker Und Grenzwächter Im Mittelalterlichen Ungarn; Franz Steiner Verlag GMBH: Wiesbaden, Germany, 1972. [Google Scholar]
- Köpeczi, B. History of Transylvania I; Akadémiai Kiadó: Budapest, Hungary, 1994; pp. 178–179. [Google Scholar]
- Kristó, G. Early Transylvania (895–1324); Lucidus Kiadó: Budapest, Hungary, 2003. [Google Scholar]
- Engel, P. The Realm of St. Stephen: A History of Medieval Hungary, 895–1526; Tauris: London, UK, 2011. [Google Scholar]
- Bárth, J. A Csíkszentmiklósi Havashasználat És a Tatros-Völgy Korai Népessége (The Usage of the Csikszentmiklos Mountains and the Early Population of the Tatros Valley). In A Csíki Székely Múzeum Évkönyve; Csíki Székely Múzeum: Csíkszereda, Romania, 2006; pp. 17–36. [Google Scholar]
- Egyed, B.; Füredi, S.; Padar, Z. Population Genetic Study in Two Transylvanian Populations Using Forensically Informative Autosomal and Y-Chromosomal STR Markers. Forensic Sci. Int. 2006, 164, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Egyed, B.; Brandstätter, A.; Irwin, J.A.; Pádár, Z.; Parsons, T.J.; Parson, W. Mitochondrial Control Region Sequence Variations in the Hungarian Population: Analysis of Population Samples from Hungary and from Transylvania (Romania). Forensic Sci. Int. Genet. 2007, 1, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Tömöry, G.; Csányi, B.; Bogácsi-Szabó, E.; Kalmár, T.; Czibula, Á.; Csősz, A.; Priskin, K.; Mende, B.; Langó, P.; Downes, C.S.; et al. Comparison of Maternal Lineage and Biogeographic Analyses of Ancient and Modern Hungarian Populations. Am. J. Phys. Anthropol. 2007, 132, 535–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csányi, B.; Bogácsi-Szabó, E.; Tömöry, G.; Czibula, Á.; Priskin, K.; Csõsz, A.; Mende, B.; Langó, P.; Csete, K.; Zsolnai, A.; et al. Y-Chromosome Analysis of Ancient Hungarian and Two Modern Hungarian-Speaking Populations from the Carpathian Basin. Ann. Hum. Genet. 2008, 72, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Brandstätter, A.; Egyed, B.; Zimmermann, B.; Duftner, N.; Padar, Z.; Parson, W. Migration Rates and Genetic Structure of Two Hungarian Ethnic Groups in Transylvania, Romania. Ann. Hum. Genet. 2007, 71, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Bíró, A.; Fehér, T.; Bárány, G.; Pamjav, H. Testing Central and Inner Asian Admixture among Contemporary Hungarians. Forensic Sci. Int. Genet. 2015, 15, 121–126. [Google Scholar] [CrossRef]
- Ádám, V.; Bánfai, Z.; Sümegi, K.; Büki, G.; Szabó, A.; Magyari, L.; Miseta, A.; Kásler, M.; Melegh, B. Genome-Wide Marker Data-Based Comparative Population Analysis of Szeklers From Korond, Transylvania, and From Transylvania Living Non-Szekler Hungarians. Front. Genet. 2022, 13, 841769. [Google Scholar] [CrossRef]
- Malyarchuk, B.; Derenko, M.; Denisova, G.; Litvinov, A.; Rogalla, U.; Skonieczna, K.; Grzybowski, T.; Pentelényi, K.; Guba, Z.; Zeke, T.; et al. Whole Mitochondrial Genome Diversity in Two Hungarian Populations. Mol. Genet. Genom. 2018, 293, 1255–1263. [Google Scholar] [CrossRef]
- Völgyi, A.; Zalán, A.; Szvetnik, E.; Pamjav, H. Hungarian Population Data for 11 Y-STR and 49 Y-SNP Markers. Forensic Sci. Int. Genet. 2009, 3, 27–28. [Google Scholar] [CrossRef]
- Pamjav, H.; Fóthi, Á.; Fehér, T.; Fóthi, E. A Study of the Bodrogköz Population in North-Eastern Hungary by Y Chromosomal Haplotypes and Haplogroups. Mol. Genet. Genom. 2017, 292, 883–894. [Google Scholar] [CrossRef]
- Cocoş, R.; Schipor, S.; Hervella, M.; Cianga, P.; Popescu, R.; Banescu, C.; Constantinescu, M.; Martinescu, A.; Raicu, F. Genetic Affinities among the Historical Provinces of Romania and Central Europe as Revealed by an MtDNA Analysis. BMC Genet. 2017, 18, 20. [Google Scholar] [CrossRef] [Green Version]
- Jakó, Z. Erdélyi Okmánytár. Codex Diplomaticus Transsylvaniae II; Jakó, Z., Ed.; Magyar Országos Levéltár: Budapest, Hungary, 2004; Volume 1, p. 413. [Google Scholar]
- MAPSWIRE. Available online: https://mapswire.com/europe/physical-maps/ (accessed on 27 October 2022).
