A Genome-Wide View of the Transcriptome Dynamics of Fresh-Cut Potato Tubers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Determination of Browning Degree of Potato Tubers
2.2. RNA Extraction, Library Construction and Sequencing
2.3. De Novo Assembly, DEG Analysis and Annotation
2.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Determination of Enzyme Activity and H2O2 Content
2.6. Statistical Analysis
3. Results
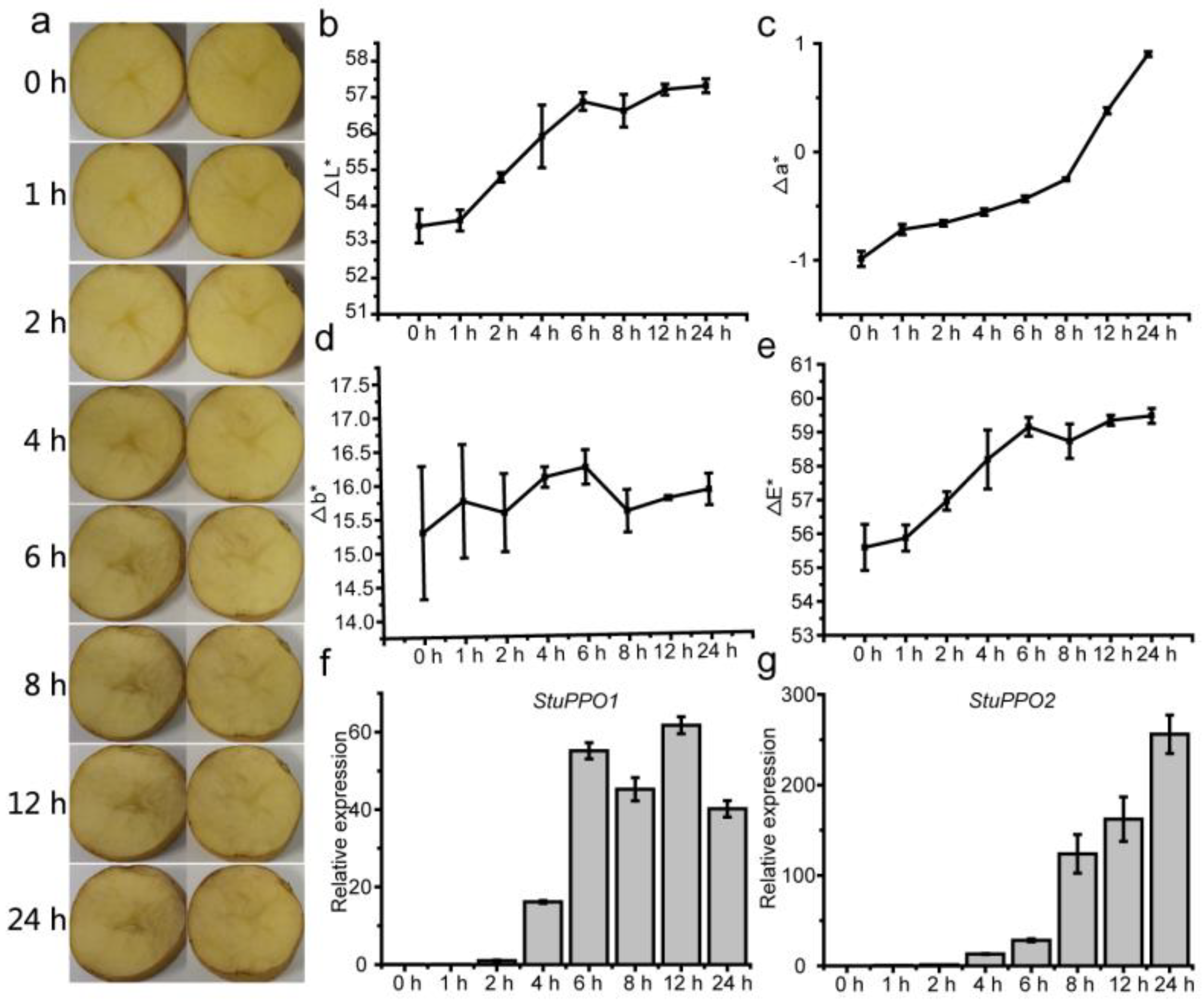
3.1. Browning Characteristic of Potato Tubers at Different Times after Cutting
3.2. Transcriptome Data Generation and Processing
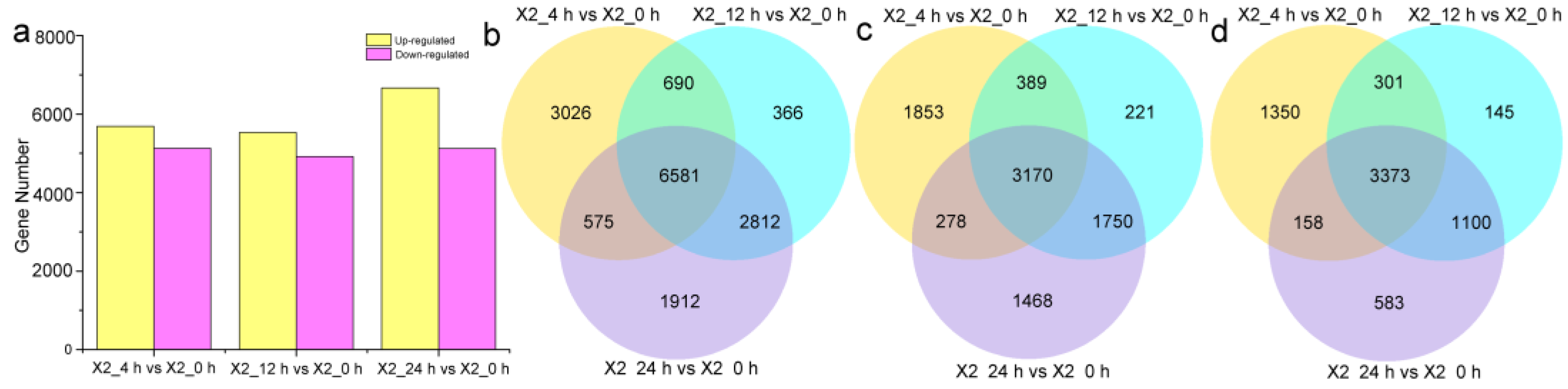
3.3. DEG Analysis of Potato Tubers after Cutting
3.4. GO and KEGG Enrichment of DEGs in Response to Cut-Wounding
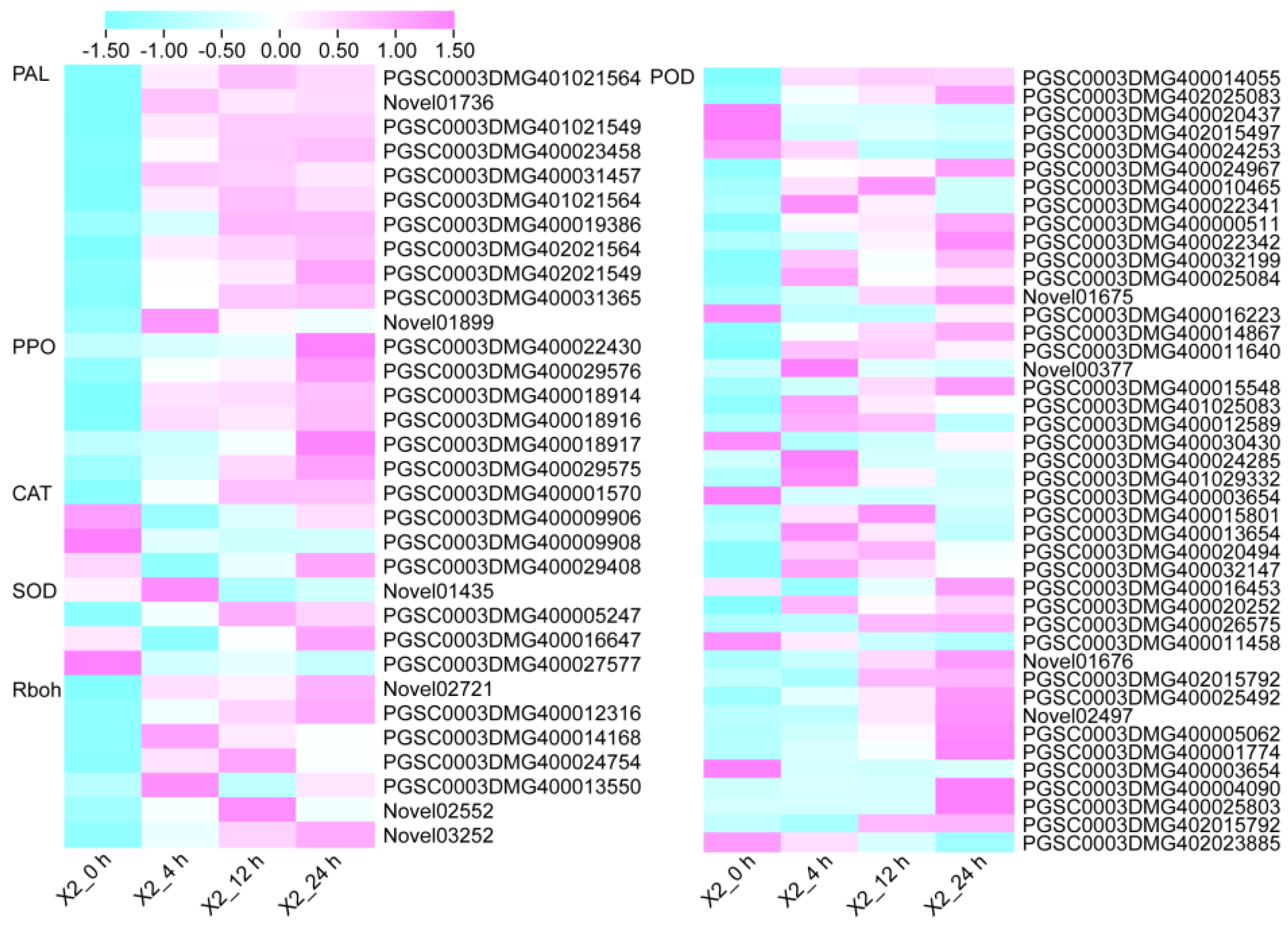
3.5. Antioxidant Enzymes and Enzymatic Browning-Related Genes in Response to Cut-Wounding
3.6. DEGs with a |Log2FC| ≥ 6
3.7. Transcription Factor Genes Identified in Response to Cut-Wounding
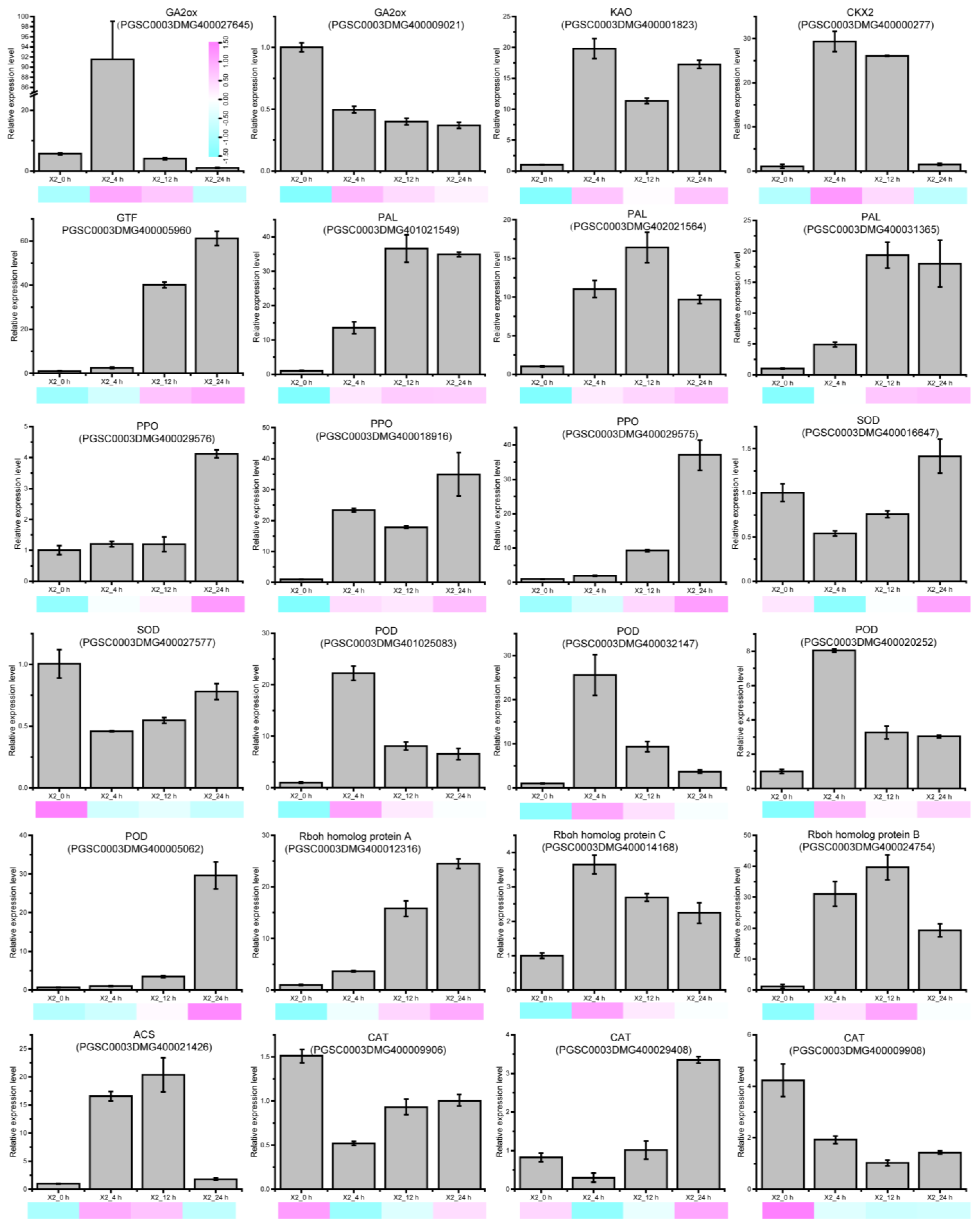
3.8. The Expression of Signal Molecular and Antioxidant Enzyme-Related DEGs
3.9. Enzyme Activity and H2O2 Content of Potato at Different Time after Cutting
4. Discussion
4.1. DEGs Identified at Different Times after Cutting
4.2. Enzymatic Browning-Related Genes
4.3. Signaling Molecules Biosynthesis Related-Genes Involved in Wound-Response
4.4. Transcription Factor Genes Identified in Response to Cut-Wounding
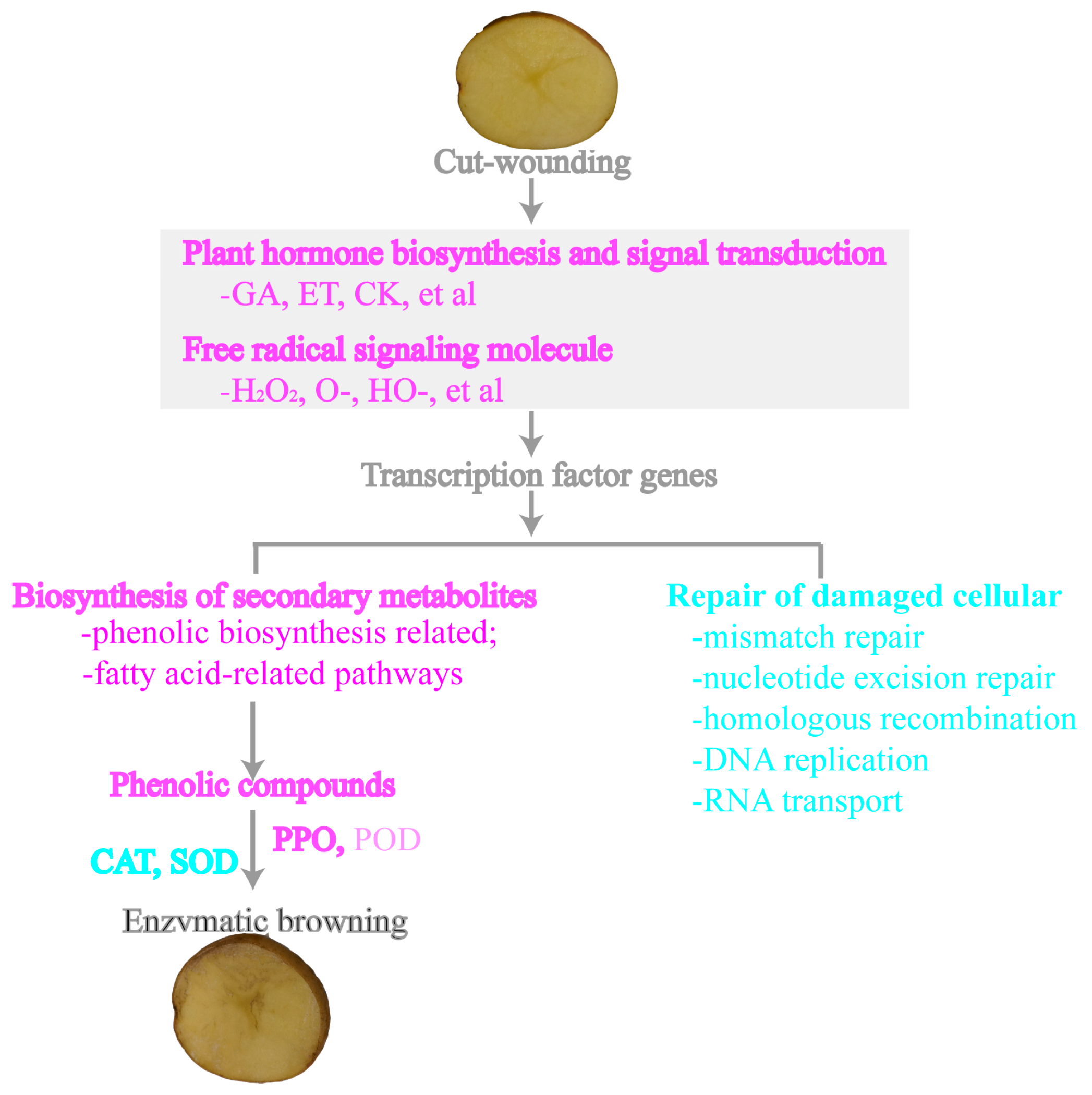
4.5. Hypothetical Model of Potato Tubers in Response to Cut-Wounding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hamdan, N.; Lee, C.H.; Wong, S.L.; Fauzi, C.E.N.C.A.; Zamri, N.M.A.; Lee, T.H. Prevention of Enzymatic Browning by Natural Extracts and Genome-Editing: A Review on Recent Progress. Molecules 2022, 27, 1101. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, L.; Kulig, B.; Moscetti, R.; Massantini, R.; Pawelzik, E.; Hensel, O.; Sturm, B. Optimisation of Physical and Chemical Treatments to Control Browning Development and Enzymatic Activity on Fresh-cut Apple Slices. Foods 2020, 9, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozturk, B.; Seyhan, F.; Ozdemir, I.S.; Karadeniz, B.; Bahar, B.; Ertas, E.; Ilgaz, S. Change of enzyme activity and quality during the processing of Turkish green tea. LWT-Food Sci. Technol. 2016, 65, 318–324. [Google Scholar] [CrossRef]
- Lim, W.Y.; Cheun, C.F.; Wong, C.W. Inhibition of enzymatic browning in sweet potato (Ipomoea batatas (L.)) with chemical and natural anti-browning agents. J. Food Process. Preserv. 2019, 43, e14195. [Google Scholar] [CrossRef]
- Jiang, Y.; Duan, X.; Joyce, D.; Zhang, Z.; Li, J. Advances in understanding of enzymatic browning in harvested litchi fruit. Food Chem. 2004, 88, 443–446. [Google Scholar] [CrossRef]
- Fortea, M.I.; López-Miranda, S.; Serrano-Martínez, A.; Carreño, J.; Núñez-Delicado, E. Kinetic characterisation and thermal inactivation study of polyphenol oxidase and peroxidase from table grape (Crimson Seedless). Food Chem. 2009, 113, 1008–1014. [Google Scholar] [CrossRef]
