Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out
Abstract
1. Introduction
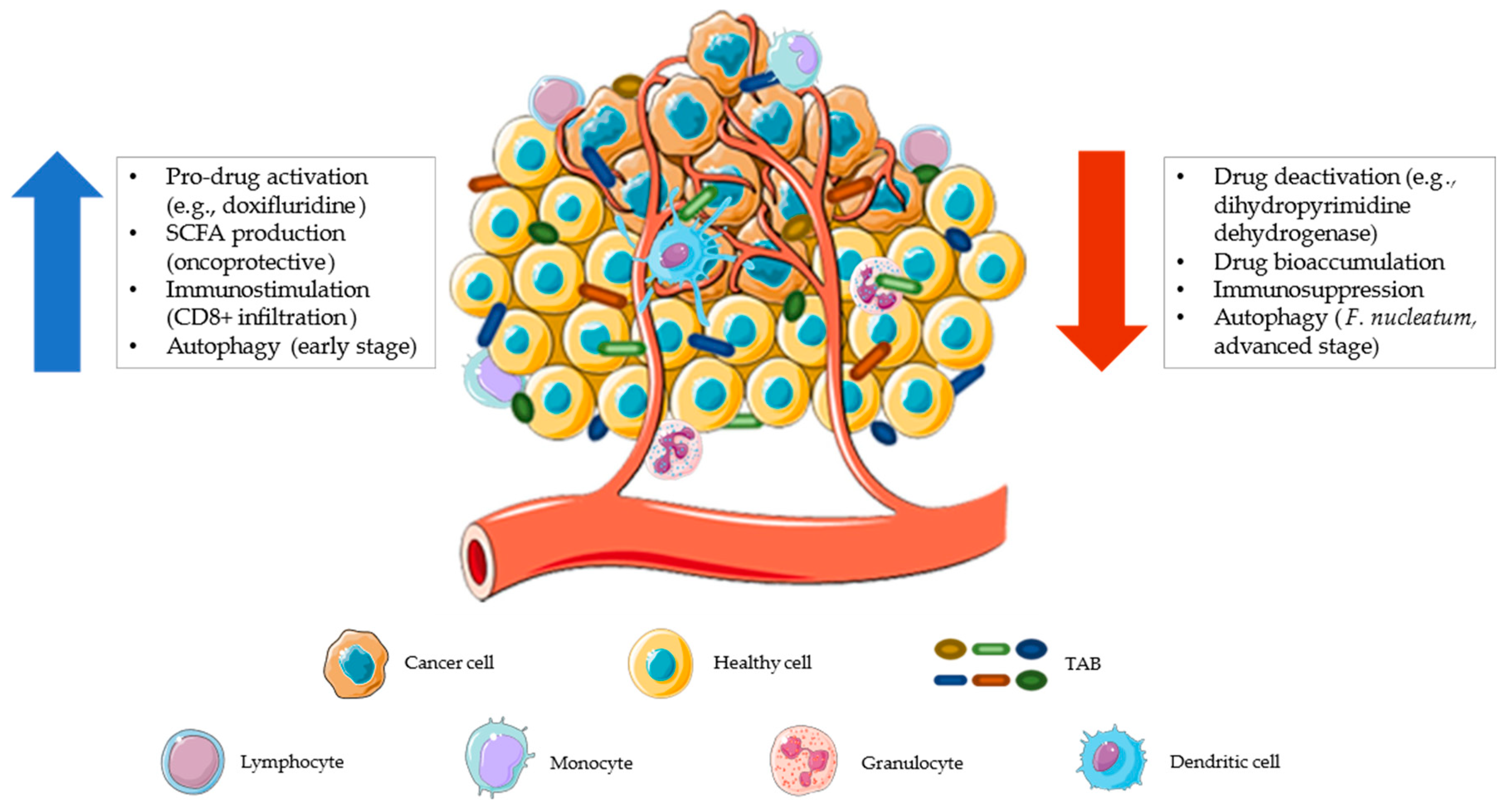
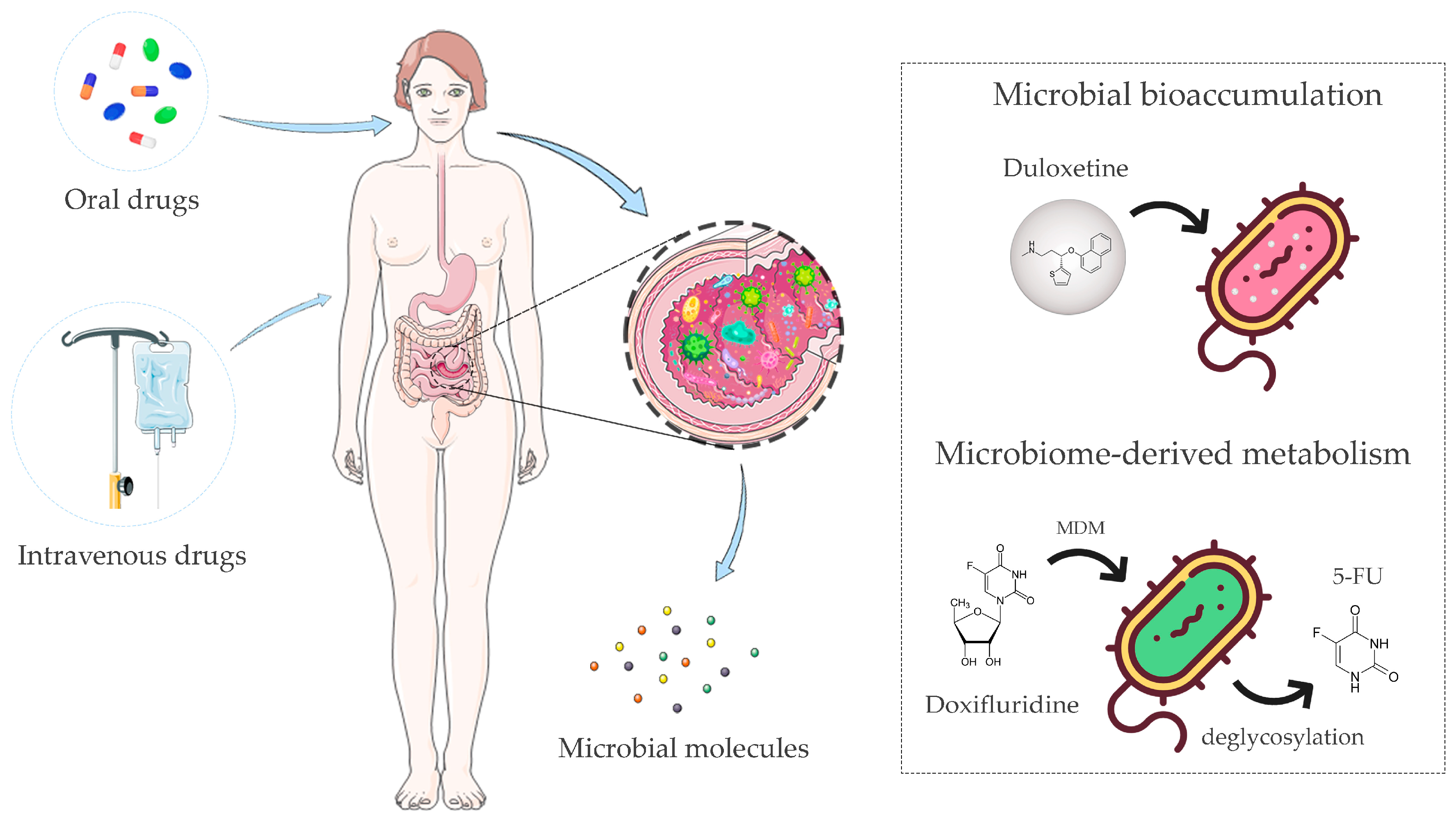
2. Pharmacomicrobiomics and Toxicomicrobiomics
3. Gut Microbiome Impact on Immuno-Chemotherapeutics
3.1. Untargeted Traditional Chemotherapy
3.2. Targeted Immuno-Chemotherapy
3.3. Monoclonal Antibodies
3.4. Small-Molecule Inhibitors
4. Modulation of the Gut Microbiota to Improve the Therapeutic Outcome
4.1. Prebiotics
4.2. Probiotics
4.3. Antibiotics
4.4. FMT
5. Bacterial-Based Anticancer Technologies
6. Multi-Omics Strategy to Study Drug–Microbiota Interactions and Derive Personalized Precision Medicine Models
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut Microbiota: Role in Pathogen Colonization, Immune Responses, and Inflammatory Disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Rizkallah, M.; Saad, R.; Aziz, R. The Human Microbiome Project, Personalized Medicine and the Birth of Pharmacomicrobiomics. Curr. Pharm. Person. Med. 2012, 8, 182–193. [Google Scholar] [CrossRef]
- Spanogiannopoulos, P.; Bess, E.N.; Carmody, R.N.; Turnbaugh, P.J. The Microbial Pharmacists within Us: A Metagenomic View of Xenobiotic Metabolism. Nat. Rev. Microbiol. 2016, 14, 273–287. [Google Scholar] [CrossRef]
- Pant, A.; Maiti, T.K.; Mahajan, D.; Das, B. Human Gut Microbiota and Drug Metabolism. Microb. Ecol. 2022, 1, 1–15. [Google Scholar] [CrossRef]
- Lindell, A.E.; Zimmermann-Kogadeeva, M.; Patil, K.R. Multimodal Interactions of Drugs, Natural Compounds and Pollutants with the Gut Microbiota. Nat. Rev. Microbiol. 2022, 20, 431–443. [Google Scholar] [CrossRef]
- Barone, M.; Rampelli, S.; Biagi, E.; Bertozzi, S.M.; Falchi, F.; Cavalli, A.; Armirotti, A.; Brigidi, P.; Turroni, S.; Candela, M. Searching for New Microbiome-Targeted Therapeutics through a Drug Repurposing Approach. J. Med. Chem. 2021, 64, 17277–17286. [Google Scholar] [CrossRef]
- Roy, S.; Trinchieri, G. Microbiota: A Key Orchestrator of Cancer Therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The Microbiome, Cancer, and Cancer Therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Peled, J.U.; Gomes, A.L.C.; Devlin, S.M.; Littmann, E.R.; Taur, Y.; Sung, A.D.; Weber, D.; Hashimoto, D.; Slingerland, A.E.; Slingerland, J.B.; et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N. Engl. J. Med. 2020, 382, 822–834. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, F.; Barone, M.; Tavella, T.; Rampelli, S.; Brigidi, P.; Turroni, S. Host Microbiomes in Tumor Precision Medicine: How Far Are We? Curr. Med. Chem. 2022, 29, 3202–3230. [Google Scholar] [CrossRef] [PubMed]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The Sequence of the Human Genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Mahurkar, A.; Zhou, W.; Buck, G.A.; Snyder, M.P.; Strauss, J.F.; Weinstock, G.M.; et al. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef]
- Abdelsalam, N.A.; Ramadan, A.T.; ElRakaiby, M.T.; Aziz, R.K. Toxicomicrobiomics: The Human Microbiome vs. Pharmaceutical, Dietary, and Environmental Xenobiotics. Front. Pharmacol. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Hitchings, R.; Kelly, L. Predicting and Understanding the Human Microbiome’s Impact on Pharmacology. Trends Pharmacol. Sci. 2019, 40, 495–505. [Google Scholar] [CrossRef]
- Klünemann, M.; Andrejev, S.; Blasche, S.; Mateus, A.; Phapale, P.; Devendran, S.; Vappiani, J.; Simon, B.; Scott, T.A.; Kafkia, E.; et al. Bioaccumulation of Therapeutic Drugs by Human Gut Bacteria. Nature 2021, 597, 533–538. [Google Scholar] [CrossRef]
- Collins, S.L.; Patterson, A.D. The Gut Microbiome: An Orchestrator of Xenobiotic Metabolism. Acta Pharm. Sin. B 2020, 10, 19–32. [Google Scholar] [CrossRef]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping Human Microbiome Drug Metabolism by Gut Bacteria and Their Genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Baillie, R.A.; Zhu, H.; Zeng, Z.B.; Wiest, M.M.; Nguyen, U.T.; Wojnoonski, K.; Watkins, S.M.; Trupp, M.; Krauss, R.M. Enteric Microbiome Metabolites Correlate with Response to Simvastatin Treatment. PLoS ONE 2011, 6, e25482. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Zhao, H.; Keerthisinghe, T.P.; Yu, Y.; Chen, D.; Huang, Y.; Fang, M. Gut Microbial Metabolite P-Cresol Alters Biotransformation of Bisphenol A: Enzyme Competition or Gene Induction? J. Hazard. Mater. 2022, 426, 128093. [Google Scholar] [CrossRef] [PubMed]
- Clayton, T.A.; Baker, D.; Lindon, J.C.; Everett, J.R.; Nicholson, J.K. Pharmacometabonomic Identification of a Significant Host-Microbiome Metabolic Interaction Affecting Human Drug Metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 14728–14733. [Google Scholar] [CrossRef] [PubMed]