- Fendt, L.; Zimmermann, B.; Daniaux, M.; Parson, W. Sequencing Strategy for the Whole Mitochondrial Genome Resulting in High Quality Sequences. BMC Genom. 2009, 10, 139. [Google Scholar] [CrossRef] [Green Version]
- Gerber, D.; Szeifert, B.; Székely, O.; Egyed, B.; Gyuris, B.; Giblin, J.I.; Horváth, A.; Palcsu, L.; Köhler, K.; Kulcsár, G.; et al. Interdisciplinary Analyses of Bronze Age Communities from Western Hungary Reveal Complex Population Histories. bioRxiv 2022. [Google Scholar] [CrossRef]
- SeqPrep. Available online: https://github.com/jstjohn/SeqPrep (accessed on 15 September 2021).
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Weissensteiner, H.; Pacher, D.; Kloss-Brandstätter, A.; Forer, L.; Specht, G.; Bandelt, H.J.; Kronenberg, F.; Salas, A.; Schönherr, S. HaploGrep 2: Mitochondrial Haplogroup Classification in the Era of High-Throughput Sequencing. Nucleic Acids Res. 2016, 44, W58–W63. [Google Scholar] [CrossRef]
- van Oven, M.; Kayser, M. Updated Comprehensive Phylogenetic Tree of Global Human Mitochondrial DNA Variation. Hum. Mutat. 2009, 30, 386–394. [Google Scholar] [CrossRef]
- Phylotree. Available online: https://www.phylotree.org/ (accessed on 27 October 2022).
- Weissensteiner, H.; Forer, L.; Fuchsberger, C.; Schöpf, B.; Kloss-Brandstätter, A.; Specht, G.; Kronenberg, F.; Schönherr, S. MtDNA-Server: Next-Generation Sequencing Data Analysis of Human Mitochondrial DNA in the Cloud. Nucleic Acids Res. 2016, 44, W64–W69. [Google Scholar] [CrossRef]
- International Society of Genetic Genealogy. International Society of Genetic Genealogy. Y-DNA Haplogroup Tree 2019, Version: 15.73. 2020. Available online: https://isogg.org/tree/ (accessed on 27 October 2022).
- Leigh, J.W.; Bryant, D. POPART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. Sea View Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansour, A. Phylip and Phylogenetics. Genes Genomes Genom. 2009, 3, 46–49. [Google Scholar]
- Rambaut, A. Figtree v 1.4.2. 2014. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 27 October 2022).
- R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org (accessed on 27 October 2022).
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Network V10.1.0.0. Available online: http://www.fluxus-engineering.com (accessed on 27 October 2022).