- Jiang, Y. Role of anthocyanins, polyphenol oxidase and phenols in lychee pericarp browning. J. Sci. Food Agric. 2000, 80, 305–310. [Google Scholar] [CrossRef]
- Escalante-Minakata, P.; Ibarra-Junquera, V.; Ornelas-Paz, J.D.J.; García-Ibáñez, V.; Virgen-Ortíz, J.J.; González-Potes, A.; Pérez-Martínez, J.D.; Orozco-Santos, M. Comparative study of the banana pulp browning process of ‘Giant Dwarf’ and FHIA-23 during fruit ripening based on image analysis and the polyphenol oxidase and peroxidase biochemical properties. 3 Biotech. 2017, 8, 30. [Google Scholar] [CrossRef]
- Boeckx, T.; Winters, A.; Webb, K.J.; Kingston-Smith, A.H. Detection of potential chloroplastic substrates for polyphenol oxidase suggests a role in undamaged leaves. Front. Plant Sci. 2017, 8, 237. [Google Scholar] [CrossRef] [Green Version]
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry. 2006, 67, 2318–2331. [Google Scholar] [CrossRef]
- Chumyam, A.; Faiyue, B.; Saengnil, K. Reduction of enzymatic browning of fresh-cut guava fruit by exogenous hydrogen peroxide-activated peroxiredoxin/thioredoxin system. Sci. Hortic. 2019, 255, 260–268. [Google Scholar] [CrossRef]
- Dong, T.; Cao, Y.; Li, G.; Zhu, Z.; Zhang, S.; Jiang, C.Z.; Wang, Q. A novel aspartic protease inhibitor inhibits the enzymatic browning of potatoes. Postharvest Biol. Technol. 2021, 172, 11135. [Google Scholar] [CrossRef]
- Gao, H.; Zeng, Q.; Ren, Z.; Li, P.; Xu, X. Effect of exogenous γ-aminobutyric acid treatment on the enzymatic browning of fresh-cut potato during storage. J. Food Sci. Tech. 2018, 55, 5035–5044. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, Q.; Lu, Y.; Li, Y.; Li, T.; Zhou, B.; Qiao, L. Effect of purslane (Portulaca oleracea L.) extract on anti-browning of fresh-cut potato slices during storage. Food Chemi. 2019, 283, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Yun, Z.; Gao, H.J.; Chen, X.; Chen, Z.; Zhang, Z.; Li, T.; Qu, H.; Jiang, Y. Effects of hydrogen water treatment on antioxidant system of litchi fruit during the pericarp browning. Food Chem. 2021, 336, 127618. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alexandria J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Singh, R.K.; Moehninsi; Navarre, D.A. R2R3-MYB transcription factors, StmiR858 and sucrose mediate potato flavonol biosynthesis. Hortic. Res. 2021, 8, 25. [Google Scholar] [CrossRef]
- Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Controlled abiotic stresses revisited: From homeostasis through hormesis to extreme stresses and the impact on nutraceuticals and quality during pre-and postharvest applications in horticultural crops. J. Agric. Food Chem. 2020, 68, 11877–11879. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.X.; Zeng, L.; Suo, H.C.; Li, C.C.; Shan, J.W.; Liu, J.T.; Luo, H.M.; Li, X.B.; Xiong, X.Y. Characteristics and differences of polyphenol oxidase, peroxidase activities and polyphenol content in different potato (solanum tuberosum) tubers. Appl. Ecol. Env. Res. 2020, 18, 8171–8187. [Google Scholar] [CrossRef]