- Dawson, L.F.; Donahue, E.H.; Cartman, S.T.; Barton, R.H.; Bundy, J.; McNerney, R.; Minton, N.P.; Wren, B.W. The Analysis of Para-Cresol Production and Tolerance in Clostridium Difficile 027 and 012 Strains. BMC Microbiol. 2011, 11, 86. [Google Scholar] [CrossRef]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.A.; Mani, S.; et al. Alleviating Cancer Drug Toxicity by Inhibiting a Bacterial Enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef]
- Wastyk, H.C.; Fragiadakis, G.K.; Perelman, D.; Dahan, D.; Merrill, B.D.; Yu, F.B.; Topf, M.; Gonzalez, C.G.; Van Treuren, W.; Han, S.; et al. Gut-Microbiota-Targeted Diets Modulate Human Immune Status. Cell 2021, 184, 4137–4153.e14. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Javdan, B.; Lopez, J.G.; Chankhamjon, P.; Lee, Y.C.J.; Hull, R.; Wu, Q.; Wang, X.; Chatterjee, S.; Donia, M.S. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 2020, 181, 1661–1679.e22. [Google Scholar] [CrossRef]
- Zimmermann-Kogadeeva, M.; Zimmermann, M.; Goodman, A.L. Insights from Pharmacokinetic Models of Host-Microbiome Drug Metabolism. Gut Microbes 2020, 11, 587–596. [Google Scholar] [CrossRef]
- Zimmermann, M.; Patil, K.R.; Typas, A.; Maier, L. Towards a Mechanistic Understanding of Reciprocal Drug–Microbiome Interactions. Mol. Syst. Biol. 2021, 17, e10116. [Google Scholar] [CrossRef]
- Campbell, J.M.; Fahey, G.C.; Wolf, B.W. Selected Indigestible Oligosaccharides Affect Large Bowel Mass, Cecal and Fecal Short-Chain Fatty Acids, PH and Microflora in Rats. J. Nutr. 1997, 127, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J. Lipid Res. 2013, 54, 2325. [Google Scholar] [CrossRef]
- Abuhelwa, A.Y.; Foster, D.J.R.; Upton, R.N. A Quantitative Review and Meta-Models of the Variability and Factors Affecting Oral Drug Absorption-Part I: Gastrointestinal PH. AAPS J. 2016, 18, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Stillhart, C.; Vučićević, K.; Augustijns, P.; Basit, A.W.; Batchelor, H.; Flanagan, T.R.; Gesquiere, I.; Greupink, R.; Keszthelyi, D.; Koskinen, M.; et al. Impact of Gastrointestinal Physiology on Drug Absorption in Special Populations––An UNGAP Review. Eur. J. Pharm. Sci. 2020, 147, 105280. [Google Scholar] [CrossRef] [PubMed]
- Firrman, J.; Liu, L.S.; Mahalak, K.; Tanes, C.; Bittinger, K.; Tu, V.; Bobokalonov, J.; Mattei, L.; Zhang, H.; Van Den Abbeele, P. The Impact of Environmental PH on the Gut Microbiota Community Structure and Short Chain Fatty Acid Production. FEMS Microbiol. Ecol. 2022, 98, fiac038. [Google Scholar] [CrossRef]
- He, Y.; Fu, L.; Li, Y.; Wang, W.; Gong, M.; Zhang, J.; Dong, X.; Huang, J.; Wang, Q.; Mackay, C.R.; et al. Gut Microbial Metabolites Facilitate Anticancer Therapy Efficacy by Modulating Cytotoxic CD8+ T Cell Immunity. Cell Metab. 2021, 33, 988–1000.e7. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, S.Y.; Yoon, Y.; Park, C.; Sohn, J.; Jeong, J.J.; Jeon, B.N.; Jang, M.; An, C.; Lee, S.; et al. Bifidobacterium Bifidum Strains Synergize with Immune Checkpoint Inhibitors to Reduce Tumour Burden in Mice. Nat. Microbiol. 2021, 6, 277–288. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The Intestinal Microbiota Modulates the Anticancer Immune Effects of Cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Kocahan, S.; Dogan, Z.; Erdemli, E.; Taskin, E. Protective Effect of Quercetin Against Oxidative Stress-Induced Toxicity Associated With Doxorubicin and Cyclophosphamide in Rat Kidney and Liver Tissue. Iran. J. Kidney Dis. 2017, 11, 124–131. [Google Scholar]
- Desai, V.G.; Herman, E.H.; Moland, C.L.; Branham, W.S.; Lewis, S.M.; Davis, K.J.; George, N.I.; Lee, T.; Kerr, S.; Fuscoe, J.C. Development of Doxorubicin-Induced Chronic Cardiotoxicity in the B6C3F1 Mouse Model. Toxicol. Appl. Pharmacol. 2013, 266, 109–121. [Google Scholar] [CrossRef]
- Manno, R.A.; Grassetti, A.; Oberto, G.; Nyska, A.; Ramot, Y. The Minipig as a New Model for the Evaluation of Doxorubicin-Induced Chronic Toxicity. J. Appl. Toxicol. 2016, 36, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Wuri, J.; Zheng, Z.; Li, W.; Yan, T. Microbiota Modulate Doxorubicin Induced Cardiotoxicity. Eur. J. Pharm. Sci. 2021, 166, 105977. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wei, S.; Jiang, C.; Xiao, Z.; Liu, J.; Peng, W.; Zhang, B.; Li, W. Involvement of Abnormal Gut Microbiota Composition and Function in Doxorubicin-Induced Cardiotoxicity. Front. Cell. Infect. Microbiol. 2022, 12, 155. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Culp, E.; Perry, J.; Lau, J.T.; Macneil, L.T.; Surette, M.G.; Wright, G.D. Transformation of the Anticancer Drug Doxorubicin in the Human Gut Microbiome. ACS Infect. Dis. 2018, 4, 68–76. [Google Scholar] [CrossRef]
