Abstract
Biosurfactants are amphipathic molecules capable of lowering interfacial and superficial tensions. Produced by living organisms, these compounds act the same as chemical surfactants but with a series of improvements, the most notable being biodegradability. Biosurfactants have a wide diversity of categories. Within these, lipopeptides are some of the more abundant and widely known. Protein-containing biosurfactants are much less studied and could be an interesting and valuable alternative. The harsh temperature, pH, and salinity conditions that target organisms can sustain need to be understood for better implementation. Here, we will explore biotechnological applications via lipopeptide and protein-containing biosurfactants. Also, we discuss their natural role and the organisms that produce them, taking a glimpse into the possibilities of research via meta-omics and machine learning.
1. Introduction
Biosurfactants are amphipathic molecules synthesized by plants, animals, and microbes, that reduce interfacial and superficial tensions in aqueous solutions and hydrocarbon mixtures [1]. Due to these properties, biosurfactants alter how other molecules interact, increasing the solubility of substances [2,3]. The hydrophilic portion—the head—will usually be a hydrocarbon, while the hydrophobic portion—the tail—can be non-ionic, positively or negatively charged, or amphoteric [4]. These characteristics allow biosurfactants to form aggregates called micelles, which gather when there is an increase in amphiphilic concentration in a liquid beyond a limit, known as critical micelle concentration (CMC) [5].
Most chemically produced surfactants are petroleum-based, and the hydrophobic parts consist of paraffins, olefins, alkylbenzenes, alkylphenols, and alcohols and the hydrophilic domain is usually a sulfate, or a sulphonate [6,7,8]. Some examples of widely used surfactants include sodium n-dodecyl sulfate, and Triton X100 [2]. They pose occupational and environmental risks, as surfactants can be extremely dangerous to the environment if not carefully applied. For example, the use of dispersants, which are a type of chemical surfactant, during oil spills has a series of documented issues and concerns [9,10]. Some studies even described dispersants being capable of diminishing microbial degradation activity [11,12]. Therefore, there is a concern regarding their usage, and research to find less aggressive alternatives is growing [13].
In contrast to the artificial surfactants, biosurfactants are eco-friendly alternatives that offer many advantages when compared to synthetic surfactants, such as surface and interfacial activity, resistance to temperature, pH and ionic force, low toxicity, availability, specificity, biocompatibility, and biodegradability [14,15,16]. Due to their varied structural diversity and composition, the biosurfactants themselves are used in various applications. Applicable areas include bioremediation, medicine, food industry, and industrial processes [17]. Though biosurfactants possess valuable capabilities, large-scale industrial applications are hindered by high costs and low efficiency of their production and recovery processes [13]. Nevertheless, the demand for biosurfactants is increasing, and both the industry and consumers are willing to pay for an alternative that is more beneficial in the long run [18]. Several strategies for optimal growth and production have been researched over the years, and this scenario is gradually changing [3,19,20,21].
There are several classes of compounds among biosurfactants, as they have a vast structural diversity. They can be categorized as glycolipids, lipopeptides and lipoproteins, fatty acids, phospholipids, natural lipids, polymeric and particulate [22,23,24,25].
Regarding lipopeptides and lipoprotein biosurfactants, the leading producers are fungi, bacteria, and yeast [26] They act to enhance mobility, decrease viscosity, facilitate solubilization, and act as metal-sequestering agents [26]. Additionally, they can disrupt biological membranes, making them potential agents to be used as hemolytic, antiviral, antibacterial, and anti-carcinogenic molecules [26]. One of the main interests in biosurfactant research is employing them as surface-active compounds in bioremediation strategies as alternatives for traditional chemical methods. This review aims to present what is known about these proteic compounds, be it how they provide advantages to their producers, their potential for biotechnological applications, and how novel biosurfactants can be discovered and produced. While also going over the different production techniques and novel research methods using bioinformatic approaches such as omics technology, molecular dynamics, and machine learning.
2. Biosurfactant-Producing Microorganisms
Microorganisms like bacteria, filamentous fungi, and yeasts can synthesize biosurfactants of varied molecular structures and surface properties [27,28,29]. Two of the most researched biosurfactant-producing microorganisms are the Pseudomonas and Bacillus genres. Table 1 shows a list of some microorganisms of interest and their produced biosurfactants. Some studies on the potential of lipoproteins in facilitating microbial hydrocarbon degradation have been conducted [30,31,32,33]. Interestingly to note is the study in which a hydrocarbon-degrading consortium of microorganisms was constructed with samples collected from uncontaminated soil [30]. It highlights the importance of how microbial interaction between different species might dictate the fate of pollutant degradation and offers new insights into bioaugmentation applications.