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Polzin, T.; Daneshmand, V.S. On Steiner Trees and Minimum Spanning Trees in Hypergraphs. Oper. Res. Lett. 2003, 31, 12–20. [Google Scholar] [CrossRef]
- Achilli, A.; Rengo, C.; Magri, C.; Battaglia, V.; Olivieri, A.; Scozzari, R.; Cruciani, F.; Zeviani, M.; Briem, E.; Carelli, V.; et al. The Molecular Dissection of MtDNA Haplogroup H Confirms That the Franco-Cantabrian Glacial Refuge Was a Major Source for the European Gene Pool. Am. J. Hum. Genet. 2004, 75, 910–918. [Google Scholar] [CrossRef]
- Neparáczki, E.; Kocsy, K.; Tóth, G.E.; Maróti, Z.; Kalmár, T.; Bihari, P.; Nagy, I.; Pálfi, G.; Molnár, E.; Raskó, I.; et al. Revising MtDNA Haplotypes of the Ancient Hungarian Conquerors with next Generation Sequencing. PLoS ONE 2017, 12, e0174886. [Google Scholar] [CrossRef] [Green Version]
- Csáky, V.; Gerber, D.; Szeifert, B.; Egyed, B.; Stégmár, B.; Botalov, S.G.; Grudochko, I.V.; Matveeva, N.P.; Zelenkov, A.S.; Sleptsova, A.V.; et al. Early Medieval Genetic Data from Ural Region Evaluated in the Light of Archaeological Evidence of Ancient Hungarians. Sci. Rep. 2020, 10, 19137. [Google Scholar] [CrossRef]
- Maár, K.; Varga, G.I.B.; Kovács, B.; Schütz, O.; Maróti, Z.; Kalmár, T.; Nyerki, E.; Nagy, I.; Latinovics, D.; Tihanyi, B.; et al. Maternal Lineages from 10-11th Century Commoner Cemeteries of the Carpathian Basin. Genes 2021, 12, 460. [Google Scholar] [CrossRef]
- Neparáczki, E.; Maróti, Z.; Kalmár, T.; Kocsy, K.; Maár, K.; Bihari, P.; Nagy, I.; Fóthi, E.; Pap, I.; Kustár, Á.; et al. Mitogenomic Data Indicate Admixture Components of Central-Inner Asian and Srubnaya Origin in the Conquering Hungarians. PLoS ONE 2018, 13, e0205920. [Google Scholar] [CrossRef]
- Szeifert, B.; Stashenkov, D.A.; Khokhlov, A.A.; Sitdikov, A.G.; Gazimzyanov, I.R.; Volkova, E.V.; Matveeva, N.P.; Zelenkov, A.S.; Poshekhonova, O.E.; Sleptsova, A.V.; et al. Tracing Genetic Connections of Ancient Hungarians to the 6-14th Century Populations of the Volga-Ural Region. Hum. Mol. Genet. 2022, 31, 3266–3280. [Google Scholar] [CrossRef]
- Skandakov, I.E.; Danchenko, E.M. Burial Mound Ust-Tara-VII in the Southern Taiga of the Irtysh Region. Humanit. Knowl. Ser. Continuity Yearb. Collect. Sci. Pap. 1999, 3, 160–186. [Google Scholar]
- De Barros Damgaard, P.; Marchi, N.; Rasmussen, S.; Peyrot, M.; Renaud, G.; Korneliussen, T.; Moreno-Mayar, J.V.; Pedersen, M.W.; Goldberg, A.; Usmanova, E.; et al. 137 Ancient Human Genomes from across the Eurasian Steppes. Nature 2018, 557, 369–374. [Google Scholar] [CrossRef]
- Purps, J.; Siegert, S.; Willuweit, S.; Nagy, M.; Alves, C.; Salazar, R.; Angustia, S.M.T.; Santos, L.H.; Anslinger, K.; Bayer, B.; et al. A Global Analysis of Y-Chromosomal Haplotype Diversity for 23 STR Loci. Forensic Sci. Int. Genet. 2014, 12, 12–23. [Google Scholar] [CrossRef]
- Neparáczki, E.; Maróti, Z.; Kalmár, T.; Maár, K.; Nagy, I.; Latinovics, D.; Kustár, Á.; Pálfi, G.; Molnár, E.; Marcsik, A.; et al. Y-Chromosome Haplogroups from Hun, Avar and Conquering Hungarian Period Nomadic People of the Carpathian Basin. Sci. Rep. 2019, 9, 16569. [Google Scholar] [CrossRef] [Green Version]
- Fóthi, E.; Gonzalez, A.; Fehér, T.; Gugora, A.; Fóthi, Á.; Biró, O.; Keyser, C. Genetic Analysis of Male Hungarian Conquerors: European and Asian Paternal Lineages of the Conquering Hungarian Tribes. Archaeol. Anthropol. Sci. 2020, 12, 31. [Google Scholar] [CrossRef] [Green Version]