- Bailly, C. Irinotecan: 25 Years of Cancer Treatment. Pharmacol. Res. 2019, 148, 104398. [Google Scholar] [CrossRef]
- Kciuk, M.; Marciniak, B.; Kontek, R. Irinotecan—Still an Important Player in Cancer Chemotherapy: A Comprehensive Overview. Int. J. Mol. Sci. 2020, 21, 4919. [Google Scholar] [CrossRef]
- Yue, B.; Gao, R.; Wang, Z.; Dou, W. Microbiota-Host-Irinotecan Axis: A New Insight Toward Irinotecan Chemotherapy. Front. Cell. Infect. Microbiol. 2021, 11, 710945. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Stringer, A.M.; Gibson, R.J.; Logan, R.M.; Bowen, J.M.; Yeoh, A.S.J.; Keefe, D.M.K. Faecal Microflora and β-Glucuronidase Expression Are Altered in an Irinotecan-Induced Diarrhoea Model in Rats. Cancer Biol. Ther. 2008, 7, 1919–1925. [Google Scholar] [CrossRef]
- Lin, X.B.; Dieleman, L.A.; Ketabi, A.; Bibova, I.; Sawyer, M.B.; Xue, H.; Field, C.J.; Baracos, V.E.; Gänzle, M.G. Irinotecan (CPT-11) Chemotherapy Alters Intestinal Microbiota in Tumour Bearing Rats. PLoS ONE 2012, 7, e39764. [Google Scholar] [CrossRef]
- Spanogiannopoulos, P.; Kyaw, T.S.; Guthrie, B.G.H.; Bradley, P.H.; Lee, J.V.; Melamed, J.; Malig, Y.N.A.; Lam, K.N.; Gempis, D.; Sandy, M.; et al. Host and Gut Bacteria Share Metabolic Pathways for Anti-Cancer Drug Metabolism. Nat. Microbiol. 2022, 7, 1605–1620. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Zhang, S.; Li, H.; Yang, F.; Mushtaq, N.; Ullah, S.; Shi, Y.; An, C.; Xu, J. The Influence of Gut Microbiota Dysbiosis to the Efficacy of 5-Fluorouracil Treatment on Colorectal Cancer. Biomed. Pharmacother. 2018, 108, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Letertre, M.P.M.; Munjoma, N.; Wolfer, K.; Pechlivanis, A.; McDonald, J.A.K.; Hardwick, R.N.; Cherrington, N.J.; Coen, M.; Nicholson, J.K.; Hoyles, L.; et al. A Two-Way Interaction between Methotrexate and the Gut Microbiota of Male Sprague-Dawley Rats. J. Proteome Res. 2020, 19, 3326–3339. [Google Scholar] [CrossRef]
- Buchen, S.; Ngampolo, D.; Melton, R.G.; Hasan, C.; Zoubek, A.; Henze, G.; Bode, U.; Fleischhack, G. Carboxypeptidase G2 Rescue in Patients with Methotrexate Intoxication and Renal Failure. Br. J. Cancer 2005, 92, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.R.; Alexander, M.; Deshpande, I.; Stapleton-Gray, K.; Rimal, B.; Patterson, A.D.; Ubeda, C.; Scher, J.U.; Turnbaugh, P.J. Methotrexate Impacts Conserved Pathways in Diverse Human Gut Bacteria Leading to Decreased Host Immune Activation. Cell Host Microbe 2021, 29, 362–377.e11. [Google Scholar] [CrossRef]
- Artacho, A.; Isaac, S.; Nayak, R.; Flor-Duro, A.; Alexander, M.; Koo, I.; Manasson, J.; Smith, P.B.; Rosenthal, P.; Homsi, Y.; et al. The Pretreatment Gut Microbiome Is Associated With Lack of Response to Methotrexate in New-Onset Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 931–942. [Google Scholar] [CrossRef]
- Kazazi-Hyseni, F.; Beijnen, J.H.; Schellens, J.H.M. Bevacizumab. Oncologist 2010, 15, 819–825. [Google Scholar] [CrossRef]
- Iqbal, N.; Iqbal, N. Imatinib: A Breakthrough of Targeted Therapy in Cancer. Chemother. Res. Pract. 2014, 2014, 357027. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z. The History and Advances in Cancer Immunotherapy: Understanding the Characteristics of Tumor-Infiltrating Immune Cells and Their Therapeutic Implications. Cell. Mol. Immunol. 2020, 17, 807–821. [Google Scholar] [CrossRef]
- Zhou, C.B.; Zhou, Y.L.; Fang, J.Y. Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer 2021, 7, 647–660. [Google Scholar] [CrossRef]
- Nishimura, C.D.; Pulanco, M.C.; Cui, W.; Lu, L.; Zang, X. PD-L1 and B7-1 Cis-Interaction: New Mechanisms in Immune Checkpoints and Immunotherapies. Trends Mol. Med. 2021, 27, 207–219. [Google Scholar] [CrossRef]
- Khan, M.; Arooj, S.; Wang, H. Soluble B7-CD28 Family Inhibitory Immune Checkpoint Proteins and Anti-Cancer Immunotherapy. Front. Immunol. 2021, 12, 651634. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Vétizou, M.; Daillère, R.; Roberti, M.P.; Yamazaki, T.; Routy, B.; Lepage, P.; Boneca, I.G.; Chamaillard, M.; Kroemer, G.; et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016, 44, 1255–1269. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, S.; Zheng, B.; Qiu, X.; Wang, H.; Chen, L. Modulation of Gut Microbiota to Enhance Effect of Checkpoint Inhibitor Immunotherapy. Front. Immunol. 2021, 12, 2554. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut Microbiome Modulates Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic Short Chain Fatty Acids Limit Antitumor Effect of CTLA-4 Blockade in Hosts with Cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small Molecules in Targeted Cancer Therapy: Advances, Challenges, and Future Perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Sun, G.; Rong, D.; Li, Z.; Sun, G.; Wu, F.; Li, X.; Cao, H.; Cheng, Y.; Tang, W.; Sun, Y. Role of Small Molecule Targeted Compounds in Cancer: Progress, Opportunities, and Challenges. Front. Cell Dev. Biol. 2021, 9, 2043. [Google Scholar] [CrossRef]