Akin to other surface-active molecules, biosurfactants contain one or several lipophilic and hydrophilic moieties. Biosurfactants are categorized by their chemical composition, mechanism of action, molecular weight, physicochemical properties, and microbial origin [28,34]. According to their molecular weight, biosurfactants can be divided into low molecular weight compounds that include phospholipids, glycolipids, and lipopeptides and into high molecular weight that contains amphipathic polysaccharides, lipoproteins, lipopolysaccharides, or more complex mixtures of those [24,25,28]. The low molecular weight molecules, such as rhamnolipids produced by Pseudomonas efficiently lower surface tension and interfacial tension, while high molecular weight polymers, like the surfactin produced by Bacillus subtilis, bind tightly to surfaces [35]. We will focus on the peculiarities of the lipopeptide and protein-containing biosurfactants. The lipophilic moiety is a protein or a peptide with hydrophobic side chains or a hydrocarbon chain of a fatty acid. The hydrophilic moiety is an ester, a hydroxy, phosphate, or carboxylate group, or a sugar carbohydrate [28]. In addition, there are highly surface-active globular proteins, such as hydrophobins, an amphiphilic protein that will bediscussed later.


Table 1.
Properties and characteristics of the main lipopeptide and protein-containing biosurfactants.
Table 1.
Properties and characteristics of the main lipopeptide and protein-containing biosurfactants.
| Biosurfactant/Bioemulsifier | Class | Producing Species | Reported Genes | Ionic Charge | Molecular Weigth (KDa) | CMC (mg/L) | Superficial Tension (mN/m) | Potential Application | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Alasan | Polymeric | Acinetobacter calcoaceticusAcinetobacter radioresistens | aln A-C | Negative | 1000 | - | 41.6 | Emulsification, solubilization activity | [36,37,38,39,40] |
| Hydrophobin | Globular protein | Lecanicillium lecaniiTrichoderma reeseiSchizophyllum commune | HFBI, HFBII | - | <20 | - | 25–45 | Drug solubilization biomineralization | [41,42] |
| Arthrofactin | Lipopeptide | Pseudomonas sp. | arfA–C | - | 1.354 | 13.5 | 72–24 | Antifungal | [43,44] |