- Stanciu, F.; Cuţăr, V.; Pîrlea, S.; Stoian, V.; Stoian, I.M.; Sevastre, O.; Popescu, O.R. Population Data for Y-Chromosome Haplotypes Defined by 17 STRs in South-East Romania. Leg. Med. 2010, 12, 259–264. [Google Scholar] [CrossRef]
- Mendez, F.L.; Karafet, T.M.; Krahn, T.; Ostrer, H.; Soodyall, H.; Hammer, M.F. Increased Resolution of Y Chromosome Haplogroup T Defines Relationships among Populations of the Near East, Europe, and Africa. Hum. Biol. 2011, 83, 39–53. [Google Scholar] [CrossRef]
- Papac, L.; Ernée, M.; Dobeš, M.; Langová, M.; Rohrlach, A.B.; Aron, F.; Neumann, G.U.; Spyrou, M.A.; Rohland, N.; Velemínský, P.; et al. Dynamic Changes in Genomic and Social Structures in Third Millennium BCE Central Europe. Sci. Adv. 2021, 7, eabi6941. [Google Scholar] [CrossRef]
- Költő, L. Honfoglalás Kori Tegezes Sír Vörsön. In A Herman Ottó Múzeum Évkönyve; Herman Ottó Múzeum: Miskolc, Hungary, 1993; Volume 30–31/2, pp. 433–445. [Google Scholar]
- Maróti, Z.; Neparáczki, E.; Schütz, O.; Maár, K.; Varga, G.I.B.; Kovács, B.; Kalmár, T.; Nyerki, E.; Nagy, I.; Latinovics, D.; et al. The Genetic Origin of Huns, Avars, and Conquering Hungarians. Curr. Biol. 2022, 32, 2858–2870.E7. [Google Scholar] [CrossRef] [PubMed]
- Grugni, V.; Raveane, A.; Colombo, G.; Nici, C.; Crobu, F.; Ongaro, L.; Battaglia, V.; Sanna, D.; Al-Zahery, N.; Fiorani, O.; et al. Y-Chromosome and Surname Analyses for Reconstructing Past Population Structures: The Sardinian Population as a Test Case. Int. J. Mol. Sci. 2019, 20, 5763. [Google Scholar] [CrossRef] [PubMed]
- Rootsi, S.; Magri, C.; Kivisild, T.; Benuzzi, G.; Help, H.; Bermisheva, M.; Kutuev, I.; Barać, L.; Peričić, M.; Balanovsky, O.; et al. Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe. Am. J. Hum. Genet. 2004, 75, 128–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margaryan, A.; Lawson, D.J.; Sikora, M.; Racimo, F.; Rasmussen, S.; Moltke, I.; Cassidy, L.M.; Jørsboe, E.; Ingason, A.; Pedersen, M.W.; et al. Population Genomics of the Viking World. Nature 2020, 585, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, Y.P. Volzhskie Bolgari, Ugri i Finni v IX–XIV vv [Volga Bulgars, Ugrians and Finns: Problems of Interaction]. In Problemy Vzaimodeistviya; Kazan, Russia, 2007. [Google Scholar]
- Berger, B.; Niederstätter, H.; Erhart, D.; Gassner, C.; Schennach, H.; Parson, W. High Resolution Mapping of Y Haplogroup G in Tyrol (Austria). Forensic Sci. Int. Genet. 2013, 7, 529–536. [Google Scholar] [CrossRef]
- Lipson, M.; Szécsényi-Nagy, A.; Mallick, S.; Pósa, A.; Stégmár, B.; Keerl, V.; Rohland, N.; Stewardson, K.; Ferry, M.; Michel, M.; et al. Parallel Palaeogenomic Transects Reveal Complex Genetic History of Early European Farmers. Nature 2017, 551, 368–372. [Google Scholar] [CrossRef]
- Rootsi, S.; Myres, N.M.; Lin, A.A.; Järve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Varendi, K.; Sahakyan, H.; et al. Distinguishing the Co-Ancestries of Haplogroup G Y-Chromosomes in the Populations of Europe and the Caucasus. Eur. J. Hum. Genet. 2012, 20, 1275–1282. [Google Scholar] [CrossRef] [Green Version]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Genome-Wide Patterns of Selection in 230 Ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef] [Green Version]
- Skourtanioti, E.; Erdal, Y.S.; Frangipane, M.; Balossi Restelli, F.; Yener, K.A.; Pinnock, F.; Matthiae, P.; Özbal, R.; Schoop, U.-D.; Guliyev, F.; et al. Genomic History of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell 2020, 181, 1158–1175.e28. [Google Scholar] [CrossRef]