- Sochacka-Ćwikła, A.; Mączyński, M.; Regiec, A. FDA-Approved Small Molecule Compounds as Drugs for Solid Cancers from Early 2011 to the End of 2021. Molecules 2022, 27, 2259. [Google Scholar] [CrossRef] [PubMed]
- Heshiki, Y.; Vazquez-Uribe, R.; Li, J.; Ni, Y.; Quainoo, S.; Imamovic, L.; Li, J.; Sørensen, M.; Chow, B.K.C.; Weiss, G.J.; et al. Predictable Modulation of Cancer Treatment Outcomes by the Gut Microbiota. Microbiome 2020, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Vasilyeva, A.; Durmus, S.; Li, L.; Wagenaar, E.; Hu, S.; Gibson, A.A.; Panetta, J.C.; Mani, S.; Sparreboom, A.; Baker, S.D.; et al. Hepatocellular Shuttling and Recirculation of Sorafenib-Glucuronide Is Dependent on Abcc2, Abcc3, and Oatp1a/1b. Cancer Res. 2015, 75, 2729–2736. [Google Scholar] [CrossRef] [PubMed]
- Pollet, R.M.; D’Agostino, E.H.; Walton, W.G.; Xu, Y.; Little, M.S.; Biernat, K.A.; Pellock, S.J.; Patterson, L.M.; Creekmore, B.C.; Isenberg, H.N.; et al. An Atlas of β-Glucuronidases in the Human Intestinal Microbiome. Structure 2017, 25, 967–977.e5. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, L.; Xu, Y.; Yu, H.; Chen, Y.; Zhao, H.; Lei, J.; Zhou, Y.; Zhang, J.; Wang, J.; et al. Microbiota-Derived SSL6 Enhances the Sensitivity of Hepatocellular Carcinoma to Sorafenib by down-Regulating Glycolysis. Cancer Lett. 2020, 481, 32–44. [Google Scholar] [CrossRef]
- Weng, M.L.; Chen, W.K.; Chen, X.Y.; Lu, H.; Sun, Z.R.; Yu, Q.; Sun, P.F.; Xu, Y.J.; Zhu, M.M.; Jiang, N.; et al. Fasting Inhibits Aerobic Glycolysis and Proliferation in Colorectal Cancer via the Fdft1-Mediated AKT/MTOR/HIF1α Pathway Suppression. Nat. Commun. 2020, 11, 1869. [Google Scholar] [CrossRef]
- Kojima, Y.; Volkmer, J.P.; McKenna, K.; Civelek, M.; Lusis, A.J.; Miller, C.L.; Direnzo, D.; Nanda, V.; Ye, J.; Connolly, A.J.; et al. CD47-Blocking Antibodies Restore Phagocytosis and Prevent Atherosclerosis. Nature 2016, 536, 86–90. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H. Gut Microbiota Modulation: A Tool for the Management of Colorectal Cancer. J. Transl. Med. 2022, 20, 178. [Google Scholar] [CrossRef]
- Fong, W.; Li, Q.; Yu, J. Gut Microbiota Modulation: A Novel Strategy for Prevention and Treatment of Colorectal Cancer. Oncogene 2020, 39, 4925–4943. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Peris, P.G.; Velasco, C.; Lozano, M.A.; Moreno, Y.; Paron, L.; de la Cuerda, C.; Bretón, I.; Camblor, M.; García-Hernández, J.; Guarner, F.; et al. Effect of a Mixture of Inulin and Fructo-Oligosaccharide on Lactobacillus and Bifidobacterium Intestinal Microbiota of Patients Receiving Radiotherapy: A Randomised, Double-Blind, Placebo-Controlled Trial. Nutr. Hosp. 2012, 27, 1908–1915. [Google Scholar] [CrossRef]
- Abot, A.; Brochot, A.; Pomié, N.; Wemelle, E.; Druart, C.; Régnier, M.; Delzenne, N.M.; de Vos, W.M.; Knauf, C.; Cani, P.D. Camu-Camu Reduces Obesity and Improves Diabetic Profiles of Obese and Diabetic Mice: A Dose-Ranging Study. Metabolites 2022, 12, 301. [Google Scholar] [CrossRef] [PubMed]
- Messaoudene, M.; Pidgeon, R.; Richard, C.; Ponce, M.; Diop, K.; Benlaifaoui, M.; Nolin-Lapalme, A.; Cauchois, F.; Malo, J.; Belkaid, W.; et al. A Natural Polyphenol Exerts Antitumor Activity and Circumvents Anti–PD-1 Resistance through Effects on the Gut Microbiota. Cancer Discov. 2022, 12, 1070–1087. [Google Scholar] [CrossRef] [PubMed]
- Masetti, R.; Biagi, E.; Zama, D.; Muratore, E.; D’Amico, F.; Leardini, D.; Turroni, S.; Prete, A.; Brigidi, P.; Pession, A. Early Modifications of the Gut Microbiome in Children with Hepatic Sinusoidal Obstruction Syndrome after Hematopoietic Stem Cell Transplantation. Sci. Rep. 2021, 11, 14307–14312. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Lindner, S.; Gomes, A.L.C.; Devlin, S.M.; Shah, G.L.; Sung, A.D.; Sauter, C.S.; Landau, H.J.; Dahi, P.B.; Perales, M.A.; et al. Fecal Microbiota Diversity Disruption and Clinical Outcomes after Auto-HCT: A Multicenter Observational Study. Blood 2021, 137, 1527–1537. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The Pros, Cons, and Many Unknowns of Probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and Prebiotics in Intestinal Health and Disease: From Biology to the Clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef]