- Zhao, L.; Lu, H.; Meng, Q.; Wang, J.; Wang, W.; Yang, L.; Lin, L. Profilings of microRNAs in the liver of common carp (Cyprinus carpio) infected with Flavobacterium columnare. Int. J. Mol. Sci. 2016, 17, 566. [Google Scholar] [CrossRef]
- Fu, Y.W.; Liang, X.X.; Li, D.H.; Gao, H.; Wang, Y.D.; Li, W.T.; Xu, K.; Hu, F.Z. Effect of dietary tryptophan on growth, intestinal microbiota, and intestinal gene expression in an improved triploid Crucian Carp. Front. Nutr. 2021, 8, 322. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diambra, L.A. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zytnicki, M. Mmquant: How to count multi-mapping reads? BMC Bioinf. 2017, 18, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Anders, S. Analysing RNA-Seq data with the DESeq package. Mol. Biol. 2010, 43, 1–17. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.-F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Chi, M.; Bhagwat, B.; Lane, W.D.; Tang, G.; Su, Y.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, X. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 2014, 14, 62. [Google Scholar] [CrossRef] [Green Version]
- Major, I.T.; Constabel, C.P. Molecular analysis of poplar defense against herbivory: Comparison of wound- and insect elicitor-induced gene expression. New Phytol. 2006, 172, 617–635. [Google Scholar] [CrossRef]
- Smith, C.M.; Rodriguez-Buey, M.; Karlsson, J.; Campbell, M.M. The Response of the poplar transcriptome to wounding and subsequent infection by a viral pathogen. New Phytol. 2004, 164, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Wen, X.; Xu, Y.; Zhang, M.; Zhou, L.; Liu, D.; Li, X.; Wang, J. Phosphoinositide signaling plays a key role in the regulation of cell wall reconstruction during the postharvest morphological development of Dictyophora indusiata. Food Chem. 2021, 346, 128890. [Google Scholar] [CrossRef] [PubMed]
- Razem, F.A.; Bernards, M.A. Hydrogen peroxide is required for poly(phenolic) domain formation during wound-induced suberization. J. Agric. Food Chem. 2002, 50, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Suprasanna, P.; Variyar, P.S.; Sharma, A. γ irradiation inhibits wound induced browning in shredded cabbage. Food Chem. 2015, 173, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, S.J.; Delude, C.; Domergue, F.; Rowland, O. Suberin: Biosynthesis, regulation, and polymer assembly of a protective extracellular barrier. Plant Cell Rep. 2015, 34, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Han, S.; He, D.; Cao, G.; Fang, K.; Xiao, X.; Yi, J.; Wan, X. The accumulation of phenolic compounds and increased activities of related enzymes contribute to early defense against walnut blight. Physiol. Mol. Plant Pathol. 2019, 108, 101433. [Google Scholar] [CrossRef]
- Saltveit, M.E. Wound induced changes in phenolic metabolism and tissue browning are altered by heat shock. Postharvest Biol. Technol. 2000, 21, 61–69. [Google Scholar] [CrossRef]