| Fengycins | Lipopeptide | B. subtilis | fenA–E gene cluster | Negative | 1.463 | - | 21 | Antifungal, antimicrobial | [45,46,47,48] |
| Iturin | Lipopeptide | B. subtilisBacillus pumilus | ituA–C | Neutral | 1.043 | - | 30–37.5 | Antimicrobial, biopesticides | [36,40,49,50] |
| Lichenysin | Lipopeptide | Bacillus licheniformisB. subtilis | licA–D | Negative | 0.993–1.049 | 10–22 | 27 | Oil recovery, hemolytic, chelating agent | [40,51,52,53] |
| Serrawettin | Lipopeptide | Serratia marcescensSerratia surfactantfaciens | pswP | Neutral | 0.541–0.731 | - | 28–33.9 | Oil recovery, antimicrobial, antitumoral | [4,40,49,54,55] |
| Surfactin | Lipopeptide | B. subtilis | srfA–D | Negative | 1.007–1.035 | 20–40 | 22–27.9 | Oil recovery, antibacterial, antitumoral, antiviral, anticoagulant | [4,36,40,52] |
| Syringomycin | Lipopeptide | Pseudomonas syringae B301D | syrB1, syrE, syrB2, syrC, syrP | Positive | 1.225 | 1250 | 33 | Antibacterial | [43,56,57] |
| Syringopeptin | Lipopeptide | P. syringae B301D | sypA–C | Positive | 2.399 | 820 | 40.2 | Antibacterial | [43,56,57,58] |
| Viscosin | Lipopeptide | Pseudomonas fluorescens | viscA–C | - | 1.126 | 10–15 | 26.5–28 | Antitumoral, antibacterial | [59,60,61] |
2.1. Lipopeptides
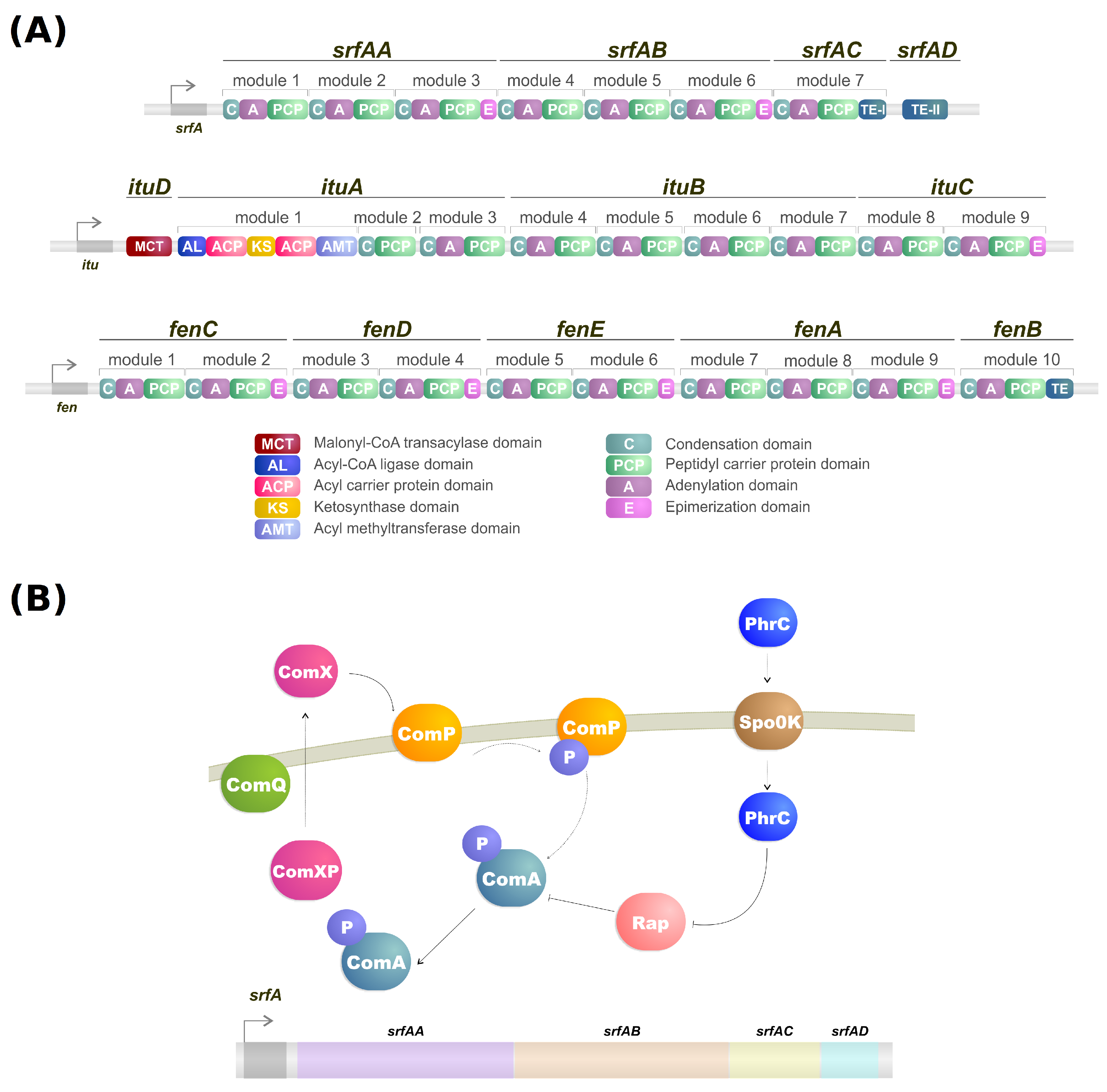
Lipopeptides are a class of biosurfactants with high industrial interest. They are linear or cyclic oligopeptides acylated with fatty acids of different length and composition which are synthesized by the nonribosomal peptide synthetases (NRPSs) [62] and commonly secreted by bacterial genres such as Bacillus, Streptomyces and Pseudomonas [25]. Microorganisms of the genus Bacillus are some of the more well-known lipopeptide producers, and their genome contains a large operon (srfA) composed of four open reading frames which encode a peptide synthetase responsible for the surfactin biosynthesis (Figure 1A,B) [63,64]. Lipopeptides have varying emulsification properties according to temperature, pressure, pH, structure and stability of the solution [65]. For example, the emulsifying activity of surfactin produced by B. subtilis can change based on pH. With a pH above 7, it forms a stable emulsion with kerosene, but when the pH drops to 3 the emulsion does not form [66].