- Górska, A.; Przystupski, D.; Niemczura, M.J.; Kulbacka, J. Probiotic Bacteria: A Promising Tool in Cancer Prevention and Therapy. Curr. Microbiol. 2019, 76, 939–949. [Google Scholar] [CrossRef]
- Gianotti, L.; Morelli, L.; Galbiati, F.; Rocchetti, S.; Coppola, S.; Beneduce, A.; Gilardini, C.; Zonenschain, D.; Nespoli, A.; Braga, M. A Randomized Double-Blind Trial on Perioperative Administration of Probiotics in Colorectal Cancer Patients. World J. Gastroenterol. 2010, 16, 167–175. [Google Scholar] [CrossRef]
- Consoli, M.L.D.; Da Silva, R.S.; Nicoli, J.R.; Bruña-Romero, O.; Da Silva, R.G.; De Vasconcelos Generoso, S.; Correia, M.I.T.D. Randomized Clinical Trial. J. Parenter. Enter. Nutr. 2016, 40, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Zaharuddin, L.; Mokhtar, N.M.; Muhammad Nawawi, K.N.; Raja Ali, R.A. A Randomized Double-Blind Placebo-Controlled Trial of Probiotics in Post-Surgical Colorectal Cancer. BMC Gastroenterol. 2019, 19, 131. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, Y.; Lee, S.; Kim, Y.; Jeon, B.; Kim, H.; Park, H. Live Biotherapeutic Lactococcus Lactis GEN3013 Enhances Antitumor Efficacy of Cancer Treatment via Modulation of Cancer Progression and Immune System. Cancers 2022, 14, 4083. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, M.; Dong, W.; Deng, B.; Wang, S.; Zhang, Y.; Wang, S.; Luo, S.; Wang, W.; Qi, Y.; et al. Secondary Bile Acid-Induced Dysbiosis Promotes Intestinal Carcinogenesis. Int. J. Cancer 2017, 140, 2545–2556. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Baxter, N.T.; Iverson, K.D.; Sadler, W.D.; Petrosino, J.F.; Chen, G.Y.; Schloss, P.D. The Gut Microbiome Modulates Colon Tumorigenesis. MBio 2013, 4, e00692-13. [Google Scholar] [CrossRef]
- Destefano Shields, C.E.; Van Meerbeke, S.W.; Housseau, F.; Wang, H.; Huso, D.L.; Casero, R.A.; O’Hagan, H.M.; Sears, C.L. Reduction of Murine Colon Tumorigenesis Driven by Enterotoxigenic Bacteroides Fragilis Using Cefoxitin Treatment. J. Infect. Dis. 2016, 214, 122–129. [Google Scholar] [CrossRef]
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 731. [Google Scholar] [CrossRef]
- Taylor, M.R.; Flannigan, K.L.; Rahim, H.; Mohamud, A.; Lewis, I.A.; Hirota, S.A.; Greenway, S.C. Vancomycin Relieves Mycophenolate Mofetil–Induced Gastrointestinal Toxicity by Eliminating Gut Bacterial β-Glucuronidase Activity. Sci. Adv. 2019, 5, eaax2358. [Google Scholar] [CrossRef]
- Kim, A.H.J.; Lee, Y.; Kim, E.; Ji, S.C.; Chung, J.Y.; Cho, J.Y. Assessment of Oral Vancomycin-Induced Alterations in Gut Bacterial Microbiota and Metabolome of Healthy Men. Front. Cell. Infect. Microbiol. 2021, 11, 412. [Google Scholar] [CrossRef]
- Tzeng, A.; Sangwan, N.; Jia, M.; Liu, C.C.; Keslar, K.S.; Downs-Kelly, E.; Fairchild, R.L.; Al-Hilli, Z.; Grobmyer, S.R.; Eng, C. Human Breast Microbiome Correlates with Prognostic Features and Immunological Signatures in Breast Cancer. Genome Med. 2021, 13, 60. [Google Scholar] [CrossRef]
- Molska, M.; Reguła, J.; Zielińska-Dawidziak, M.; Tomczak, A.; Świeca, M. Starch and Protein Analysis in Buckwheat (Fagopyrum Esculentum Moench) Sprouts Enriched with Probiotic Yeast. LWT 2022, 168, 113903. [Google Scholar] [CrossRef]
- Terveer, E.M.; van Beurden, Y.H.; Goorhuis, A.; Mulder, C.J.J.; Kuijper, E.J.; Keller, J.J. Working Group of The Netherlands Donor Feces Bank Faecal Microbiota Transplantation in Clinical Practice. Gut 2018, 67, 196. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Wu, J.; Jin, D.; Wang, B.; Cao, H. Fecal Microbiota Transplantation in Cancer Management: Current Status and Perspectives. Int. J. Cancer 2019, 145, 2021–2031. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hua, W.; Li, C.; Chang, H.; Liu, R.; Ni, Y.; Sun, H.; Li, Y.; Wang, X.; Hou, M.; et al. Protective Role of Fecal Microbiota Transplantation on Colitis and Colitis-Associated Colon Cancer in Mice Is Associated With Treg Cells. Front. Microbiol. 2019, 10, 2498. [Google Scholar] [CrossRef] [PubMed]
- Santoni, M.; Piva, F.; Conti, A.; Santoni, A.; Cimadamore, A.; Scarpelli, M.; Battelli, N.; Montironi, R. Re: Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Eur. Urol. 2018, 74, 521–522. [Google Scholar] [CrossRef]
- Ding, X.; Li, Q.; Li, P.; Chen, X.; Xiang, L.; Bi, L.; Zhu, J.; Huang, X.; Cui, B.; Zhang, F. Fecal Microbiota Transplantation: A Promising Treatment for Radiation Enteritis? Radiother. Oncol. 2020, 143, 12–18. [Google Scholar] [CrossRef]