- Lulai, E.C.; Neubauer, J.D. Wound-induced suberization genes are differentially expressed, spatially and temporally, during closing layer and wound periderm formation. Postharvest Biol. Tech. 2014, 90, 24–33. [Google Scholar] [CrossRef]
- Ramamurthy, M.S.; Ussuf, K.K.; Nair, P.M.; Thomas, P. Lignin biosynthesis during wound healing of potato tubers in response to γ irradiation. Postharvest Biol. Tech. 2000, 18, 267–272. [Google Scholar] [CrossRef]
- Zamira, G.; Alik, A.; Nigora, K. Induction of peroxidase as a disease resistance response in resistant (Hibiscus trionum) and susceptible (Althea armeniaca) species in the family malvaceae. Phytopathology 2007, 35, 401–413. [Google Scholar]
- Zhang, J.; Sun, X.L. Recent advances in polyphenol oxidase-mediated plant stress responses. Phytochemistry 2021, 181, 112588. [Google Scholar] [CrossRef] [PubMed]
- Munir, N.; Cheng, C.; Xia, C.; Xu, X.; Nawaz, M.A.; Iftikhar, J.; Chen, Y.; Lin, Y.; Lai, Z. RNA-Seq analysis reveals an essential role of tyrosine metabolism pathway in response to root-rot infection in Gerbera hybrida. PLoS ONE 2019, 14, 0223519. [Google Scholar] [CrossRef] [PubMed]
- Aziz, E.; Batool, R.; Akhtar, W.; Rehman, S.; Gregersen, P.L.; Mahmood, T. Expression analysis of the polyphenol oxidase gene in response to signaling molecules, herbivory and wounding in antisense transgenic tobacco plants. 3 Biotech. 2019, 9, 55. [Google Scholar] [CrossRef]
- Alam, N.B.; Ghosh, A. Comprehensive analysis and transcript profiling of Arabidopsis thaliana and Oryza sativa catalase gene family suggests their specific roles in development and stress responses. Plant Physiol. Bioch. 2018, 123, 54–64. [Google Scholar] [CrossRef]
- Nicholls, P.; Fita, I.; Loewen, P.C. Enzymology and structure of catalases. Adv. Inorg. Chem. 2000, 51, 51–106. [Google Scholar]
- Sun, J.; Li, C.; Nagendra Prasad, K.; You, X.; Li, L.; Liao, F.; Peng, H.; He, X.; Li, Z.; Zhang, Y. Membrane deterioration, enzymatic browning and oxidative stress in fresh fruits of three litchi cultivars during six-day storage. Sci. Hortic. 2012, 148, 97–103. [Google Scholar] [CrossRef]
- Suttle, J.C.; Lulai, E.C.; Huckle, L.L.; Neubauer, J.D. Wounding of potato tubers induces increases in ABA biosynthesis and catabolism and alters expression of ABA metabolic genes. J. Plant Physiol. 2013, 170, 560–566. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Rymen, B.; Lambolez, A.; Kojima, M.; Takebayashi, Y.; Heyman, J.; Watanabe, S.; Seo, M.; De Veylder, L.; et al. Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 2017, 175, 1158–1174. [Google Scholar] [CrossRef] [Green Version]
- Kumar, G.N.M.; Lulai, E.C.; Suttle, J.C.; Knowles, N.R. Age-induced loss of wound-healing ability in potato tubers is partly regulated by ABA. Planta 2010, 232, 1433–1445. [Google Scholar] [CrossRef]
- Kumar, G.N.M.; Knowles, N.R. Wound-induced superoxide production and PAL activity decline with potato tuber age and wound healing ability. Physiol. Plant. 2003, 117, 108–117. [Google Scholar] [CrossRef]