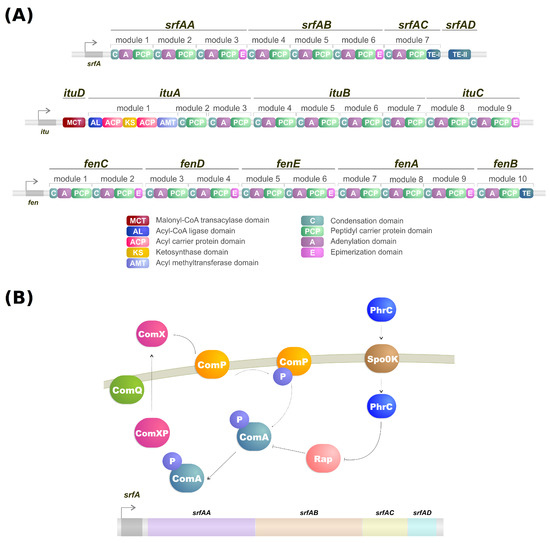
Figure 1.
(A) NRPS modules of surfactin, iturin, and fengycin. (B) Simplified model of transcriptional regulation of srfA gene from Bacillus.
The lipopeptidic biosurfactants offer a great range of natural advantages for their microbial producers. Some noteworthy examples include the secretion of polymers aiding in the structure of biofilm formation and, in turn, improving survival against adverse conditions such as antibiotic treatment and shearing [67]. Moreover, predator avoidance is another significant natural function of biosurfactants.Laboratory studies showed that the serrawetin W2 produced by S. marcescens and surfactin produced by B. subtilis induced Caenorhabditis elegans individuals to avoid them [68]. Lipopeptides were also shown to affect interaction in natural habitats, in vitro testing with lipopeptides produced by both Pseudomonas and Bacillus species exhibited lytic activity and inhibited growth against microorganisms such as viruses, mycoplasmas, bacteria, fungi, and oomycetes [62,69,70].
For commercial interests, biosurfactants must have good yield levels and cost of production. In this sense, surfactin was reported to have increased yield (36-fold), reaching 3.6 g/L, by B. subtilis ATCC 21332 when incorporated with activated carbon [71]. Compared to some other types of biosurfactants, such as the glycolipids (i.e., rhamnolipids), that reach yields of 1 g/L, surfactin shows many prospects for industrial use [63]. Another study investigated the bioremediation potential of surfactin produced by B. subtilis ATCC 21332, and compared it with rhamnolipid produced by P. aeruginosa J4 for enhanced degradation of diesel-contaminated water and soil, and showed that both had successfully reached degradation levels between 80-100% and CMCs between 45 mg/L and 50 mg/L, further confirming surfactin as an efficient lipopeptide biosurfactant [72].
2.2. Protein-Containing Biosurfactants
Surface-active proteins are a lesser-studied class of biosurfactants that still has some relevance for industrial applications. Depending on the author, lipopeptides will occasionally be grouped with lipoproteins [73]. Considered as high molecular mass bioemulsifiers, they are complex structures with multiple reactive groups exposed, turning them into effective emulsifiers as they bind tightly to hydrophobic molecules [63]. Surface tension at liquid-air interfaces is a significant barrier encountered by distinct organisms. Conquering the surface tension is an essential mechanism in several species, i.e., as sporulation of bacteria and fungi, foaming in frog nests, and evaporative cooling in horses [25,73]. These biosurfactants are divided into different groups according to structure: lipid-associated proteins, such as pulmonary surfactants [74], and non-lipid-associated globular proteins, such as hydrophobins [73]. Polymeric biosurfactants are a different category of high molecular weight biosurfactants containing protein in their composition [26]. Alasan (Table 1) is the best-known biopolymer of this group and consists of a high molecular weight polysaccharide and protein complex with solubilizing and emulsifying activity [26]. Another effective emulsifier is liposan, composed of 87% carbohydrates and 17% proteins, produced by Candida lipolytica [26,75]. It was first described in 1985 by Cirigliano and Carman [75], who showed that it is not able to reduce the surface tension of water but effectively emulsified and stabilized water-in-oil emulsion [26]. There are still few studies that explore the functionality and application of these biopolymers.
The bacterial species B. subtilis has been shown to protect plant roots from pathogenic fungi and bacteria by using lipoproteins [73]. Upon colonizing the area and forming a biofilm, it produces the lipoprotein BslA on the outer layer, thus forming a hydrophobic barrier, often called a “bacterial raincoat” [73,76,77]. This is the more common natural function of the globular biosurfactants, as other examples include the hydrophobins produced by many fungi to create a hydrophobic layer on the hydrophilic hypha or spores for protection [73,78,79,80,81,82]. Moreover, surface-active proteins have some described biotechnological applications, such as using their properties of self-assembly and reversal of surface wettability in nanodevice and medical implant coating, e.g., coating electrodes and the surface of biliary stents to prevent oxidation and fouling, respectively [73,83].
3. Lipopeptides Synthesis and Regulation
Biosurfactants are secondary metabolites produced by microorganisms, and as commented in a previous section, the lipopeptides, specifically, are produced by NRPSs.