- Dobos, A.; Gál, S.S.; Kelemen, I.; Neparáczki, E. Attila’s Europe? Structural Transformation and Strategies of Success in the European Hun Period; Rácz, Z., Szenthe, G., Eds.; Hungarian National Museum, Eötvös Loránd University: Budapest, Hungary, 2021; pp. 327–356. [Google Scholar]
- Csáky, V.; Gerber, D.; Koncz, I.; Csiky, G.; Mende, B.G.; Szeifert, B.; Egyed, B.; Pamjav, H.; Marcsik, A.; Molnár, E.; et al. Genetic Insights into the Social Organisation of the Avar Period Elite in the 7th Century AD Carpathian Basin. Sci. Rep. 2020, 10, 948. [Google Scholar] [CrossRef] [Green Version]
- Gnecchi-Ruscone, G.A.; Szécsényi-Nagy, A.; Koncz, I.; Csiky, G.; Rácz, Z.; Rohrlach, A.B.; Brandt, G.; Rohland, N.; Csáky, V.; Cheronet, O.; et al. Ancient Genomes Reveal Origin and Rapid Trans-Eurasian Migration of 7th Century Avar Elites. Cell 2022, 185, 1402–1413.e21. [Google Scholar] [CrossRef]
- Grugni, V.; Raveane, A.; Ongaro, L.; Battaglia, V.; Trombetta, B.; Colombo, G.; Capodiferro, M.R.; Olivieri, A.; Achilli, A.; Perego, U.A.; et al. Analysis of the Human Y-Chromosome Haplogroup Q Characterizes Ancient Population Movements in Eurasia and the Americas. BMC Biol. 2019, 17, 3. [Google Scholar] [CrossRef]
- Olasz, J.; Seidenberg, V.; Hummel, S.; Szentirmay, Z.; Szabados, G.; Melegh, B.; Kásler, M. DNA Profiling of Hungarian King Béla III and Other Skeletal Remains Originating from the Royal Basilica of Székesfehérvár. Archaeol. Anthropol. Sci. 2019, 11, 1345–1357. [Google Scholar] [CrossRef] [Green Version]
- Nagy, P.L.; Olasz, J.; Neparáczki, E.; Rouse, N.; Kapuria, K.; Cano, S.; Chen, H.; Di Cristofaro, J.; Runfeldt, G.; Ekomasova, N.; et al. Determination of the Phylogenetic Origins of the Árpád Dynasty Based on Y Chromosome Sequencing of Béla the Third. Eur. J. Hum. Genet. 2021, 29, 164–172. [Google Scholar] [CrossRef]
- Benkő, E.; Sándor, K.; Vásáry, I. A Székely Írás Emlékei. Corpus Monumentorum Alphabeto Siculico Exaratorum; Bölcsészettudományi Kutatóközpont: Budapest, Hungary, 2021; pp. 834–835. [Google Scholar]
- Dulias, K.; Foody, M.G.; Justeau, P.; Silva, M.; Martiniano, R.; Oteo-García, G.; Fichera, A.; Simão, R.; Gandini, F.; Meynert, A.; et al. Ancient DNA at the Edge of the World: Continental Immigration and the Persistence of Neolithic Male Lineages in Bronze Age Orkney. Proc. Natl. Acad. Sci. USA 2022, 119, e2108001119. [Google Scholar] [CrossRef]
- Myres, N.M.; Rootsi, S.; Lin, A.A.; Järve, M.; King, R.J.; Kutuev, I.; Cabrera, V.M.; Khusnutdinova, E.K.; Pshenichnov, A.; Yunusbayev, B.; et al. A Major Y-Chromosome Haplogroup R1b Holocene Era Founder Effect in Central and Western Europe. Eur. J. Hum. Genet. 2011, 19, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Langó, P. Salamon Gyűrűi”—Pajzs Alakú, Kiszélesedő, Díszített Fejű Pántgyűrűk a X. Századi Kárpát-Medencei Emlékanyagban. In Beatus Homo qui Invenit Sapientiam; Csécs, T., Takács, M., Eds.; Lekri Group Kft: Győr, Hungary, 2016; pp. 387–408. [Google Scholar]
- Kovács, L. A Kárpát-Medence Honfoglalás És Kora Árpád-Kori Szállási És Falusi Temetői. In A Honfoglás kor Kutasánák Legújabb Eredményei; Tanulmányok; Kovács László 70. születésnapjára; Szegedi Tudományegyetem: Szeged, Hungary, 2013; pp. 511–604. [Google Scholar]
- MTree v. 1.02.16072. Available online: https://www.yfull.com/mtree/ (accessed on 24 April 2022).