- Tomás-Barberán, F.A.; Espin, J.C. Phenolic compounds and related enzymes as determinants of quality in fruits and vegetables. J. Sci. Food Agric. 2001, 81, 853–876. [Google Scholar] [CrossRef]
- Zhang, T.; Poudel, A.N.; Jewell, J.B.; Kitaoka, N.; Staswick, P.; Matsuura, H.; Koo, A.J. Hormone crosstalk in wound stress response: Wound-inducible amidohydrolases can simultaneously regulate jasmonate and auxin homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 2107–2120. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Yang, F.; Na, R.; Zhang, X.; Yang, S.; Gao, J.; Fan, M.; Zhao, Y.; Zhao, J. AtROP1 negatively regulates potato resistance to Phytophthora infestans via NADPH oxidase-mediated accumulation of H2O2. BMC Plant Biol. 2014, 14, 392. [Google Scholar] [CrossRef]
- Si, T.; Wang, X.; Zhao, C.; Huang, M.; Cai, J.; Zhou, Q.; Dai, T.; Jiang, D. The Role of hydrogen peroxide in mediating the mechanical wounding-induced freezing tolerance in wheat. Front. Plant Sci. 2018, 9, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Pan, Q.H.; Yang, H.R.; Liu, Y.Y.; Huang, W.D. Relationship between H2O2 and jasmonic acid in pea leaf wounding response. Russ. J. Plant Physiol. 2008, 55, 765–775. [Google Scholar] [CrossRef]
- Cui, F.; Brosché, M.; Sipari, N.; Tang, S.; Overmyer, K. Regulation of ABA dependent wound induced spreading cell death by MYB108. New Phytol. 2013, 200, 634–640. [Google Scholar] [CrossRef]
- Zhou, Q.; Gao, B.; Li, W.F.; Mao, J.; Yang, S.J.; Li, W.; Ma, Z.H.; Zhao, X.; Chen, B.H. Effects of exogenous growth regulators and bud picking on grafting of grapevine hard branches. Sci. Hortic. 2020, 264, 109186. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Ye, M.; Li, X.W.; Lin, S.B.; Sun, X.L. The jasmonic acid pathway positively regulates the polyphenol oxidase-based defense against tea geometrid caterpillars in the tea plant (Camellia sinensis). J. Chem. Ecol. 2020, 46, 308–316. [Google Scholar] [CrossRef]
- Yun, K.Y.; Park, M.R.; Mohanty, B.; Herath, V.; Xu, F.; Mauleon, R.; Wijaya, E.; Bajic, V.B.; Bruskiewich, R.; de Los Reyes, B.G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010, 10, 16. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Chen, X.; Ren, H.; Zhang, Z.; Zhang, H.; Wang, J.; Wang, X.C.; Huang, R. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 2007, 226, 815–825. [Google Scholar] [CrossRef]
- Iakimova, E.T.; Woltering, E.J. The wound response in fresh-cut lettuce involves programmed cell death events. Protoplasma 2018, 255, 1225–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugimoto, K.; Takeda, S.; Hirochika, H. MYB-Related transcription factor NtMYB2 induced by wounding and elicitors is a regulator of the tobacco retrotransposon Tto1 and defense-related genes. Plant Cell 2000, 12, 2511–2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotech. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Czemmel, S.; Heppel, S.C.; Bogs, J. R2R3 MYB transcription factors: Key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 2012, 249, 109–118. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, L.F.; Dai, L.J.; Yang, S.G.; Tian, W.M. Characterization of HbEREBP1, a wound-responsive transcription factor gene in laticifers of Heveabrasiliensis Muell. Arg. Mol. Biol. Rep. 2012, 39, 3713–3719. [Google Scholar] [CrossRef]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef]