- Ianiro, G.; Rossi, E.; Thomas, A.M.; Schinzari, G.; Masucci, L.; Quaranta, G.; Settanni, C.R.; Lopetuso, L.R.; Armanini, F.; Blanco-Miguez, A.; et al. Faecal Microbiota Transplantation for the Treatment of Diarrhoea Induced by Tyrosine-Kinase Inhibitors in Patients with Metastatic Renal Cell Carcinoma. Nat. Commun. 2020, 11, 4333. [Google Scholar] [CrossRef]
- Zhang, T.; Lu, G.; Zhao, Z.; Liu, Y.; Shen, Q.; Li, P.; Chen, Y.; Yin, H.; Wang, H.; Marcella, C.; et al. Washed Microbiota Transplantation vs. Manual Fecal Microbiota Transplantation: Clinical Findings, Animal Studies and in Vitro Screening. Protein Cell 2020, 11, 251–266. [Google Scholar] [CrossRef]
- Zhang, T.; Ding, X.; Dai, M.; Zhang, H.; Xiao, F.; He, X.; Zhang, F.; Zhang, X. Washed Microbiota Transplantation in Patients with Respiratory Spreading Diseases: Practice Recommendations. Med. Microecol. 2021, 7, 100024. [Google Scholar] [CrossRef]
- Wardill, H.R.; Secombe, K.R.; Bryant, R.V.; Hazenberg, M.D.; Costello, S.P. Adjunctive Fecal Microbiota Transplantation in Supportive Oncology: Emerging Indications and Considerations in Immunocompromised Patients. eBioMedicine 2019, 44, 730–740. [Google Scholar] [CrossRef]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The Human Tumor Microbiome Is Composed of Tumor Type-Specific Intracellular Bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Q.; Lin, Y.; Ma, Y.; Li, X.; Liang, J.; Chen, Z.; Liu, K.; Huang, Y.; Luo, H.; Huang, R.; et al. Exploring the Emerging Role of the Gut Microbiota and Tumor Microenvironment in Cancer Immunotherapy. Front. Immunol. 2020, 11, 612202. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Helbig, L.; Fu, J.; Bian, X.; Herrmann, J.; Baumann, M.; Stewart, A.F.; Müller, R.; Li, A.; Zips, D.; et al. Expressing Cytotoxic Compounds in Escherichia Coli Nissle 1917 for Tumor-Targeting Therapy. Res. Microbiol. 2019, 170, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.C.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium Nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Song, W.; Anselmo, A.C.; Huang, L. Nanotechnology Intervention of the Microbiome for Cancer Therapy. Nat. Nanotechnol. 2019, 14, 1093–1103. [Google Scholar] [CrossRef]
- Ye, Z.; Liang, L.; Lu, H.; Shen, Y.; Zhou, W.; Li, Y. Nanotechnology-Employed Bacteria-Based Delivery Strategy for Enhanced Anticancer Therapy. Int. J. Nanomed. 2021, 16, 8069–8086. [Google Scholar] [CrossRef]
- Lou, X.; Chen, Z.; He, Z.; Sun, M.; Sun, J. Bacteria-Mediated Synergistic Cancer Therapy: Small Microbiome Has a Big Hope. Nano-Micro Lett. 2021, 13, 37. [Google Scholar] [CrossRef]
- Kasinskas, R.W.; Forbes, N.S. Salmonella Typhimurium Specifically Chemotax and Proliferate in Heterogeneous Tumor Tissue in Vitro. Biotechnol. Bioeng. 2006, 94, 710–721. [Google Scholar] [CrossRef]
- Kasinskas, R.W.; Forbes, N.S. Salmonella Typhimurium Lacking Ribose Chemoreceptors Localize in Tumor Quiescence and Induce Apoptosis. Cancer Res. 2007, 67, 3201–3209. [Google Scholar] [CrossRef]
- Nishikawa, H.; Sato, E.; Briones, G.; Chen, L.M.; Matsuo, M.; Nagata, Y.; Ritter, G.; Jäger, E.; Nomura, H.; Kondo, S.; et al. In Vivo Antigen Delivery by a Salmonella Typhimurium Type III Secretion System for Therapeutic Cancer Vaccines. J. Clin. Investig. 2006, 116, 1946–1954. [Google Scholar] [CrossRef]
- Hosseinidoust, Z.; Mostaghaci, B.; Yasa, O.; Park, B.W.; Singh, A.V.; Sitti, M. Bioengineered and Biohybrid Bacteria-Based Systems for Drug Delivery. Adv. Drug Deliv. Rev. 2016, 106, 27–44. [Google Scholar] [CrossRef]
- Angsantikul, P.; Thamphiwatana, S.; Zhang, Q.; Spiekermann, K.; Zhuang, J.; Fang, R.H.; Gao, W.; Obonyo, M.; Zhang, L. Coating Nanoparticles with Gastric Epithelial Cell Membrane for Targeted Antibiotic Delivery against Helicobacter Pylori Infection. Adv. Ther. 2018, 1, 1800016. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Tiruthani, K.; Wang, Y.; Shen, L.; Hu, M.; Dorosheva, O.; Qiu, K.; Kinghorn, K.A.; Liu, R.; Huang, L. Trapping of Lipopolysaccharide to Promote Immunotherapy against Colorectal Cancer and Attenuate Liver Metastasis. Adv. Mater. 2018, 30, e1805007. [Google Scholar] [CrossRef] [PubMed]
- Xiong, M.H.; Bao, Y.; Du, X.J.; Tan, Z.B.; Jiang, Q.; Wang, H.X.; Zhu, Y.H.; Wang, J. Differential Anticancer Drug Delivery with a Nanogel Sensitive to Bacteria-Accumulated Tumor Artificial Environment. ACS Nano 2013, 7, 10636–10645. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; He, H.; Luo, Z.; Zhou, H.; Liang, R.; Pan, H.; Ma, Y.; Cai, L. In Situ Photocatalyzed Oxygen Generation with Photosynthetic Bacteria to Enable Robust Immunogenic Photodynamic Therapy in Triple-Negative Breast Cancer. Adv. Funct. Mater. 2020, 30, 1910176. [Google Scholar] [CrossRef]