NRPSs are multidomain mega-enzymes that synthesize NRPs without using the ribosomal machinery [84]. They have a modular structure in which each part incorporates an amino acid to the peptide moiety of the lipopeptide (Figure 1B). Another characteristic is that the NRPS obeys the colinearity rule; that is, the modules are colinear with the amino acid sequence of the peptide [62]. There are two types of modules: initiation and elongation modules. Each of these modules also contains domains that perform specific tasks [62]. While typically, initiation modules have domains responsible for the amino acid election, activation, and thioesterification of the activated amino acid, the first module also contains a condensation domain in lipopeptide biosynthesis. This domain is responsible for catalyzing the N-acylation of the first amino acid of the lipopeptide, thus linking the lipid moiety to the oligopeptide [62,85]. Elongation modules contain the same domains, but this time the condensation domain is responsible for catalyzing the peptide bond between two amino acids. The condensation domain from the elongation module will generate a lipopeptide that, by the end of the assembly line, will be cleaved by a thioesterase [62].
It has been shown via analyses of the metabolic profiles of Pseudomonas and Bacillus species that a single strain may be able to produce different forms of the same biosurfactant [62]. Examples include B. subtilis strain OKB 105, which can produce 12 different surfactin analogs [86] and P. fluorescens strain SS101 that can produce up to eight analogs of the lipopeptide massetolide A [87]. These analogs are thought to be the product of flexibility in the amino acid selection and activation by the adenylation domain of the initiation module in NRPSs. Said flexibility is common in nonribosomal peptide synthesis, which may have biological purposes for the producers [62].
Several advancements have been made in the quest to elucidate biosurfactant synthesis regulation over the decades. It is known that, for example, in Pseudomonas, the GacA/GacS regulatory system is the primary regulator in lipopeptide production. A mutation in either one of the encoding genes results in the loss of this function [88,89,90].
Iturin production has a series of regulation factors. For example, the methylation of tyrosine residues in iturin can decerease yield and antibacterial activity [91,92]. Furthermore, iturin can also be regulated by controlling the expression of sigma factor A and the transcription factor ComA. Overexpressing the genes sigA and comA increased iturin yield [93].
As mentioned, the biosynthesis of surfactin is coordinated by the srfA operon, which contains four open reading frames (srfAA, srfAB, srfAC, and srfAD) as seen in Figure 1A [94]. These open reading frames are responsible for the peptide chain extension, a key step in the surfactin synthesis regulation [92]. Surfactin, much like iturin, is also strongly regulated by the transcription factor ComA, which binds to srfA, and thus controls its transcription and regulates srfA expression (Figure 1B) [92]. ComA phosphorylation is the key to activating the srfA operon transcription by two different pathways. The first involves the ComX peptide modified by ComQ that stimulates ComP autophosphorylation, triggering ComA phosphorylation (Figure 1A) [43]. Activated ComA translocates to the nucleus and promotes srfA transcription. PhrC importation is mediated by Spo0K, which interacts with Rap protein and inhibits its phosphatase activity, thus, preventing ComA dephosphorylation and facilitating ComA-induced srfA transcription (Figure 1A) [43,95].
Another regulatory system tightly related to biosurfactant production is quorum sensing, identified in a series of lipopeptide-producing species. Quorum sensing is a cell-to-cell communication process where colonies of bacteria identify population densities via detecting secreted molecules called autoinducers, tracking cell density, and adjusting gene regulation accordingly [96,97,98]. Examples include the production of serrawetin W2 in Serratia liquefaciens, with the quorum sensing genes swrI-swrR and regulated by N-butanoyl-L-homoserine lactone and N-hexanoyl-L-homoserine lactone production [99], and the production of surfactin in B. subtilis via the comA-comP genes which is regulated by competence and sporulation stimulating factor [100].
It is known that the pathways of different lipopeptides iteract with each other. For example, knocking out the sfp gene can significantly increase plipastasin production, while regulating plipastasin also increases the production of surfactin [101,102]. On the other hand, the absence of srfAB lowers surfactin production while significantly increasing the yield of iturin [103]. Furthermore, the mutation of fenC, a fengycin biosythesis gene (Figure 1A), increased iturin production while mutating ituC, an iturin biosynthesis gene (Figure 1A), increased fengycin production [104]. By observing these discoveries, we can confirm that the pathways of biosurfactant production share a strong correlation with each other, implying the use of similar precursors and regulators, while specific regulatory mechanisms remain to be explored [92].