- Van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef]
- Zhao, L.; Xu, S.; Chai, T.; Wang, T. OsAP2-1, an AP2-like gene from Oryza sativa, is required for flower development and male fertility. Sex. Plant Reprod. 2006, 19, 197–206. [Google Scholar] [CrossRef]
- Qiao, L.; Han, X.; Wang, H.; Gao, M.; Tian, J.; Lu, L.; Liu, X. Novel alternative for controlling enzymatic browning: Catalase and its application in fresh-cut potatoes. J. Food Sci. 2021, 86, 3529–3539. [Google Scholar] [CrossRef] [PubMed]



| Pathways | Gene Number | p-Value | Gene ID |
|---|---|---|---|
| Flavonoid biosynthesis | 7 | 0.000261 | PGSC0003DMG400003563, PGSC0003DMG400013684, PGSC0003DMG400029621, PGSC0003DMG400029620, PGSC0003DMG400014093, PGSC0003DMG400024643, PGSC0003DMG400019110, |
| Phenylpropanoid biosynthesis | 15 | 0.00058 | PGSC0003DMG400021152, PGSC0003DMG400010465, PGSC0003DMG400014867, PGSC0003DMG400012658, PGSC0003DMG400013684, PGSC0003DMG400030430, PGSC0003DMG400003013, PGSC0003DMG400026575, Novel01675, Novel01676, PGSC0003DMG400001774, PGSC0003DMG400016114, PGSC0003DMG400016113, PGSC0003DMG400015548, PGSC0003DMG400018446 |
| DNA replication | 6 | 0.00465 | PGSC0003DMG400024634, PGSC0003DMG403013782, PGSC0003DMG400019109, PGSC0003DMG400008308, PGSC0003DMG400008876, PGSC0003DMG400011837 |
| Zeatin biosynthesis | 5 | 0.005041 | PGSC0003DMG400008137, PGSC0003DMG400000277, PGSC0003DMG400005960, PGSC0003DMG402027210, PGSC0003DMG400027200 |
| Taurine and hypotaurine metabolism | 3 | 0.007694 | Novel02601, Novel00136, PGSC0003DMG400022764 |
| Butanoate metabolism | 4 | 0.008258 | Novel02601, Novel00136, PGSC0003DMG400022764, PGSC0003DMG400025228 |
| Pentose and glucuronate interconversions | 8 | 0.013374 | PGSC0003DMG400012640, PGSC0003DMG401019255, PGSC0003DMG402019255, PGSC0003DMG400031791, PGSC0003DMG400031816, PGSC0003DMG400015815, PGSC0003DMG400015230, PGSC0003DMG400029645 |
| Other types of O-glycan biosynthesis | 2 | 0.021717 | PGSC0003DMG400021652, PGSC0003DMG400014295 |
| Homologous recombination | 5 | 0.022726 | PGSC0003DMG400019109, PGSC0003DMG402010496, PGSC0003DMG400024634, PGSC0003DMG403013782, PGSC0003DMG400008876 |
| Phenylalanine metabolism | 9 | 0.034537 | PGSC0003DMG400026575, PGSC0003DMG400010465, PGSC0003DMG400014867, PGSC0003DMG400013684, PGSC0003DMG400030430, Novel01675, Novel01676 PGSC0003DMG400001774, PGSC0003DMG400015548 |
| Mismatch repair | 4 | 0.038656 | PGSC0003DMG400019109, PGSC0003DMG400024634, PGSC0003DMG403013782, PGSC0003DMG400008876 |
| Sesquiterpenoid and triterpenoid biosynthesis | 3 | 0.054213 | PGSC0003DMG400020939, PGSC0003DMG400017997, PGSC0003DMG400006713 |
| Alanine aspartate and glutamate metabolism | 4 | 0.058281 | Novel02601, Novel00136, PGSC0003DMG400022764, PGSC0003DMG400025228 |