- Wu, M.; Wu, W.; Duan, Y.; Li, X.; Qi, G.; Liu, B. Photosensitizer-Bacteria Biohybrids Promote Photodynamic Cancer Cell Ablation and Intracellular Protein Delivery. Chem. Mater. 2019, 31, 7212–7220. [Google Scholar] [CrossRef]
- Cheng, L.; Ke, Y.; Yu, S.; Jing, J. Co-Delivery of Doxorubicin and Recombinant Plasmid PHSP70-Plk1-ShRNA by Bacterial Magnetosomes for Osteosarcoma Therapy. Int. J. Nanomed. 2016, 11, 5277–5286. [Google Scholar] [CrossRef]
- Abedi, M.H.; Yao, M.S.; Mittelstein, D.R.; Bar-Zion, A.; Swift, M.B.; Lee-Gosselin, A.; Barturen-Larrea, P.; Buss, M.T.; Shapiro, M.G. Ultrasound-Controllable Engineered Bacteria for Cancer Immunotherapy. Nat. Commun. 2022, 13, 1–11. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, G.; Wu, W.; Wang, J.; Hu, J.; Mao, J.; Chu, P.K.; Bai, H.; Tang, G. A Hybrid Eukaryotic–Prokaryotic Nanoplatform with Photothermal Modality for Enhanced Antitumor Vaccination. Adv. Mater. 2020, 32, 1908185. [Google Scholar] [CrossRef]
- Zheng, D.W.; Dong, X.; Pan, P.; Chen, K.W.; Fan, J.X.; Cheng, S.X.; Zhang, X.Z. Phage-Guided Modulation of the Gut Microbiota of Mouse Models of Colorectal Cancer Augments Their Responses to Chemotherapy. Nat. Biomed. Eng. 2019, 3, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Chen, Y.; Jiang, H.; Nie, D. Short-Chain Fatty Acids Induced Autophagy Serves as an Adaptive Strategy for Retarding Mitochondria-Mediated Apoptotic Cell Death. Cell Death Differ. 2010, 18, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ouyang, J.; Sun, F.; Yang, J. Short-Chain Fatty Acids: A Soldier Fighting Against Inflammation and Protecting From Tumorigenesis in People With Diabetes. Front. Immunol. 2020, 11, 590685. [Google Scholar] [CrossRef]
- Hsieh, Y.H.; Peterson, C.M.; Raggio, A.; Keenan, M.J.; Martin, R.J.; Ravussin, E.; Marco, M.L. Impact of Different Fecal Processing Methods on Assessments of Bacterial Diversity in the Human Intestine. Front. Microbiol. 2016, 7, 1643. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Hayes, R.B. Environmental Influences on the Human Microbiome and Implications for Noncommunicable Disease. Annu. Rev. Public Health 2020, 42, 277–292. [Google Scholar] [CrossRef] [PubMed]
- Burr, A.H.P.; Bhattacharjee, A.; Hand, T.W. Nutritional Modulation of the Microbiome and Immune Response. J. Immunol. 2020, 205, 1479–1487. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing Functional and Translational Microbiome Research Using Meta-Omics Approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Ramírez-Puebla, S.T.; Borisy, G.G. Oral Microbiome Geography: Micron-Scale Habitat and Niche. Cell Host Microbe 2020, 28, 160–168. [Google Scholar] [CrossRef]
- Schell, S.L.; Schneider, A.M.; Nelson, A.M. Yin and Yang: A Disrupted Skin Microbiome and an Aberrant Host Immune Response in Hidradenitis Suppurativa. Exp. Dermatol. 2021, 30, 1453–1470. [Google Scholar] [CrossRef]
- Yan, Y.; Nguyen, L.H.; Franzosa, E.A.; Huttenhower, C. Strain-Level Epidemiology of Microbial Communities and the Human Microbiome. Genome Med. 2020, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.M.; Wang, P.; Teka, T.; Zhang, Y.C.; Yang, W.Z.; Zhang, Y.; Wang, T.; Liu, L.X.; Han, L.F.; Liu, C.X. 1H NMR and UHPLC/Q-Orbitrap-MS-Based Metabolomics Combined with 16S rRNA Gut Microbiota Analysis Revealed the Potential Regulation Mechanism of Nuciferine in Hyperuricemia Rats. J. Agric. Food Chem. 2020, 68, 14059–14070. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Liu, C.W.; Yang, Y.; Hsiao, Y.C.; Ru, H.; Lu, K. High-Coverage Metabolomics Uncovers Microbiota-Driven Biochemical Landscape of Interorgan Transport and Gut-Brain Communication in Mice. Nat. Commun. 2021, 12, 6000. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 313. [Google Scholar] [CrossRef]
- Jean-Pierre, F.; Henson, M.A.; O’Toole, G.A. Metabolic Modeling to Interrogate Microbial Disease: A Tale for Experimentalists. Front. Mol. Biosci. 2021, 8, 634479. [Google Scholar] [CrossRef]

| Intervention Type | Title | Status | Study Results | Conditions | Intervention | Location | NCT Number |
|---|---|---|---|---|---|---|---|
| Prebiotics | |||||||
| Prebiotics in Rectal Cancer | Recruiting | Not available | Rectal cancer | Soluble corn fiber | United States | NCT05516641 | |
| Camu-Camu Prebiotic and Immune Checkpoint Inhibition in Patients With Non-Small Cell Lung Cancer and Melanoma | Recruiting | Not available | Non-small cell lung carcinoma, melanoma | Bifidobacterium longum | Canada | NCT05303493 | |