4. Biosurfactant Toxicity
Most chemically synthesized surfactants widely available on the market resist biodegradation and accumulate in nature. On the other hand, Biosurfactants are, like all-natural products, susceptible to degradation in water and soil [14]. Other than that, they are known for being less toxic or even non-toxic to the environment when compared to their synthetic counterparts and, thus, being the better choice when degrading pollutants. Some studies were already published comparing synthetic and natural surfactants and show how biosurfactants can be less toxic or even non-toxic at all [105,106]. However, Edwards et al. (2003) determined that the difference may not be significant [107]. Still, there are issues regarding the conditions in nature and the concentration and availability of biosurfactants in said situations, as they can vary between species and compounds, and lab conditions cannot properly mimic that.
Another concerning issue about biosurfactants is that some of the most significant producing organisms discovered so far are human pathogens and, thus, offer some degree of risk in their cultivation [108,109]. For example, one of the best-known bacteria capable of producing rhamnolipids is P. aeruginosa, a pathogenic organism [63]. Other examples of pathogenic species include the lipopeptide producers S. marcescens and Inquilinus limosus KB3 [110,111]. Hence, research on viable alternatives is steadily increasing over the years. According to Marchant et al. (2014), special attention to molecular biology techniques and quality bioinformatics assessment should be encouraged when studying alternative biosurfactant producers, as many studies being published are not replicable or are offering erroneous phylogeny; creating hindrances to getting better results and finding alternatives to pathogenic organisms [63].
5. Emerging Strategies for Biosurfactant Production
Being able to implement the newly discovered compounds is needed. As mentioned before, biosurfactants still struggle to compete with synthetic surfactants on large-scale applications globally. With that in mind, recent strategies have been developed to turn biosurfactants into competitive alternatives for industrial applications.
One such application is solid-state fermentation (SSF). A strategy for biosurfactant production that is geared towards overcoming the foaming encountered in the more popular submerged fermentation (SmF), this is used not only for biosurfactants but also other bioproducts [112,113,114]. This methodology has already been applied to the peptidic biosurfactant surfactin,. By using a medium based on okara, with the addition of sugarcane bagasse as a bulking agent, SSF was employed for surfactin production by B. pumilus UFPEDA 448 [115]. Other examples include SSF of soybean flour and rice straw as a substrate for the production of an antimicrobial lipopeptide by B. amyloliquefaciens XZ-173 [116], lipopeptide production by B. subtilis SPB1 grown on a mixture of olive leaf residue flour and olive cake flour [117] and surfactin production via SSF with rapeseed cake mixed with bacterial solution [118].
When producing biosurfactants, one of the critical elements is the media constituents. They play a role in the type and quantity of biosurfactant produced [3]. A technique that exploits this is using a carrier in the growth medium, which proved to be efficient in some cases, although not much has been done in this field yet [119,120]. When evaluating the lipoprotein biosurfactant produced by Pediococcus dextrinicus SHU1593, a study showed that using molasses and date syrup improved biomass formation [121]. Another study on B. subtilis CN2 showed that carbon sources on a hydrophobic substrate yielded much better activity and quality of the biosurfactant compared to a hydrophilic substrate [122].
The market always favors lucrative processes, and with that in mind, another feasible option for biosurfactant production is the co-production with other economically necessary products in a single bioprocess. Microorganisms tend to synthesize biosurfactants and other compounds, which could be taken advantage of. Before, co-production of pectinase and biosurfactant was observed in a B. subtilis strain isolated from a fruit dump yard [123].
As commented in a previous section, there are many pathogenic biosurfactant producers. To circumvent that, another alternative that tries to solve both the large-scale production and pathogenicity problems is using non-pathogenic microorganisms via recombinant production. For example, the gram-negative bacteria S. marcescens, an opportunistic human pathogen, is capable of producing serrawettin W1, a lipopeptide biosurfactant, and the NRPS protein SwrW is what catalyzes it [124]. By amplifying this protein, a recombinant production was established using the model organism Escherichia coli and a 20-fold increase in serrawettin W1 yield were achieved when compared to S. marcescens [108]. Another successful application of heterologous expression in E. coli occurs when using it as a vector for the hydrophobin protein DewA, which is naturally produced by the fungus Aspergillus nidulans, as it efficiently secretes it and provides facilitated purification [80]. There are still some hardships when approaching this method; for example, the recombinant expression of class I hydrophobin biosurfactant HGFI in Pichia pastoris has a somewhat lower yield after purification [83,125]. Also, when some hydrophobins are expressed in the model bacteria E. coli, the vector fails to deliver as it produces the biosurfactant in inclusion bodies or is often unfolded/misfolded [83].