| Biosynthesis of unsaturated fatty acids | 4 | 0.058281 | PGSC0003DMG400002943, PGSC0003DMG400023833, PGSC0003DMG400023843, PGSC0003DMG400000984 |
| Diterpenoid biosynthesis | 3 | 0.071621 | PGSC0003DMG400027645, PGSC0003DMG400001823, PGSC0003DMG400009021 |
| Starch and sucrose metabolism | 10 | 0.081798 | PGSC0003DMG400004659, PGSC0003DMG400031816, PGSC0003DMG401019255, PGSC0003DMG402019255, PGSC0003DMG400003013, PGSC0003DMG400029892, PGSC0003DMG400031800, PGSC0003DMG400013809, PGSC0003DMG400031791, PGSC0003DMG400009109 |
| Nitrogen metabolism | 3 | 0.096402 | PGSC0003DMG400016996, Novel01004, PGSC0003DMG400008094 |
| Cutin suberine and wax biosynthesis | 3 | 0.107167 | PGSC0003DMG402023841, PGSC0003DMG400007405, PGSC0003DMG401023841 |
| Nucleotide excision repair | 4 | 0.114737 | PGSC0003DMG400019109, PGSC0003DMG400024634, PGSC0003DMG403013782, PGSC0003DMG400008876 |
| 2-Oxocarboxylic acid metabolism | 4 | 0.114737 | PGSC0003DMG400006730, PGSC0003DMG400017513, Novel03278, Novel00090 |
| Cyanoamino acid metabolism | 3 | 0.154364 | PGSC0003DMG400004660, PGSC0003DMG400003013, Novel00090 |
| Circadian rhythm-plant | 3 | 0.154364 | PGSC0003DMG400029621, PGSC0003DMG400019110, PGSC0003DMG400029620 |
| Valine leucine and isoleucine biosynthesis | 2 | 0.184382 | PGSC0003DMG400006730, Novel03278 |
| Glucosinolate biosynthesis | 1 | 0.191014 | Novel00090 |
| Carotenoid biosynthesis | 2 | 0.271484 | PGSC0003DMG400018481, PGSC0003DMG400018480 |
| Monoterpenoid biosynthesis | 1 | 0.272409 | PGSC0003DMG400038230 |
| Pyrimidine metabolism | 5 | 0.288408 | PGSC0003DMG400019109, PGSC0003DMG400007335, PGSC0003DMG400020112, PGSC0003DMG400024725, PGSC0003DMG400033050 |
| Plant hormone signal transduction | 10 | 0.289881 | PGSC0003DMG400028800, PGSC0003DMG400006480, PGSC0003DMG400028254, PGSC0003DMG400019274, PGSC0003DMG400003227, PGSC0003DMG400008950, PGSC0003DMG400005585, PGSC0003DMG400008001, PGSC0003DMG400023678, PGSC0003DMG400003228 |
| β-Alanine metabolism | 3 | 0.298071 | Novel02601, Novel00136, PGSC0003DMG400022764 |
| Fatty acid metabolism | 4 | 0.318414 | PGSC0003DMG400002943, PGSC0003DMG400023833, PGSC0003DMG400023843, PGSC0003DMG400000984 |
| Degradation of aromatic compounds | 1 | 0.32805 | PGSC0003DMG400013684 |
| Glutathione metabolism | 4 | 0.336258 | PGSC0003DMG400002171, PGSC0003DMG400002167, PGSC0003DMG400024452 PGSC0003DMG400015726 |
| SNARE interactions in vesicular transport | 2 | 0.367906 | PGSC0003DMG400016283, PGSC0003DMG400000733 |
| Vitamin B6 metabolism | 1 | 0.379449 | Novel01870 |
| One carbon pool by folate | 1 | 0.395696 | PGSC0003DMG400004660 |
| Ubiquitin mediated proteolysis | 5 | 0.464025 | Novel02424, PGSC0003DMG400031240 PGSC0003DMG400000540, PGSC0003DMG400029675, PGSC0003DMG401027032 |
| Amino sugar and nucleotide sugar metabolism | 4 | 0.466024 | PGSC0003DMG400026853, PGSC0003DMG400031800, PGSC0003DMG400004659, PGSC0003DMG400026854 |
| Stilbenoid diarylheptanoid and gingerol biosynthesis | 1 | 0.498149 | PGSC0003DMG400013684 |
| Glyoxylate and dicarboxylate metabolism | 2 | 0.542125 | PGSC0003DMG400004660, Novel01867 |