- Logan, I. Available online: http://www.ianlogan.co.uk/ (accessed on 27 October 2022).
- Underhill, P.A.; Poznik, G.D.; Rootsi, S.; Järve, M.; Lin, A.A.; Wang, J.; Passarelli, B.; Kanbar, J.; Myres, N.M.; King, R.J.; et al. The Phylogenetic and Geographic Structure of Y-Chromosome Haplogroup R1a. Eur. J. Hum. Genet. 2015, 23, 124–131. [Google Scholar] [CrossRef] [Green Version]
- Dudás, E.; Vágó-Zalán, A.; Vándor, A.; Spasheva, A.; Pomozi, P.; Pamjav, H. Genetic History of Bashkirian Mari and Southern Mansi Ethnic Groups in the Ural Region. Mol. Genet. Genomics 2019, 294, 919–930. [Google Scholar] [CrossRef]
- YHRD. Available online: https://yhrd.org/pages/tools/amova (accessed on 6 July 2021).
- Gephi—The Open Graph Viz Platform. 2022. Available online: https://github.com/gephi (accessed on 27 October 2021).
- Vanecek, T.; Vorel, F.; Sip, M. Mitochondrial DNA D-Loop Hypervariable Rogions: Czech Population Data. Int. J. Legal Med. 2004, 118, 14–18. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borbély, N.; Székely, O.; Szeifert, B.; Gerber, D.; Máthé, I.; Benkő, E.; Mende, B.G.; Egyed, B.; Pamjav, H.; Szécsényi-Nagy, A. High Coverage Mitogenomes and Y-Chromosomal Typing Reveal Ancient Lineages in the Modern-Day Székely Population in Romania. Genes 2023, 14, 133. https://doi.org/10.3390/genes14010133
Borbély N, Székely O, Szeifert B, Gerber D, Máthé I, Benkő E, Mende BG, Egyed B, Pamjav H, Szécsényi-Nagy A. High Coverage Mitogenomes and Y-Chromosomal Typing Reveal Ancient Lineages in the Modern-Day Székely Population in Romania. Genes. 2023; 14(1):133. https://doi.org/10.3390/genes14010133
Chicago/Turabian StyleBorbély, Noémi, Orsolya Székely, Bea Szeifert, Dániel Gerber, István Máthé, Elek Benkő, Balázs Gusztáv Mende, Balázs Egyed, Horolma Pamjav, and Anna Szécsényi-Nagy. 2023. "High Coverage Mitogenomes and Y-Chromosomal Typing Reveal Ancient Lineages in the Modern-Day Székely Population in Romania" Genes 14, no. 1: 133. https://doi.org/10.3390/genes14010133
APA StyleBorbély, N., Székely, O., Szeifert, B., Gerber, D., Máthé, I., Benkő, E., Mende, B. G., Egyed, B., Pamjav, H., & Szécsényi-Nagy, A. (2023). High Coverage Mitogenomes and Y-Chromosomal Typing Reveal Ancient Lineages in the Modern-Day Székely Population in Romania. Genes, 14(1), 133. https://doi.org/10.3390/genes14010133