6. Physical and Chemical Properties of Biosurfactants
Biosurfactants are bioderived or biomimetic surfactants that can act as detergents, wetting agents, emulsifiers, dispersants, and foaming agents [126,127]. These molecules have the properties of reducing water surface tension and decreasing water/oil interface tension (Figure 2) [3]. This occurs because these amphiphilic molecules can dispose of the limit of surface water and assemble into micelles, which can shield hydrophobic molecules present in the solution from the unfavorable interactions with water molecules [65]. Moreover, biosurfactants present adsorptive properties on surfaces and interfaces [128].
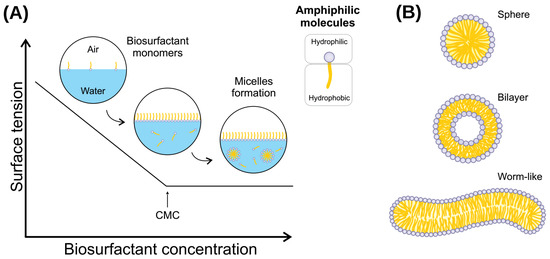
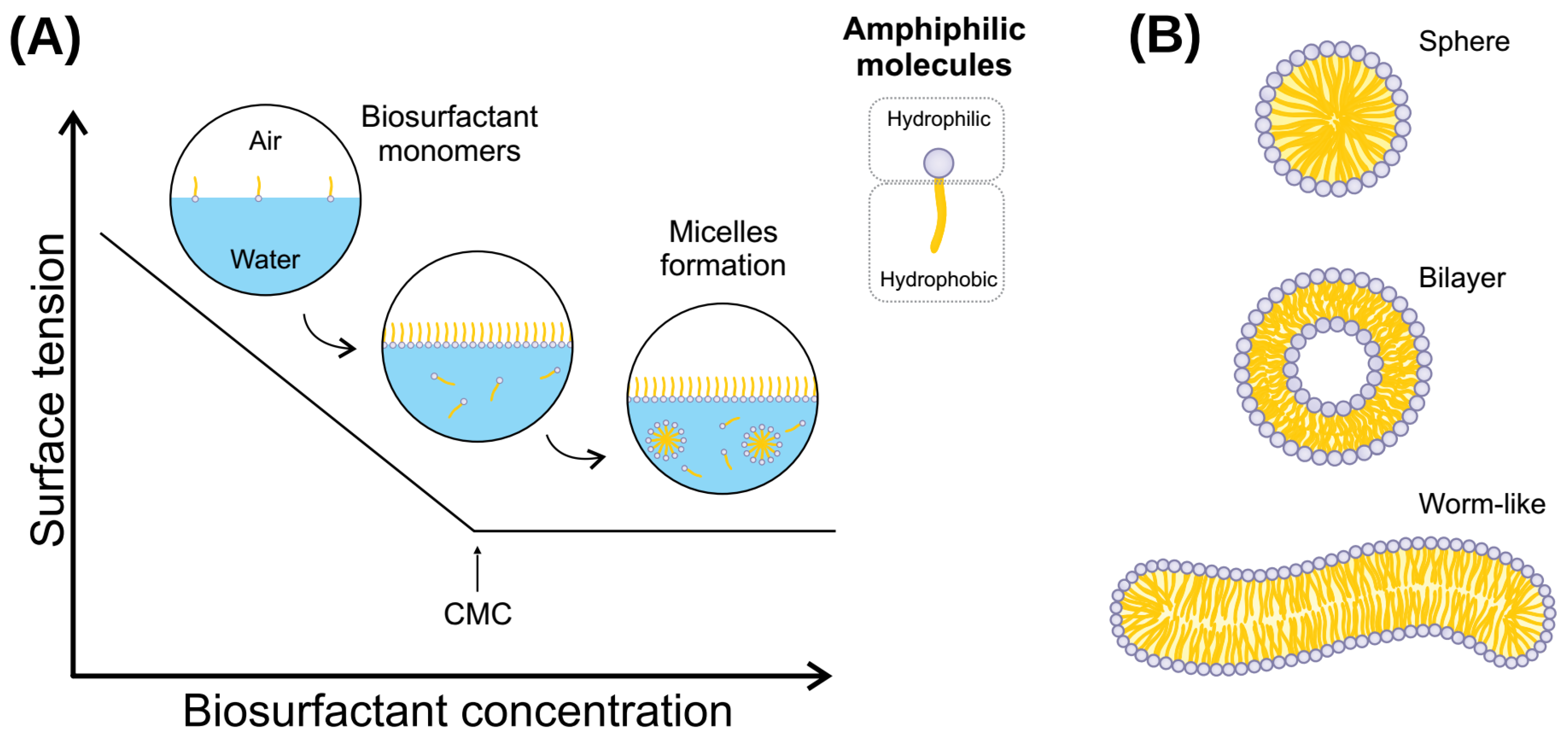
Figure 2.