| Histidine metabolism | 1 | 0.583279 | PGSC0003DMG400006731 |
| Photosynthesis-antenna proteins | 1 | 0.594203 | PGSC0003DMG400007375 |
| Pantothenate and CoA biosynthesis | 1 | 0.594203 | PGSC0003DMG400004975 |
| Glycine serine and threonine metabolism | 2 | 0.601955 | PGSC0003DMG400004660 Novel01867 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 1 | 0.615203 | PGSC0003DMG400013684 |
| Fatty acid elongation | 1 | 0.635119 | PGSC0003DMG400000984 |
| Plant-pathogen interaction | 5 | 0.654274 | PGSC0003DMG400028800, PGSC0003DMG400026914, PGSC0003DMG400013427, PGSC0003DMG400023125, PGSC0003DMG400006480 |
| Base excision repair | 1 | 0.68891 | PGSC0003DMG400019109 |
| Pyruvate metabolism | 2 | 0.709095 | PGSC0003DMG400006730, Novel03278 |
| Photosynthesis | 2 | 0.71458 | PGSC0003DMG400017556, PGSC0003DMG400017848 |
| Arginine and proline metabolism | 2 | 0.72529 | PGSC0003DMG400024452, PGSC0003DMG400017513 |
| Phagosome | 2 | 0.740718 | PGSC0003DMG400030627, PGSC0003DMG400000304 |
| Protein export | 1 | 0.755139 | Novel00948 |
| Peroxisome | 2 | 0.769364 | PGSC0003DMG400007405, PGSC0003DMG403005658 |
| Endocytosis | 3 | 0.789757 | PGSC0003DMG400011197, PGSC0003DMG400021197, PGSC0003DMG400013265 |
| Galactose metabolism | 1 | 0.812368 | PGSC0003DMG401001316 |
| Glycolysis/Gluconeogenesis | 2 | 0.855078 | PGSC0003DMG400006270, PGSC0003DMG401001316 |
| Biosynthesis of amino acids | 4 | 0.881486 | PGSC0003DMG400004660, PGSC0003DMG400006730, Novel03278 PGSC0003DMG400017513 |
| Carbon fixation in photosynthetic organisms | 1 | 0.883868 | PGSC0003DMG400029406 |
| Purine metabolism | 2 | 0.931195 | PGSC0003DMG400019109, PGSC0003DMG400007335 |
| Cysteine and methionine metabolism | 1 | 0.945021 | PGSC0003DMG400021426 |
| mRNA surveillance pathway | 1 | 0.965116 | PGSC0003DMG400003416 |
| Protein processing in endoplasmic reticulum | 2 | 0.991942 | PGSC0003DMG400011197, PGSC0003DMG400016468 |
| Carbon metabolism | 2 | 0.992669 | PGSC0003DMG400029406, PGSC0003DMG400004660 |
| Spliceosome | 1 | 0.995473 | PGSC0003DMG400011197 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Wang, W.; Shan, J.; Li, C.; Suo, H.; Liu, J.; An, K.; Li, X.; Xiong, X. A Genome-Wide View of the Transcriptome Dynamics of Fresh-Cut Potato Tubers. Genes 2023, 14, 181. https://doi.org/10.3390/genes14010181
Wang L, Wang W, Shan J, Li C, Suo H, Liu J, An K, Li X, Xiong X. A Genome-Wide View of the Transcriptome Dynamics of Fresh-Cut Potato Tubers. Genes. 2023; 14(1):181. https://doi.org/10.3390/genes14010181
Chicago/Turabian StyleWang, Li, Wanxing Wang, Jianwei Shan, Chengchen Li, Haicui Suo, Jitao Liu, Kang An, Xiaobo Li, and Xingyao Xiong. 2023. "A Genome-Wide View of the Transcriptome Dynamics of Fresh-Cut Potato Tubers" Genes 14, no. 1: 181. https://doi.org/10.3390/genes14010181
APA StyleWang, L., Wang, W., Shan, J., Li, C., Suo, H., Liu, J., An, K., Li, X., & Xiong, X. (2023). A Genome-Wide View of the Transcriptome Dynamics of Fresh-Cut Potato Tubers. Genes, 14(1), 181. https://doi.org/10.3390/genes14010181