| Study Using Prebiotics to Improve Gut Microbiome Diversity After Autologous Stem Cell Transplantation | Recruiting | Not available | Multiple myeloma, lymphoma | Resistant starch | United States | NCT05135351 | |
| Effects of Prebiotics on Gut Microbiome in Patients Undergoing HSCT (HCTDiet) | Active, not recruiting | Not available | Multiple myeloma, leukemia, lymphoma | Prebiotic food/drinks (not specified) | United States | NCT04629430 | |
| Probiotics | |||||||
| Effects of Probiotics on the Gut Microbiome and Immune System in Operable Stage I-III Breast or Lung Cancer | Recruiting | Not available | Breast and lung cancer | Probiotics (not specified) | United States | NCT04857697 | |
| Study to Investigate Efficacy of a Novel Probiotic on the Bacteriome and Mycobiome of Breast Cancer | Not yet recruiting | Not available | Breast cancer | BIOHM®: Bifidobacterium breve, Saccharomyces boulardii, Lactobacillus acidophilus, L. rhamnosus | United States | NCT04362826 | |
| Probiotics Enhance the Treatment of PD-1 Inhibitors in Patients With Liver Cancer | Recruiting | Not available | Liver cancer | Probio-M9: L. rhamnosus | China | NCT05032014 | |
| Gut Microbiome and Its Immune Modulation in Locally Advanced Rectal Cancer | Not yet recruiting | Not Available | Rectal cancer | GEN-001: Lactococcus lactis | Korea | NCT05079503 | |
| FMT | |||||||
| Gut Microbiota Reconstruction for NSCLC Immunotherapy | Not yet recruiting | Not available | Non-small-cell lung cancer | FMT (capsule) | China | NCT05008861 | |
| Role of Gut Microbiome and Fecal Transplant on Medication-Induced GI Complications in Patients With Cancer | Recruiting | Not available | Melanoma | FMT | United States | NCT03819296 | |
| Fecal Microbiota Transplantation in Patients With Malignancies Not Responding to Immune Checkpoint Inhibitor Therapy | Recruiting | Not available | Cancer | FMT | Switzerland | NCT05273255 | |
| FMT Combined With Immune Checkpoint Inhibitor and TKI in the Treatment of CRC Patients With Advanced Stage | Recruiting | Not available | Colorectal cancer | FMT | China | NCT05279677 | |
| Fecal Microbiota Transplantation With Immune Checkpoint Inhibitors in Lung Cancer | Not yet recruiting | Not available | Lung cancer | FMT, placebo antibiotics, placebo FMT | Israel | NCT05502913 | |
| Microbiota Transplant in Advanced Lung Cancer Treated With Immunotherapy | Recruiting | Not available | Lung cancer | FMT | Spain | NCT04924374 | |
| Washed Microbiota Transplantation for The Treatment of Oncotherapy-Related Intestinal Complications | Recruiting | Not available | Cancer | Washed Microbiota Transplantation | China | NCT04721041 | |
| Faecal Microbiota Transplantation After Allogeneic Stem Cell Transplantation (TMF-Allo) | Not yet recruiting | Not available | Leukemia, lymphoma, myeloma | FMT | France | NCT04935684 | |
| Antibiotics | |||||||
| Modulation of the Gut Microbiome With Pembrolizumab Following Chemotherapy in Resectable Pancreatic Cancer | Not yet recruiting | Not available | Pancreatic cancer | Ciprofloxacin, metronidazole | United States | NCT05462496 | |
| Early Termination of Empirical Antibiotics in Febrile Neutropenia in Children With Cancer | Recruiting | Not available | Cancer | Antibiotics suspension | Denmark | NCT04637464 | |
| Microbiome and Association With Implant Infections | Recruiting | Not available | Breast cancer | Cephalexin | United States | NCT05020574 | |
| The Impact of an Antibiotic (Cefazolin) Before Surgery on the Microbiome in Patients With Stage I-II Melanoma | Recruiting | Not available | Melanoma | Cefazolin | United States | NCT04875728 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conti, G.; D’Amico, F.; Fabbrini, M.; Brigidi, P.; Barone, M.; Turroni, S. Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out. Genes 2023, 14, 55. https://doi.org/10.3390/genes14010055
Conti G, D’Amico F, Fabbrini M, Brigidi P, Barone M, Turroni S. Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out. Genes. 2023; 14(1):55. https://doi.org/10.3390/genes14010055
Chicago/Turabian StyleConti, Gabriele, Federica D’Amico, Marco Fabbrini, Patrizia Brigidi, Monica Barone, and Silvia Turroni. 2023. "Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out" Genes 14, no. 1: 55. https://doi.org/10.3390/genes14010055
APA StyleConti, G., D’Amico, F., Fabbrini, M., Brigidi, P., Barone, M., & Turroni, S. (2023). Pharmacomicrobiomics in Anticancer Therapies: Why the Gut Microbiota Should Be Pointed Out. Genes, 14(1), 55. https://doi.org/10.3390/genes14010055