(A) Relationship between biosurfactant concentration and surface tension. CMC is defined by the concentration at which biosurfactant monomers form micelles and can reduce interfacial tension. (B) Different types of biosurfactant aggregates. A spherical shape occurs when the solution reaches CMC, and may become asymmetrical (such as a worm-like shape) at higher biosurfactant concentrations. Likewise, temperature or other environmental conditions induces the transition from spherical shape micelles to micellar vesicles in biosurfactant assemblies.
Biosurfactants are molecules that present a wide range of industrial applications, such as cleaning (laundry products), biofilm prevention and disruption, biocidal activity, wound healing, and various uses in the petroleum industry and oil bioremediation [127,129]. This array of applications comes from the particular chemical features that biosurfactant molecules present. From a physicochemical perspective, (bio)surfactants are amphipathic molecules that lower surface and interfacial tensions. Consequently, water-immiscible substances will increase their solubility at the surfactant-water interface [3]. The molecules will spontaneously aggregate into micelles under non-extreme conditions and at a minimum concentration. For this reason, understanding the micelle formation process is of pivotal importance for the application of biosurfactants. The emulsifying ability of each biosurfactant is the result of the micelle characteristics it will form, and the micelle characteristics are the consequence of the isolated molecule properties [130].
The CMC and the aggregation number are the two main parameters to investigate in a new biosurfactant. When biosurfactant molecules are slowly added to water, the molecules will stay only on the surface (at a very low concentration). At this point, there are no micelles in the solution. With the slow increase of molecules in the solution, it will reach a concentration in which no micelles are yet stably assembled at a certain point. Still, more biosurfactant molecules will promote the formation of thermodynamically stable micelles. This specific concentration is called CMC (Figure 2). The number of molecules that constitute a micelle formed just as CMC has been overreached is defined as the aggregation number [131]. There are different methods to determine both the CMC and aggregation number of a given biosurfactant, such as isothermal titration calorimetry [132], THP-based determination method [133], fluorimetry, conductometry, and surface tension [134]. Furthermore, CMC depends on temperature, pressure, pH, and ionic strength; thus, investigating the relationship between CMC and those parameters is utterly essential.
As stated before, CMC revolves around the concentration where micelles will assemble spontaneously. Thus, standard Gibbs free energy of micellization (Gom) is calculated relatively to CMC, as shown in Table 2. Note that ionic micelles are more sensitive to salt concentration, thus taking into account free counter-ion concentration. Moreover, the standard enthalpy of micellization (Hom) indicates if the process is endothermic or exothermic for a given biosurfactant. Many biosurfactants, such as peptide-derived ones, are ionic or zwitterionic molecules, which in most cases present an endothermic micellization process, where the negative free energy is a compensation in the system’s entropy increase (So = Som + Sosol) [135]. This observation implies that the micellization process is driven by the water molecules surrounding the biosurfactant rather than a strong interaction between the biosurfactant monomers. Thus, organisms that are dependent on this type of biosurfactant to survive should be adapted to environments where the ionic strength will not be enough to organize and restrain water molecules in such a way that will lower the solvation entropy or possess a secondary mechanism that will compensate or reduce the ionic strength. It is also important to note that when considering the effect of ions, it can not be assumed a monotonic behavior for the micellization process [136].

Table 2.
Equations relating to the physicochemical processes involved with biosurfactants.
An essential aspect of biosurfactants CMC and its aggregation number is how temperature affects the assembly process. Attempting to predict CMC as a function of temperature is not a straightforward process. That is because surfactants, in a general perspective, vary considerably in their intrinsic structural features, such as ionic and non-ionic nature, hydrophobic moiety size, polar moiety size, and the number of hydrogen bond possibilities. Whereas some studies predicted a monotonic behavior between CMC and temperature for different surfactants [137,138], new evidence suggests that the effect of the temperature on the CMC is non-monotonic and the monotonic behavior is observed only in a specific range of temperature [139] (see Table 2 for the equations). This observation is due to the hydrophobic effect between the surfactant tails and the polar solvent molecules. The transfer of the surfactant n-alkyl tail from the aqueous solution to the hydrocarbon core of the micelle is the main energetic contribution to the micellization process. This way, an increase in the length of the n-alkyl tail should increase the hydrophobic effect and lower the CMC, but other variables, such as tail deformation, should be considered. Therefore, we observe a non-monotonic relationship between n-alkyl chain length and CMC [140].
Moreover, a study about the effects of temperature on micelle formation in apolar media showed that SPANs, which are sorbitan oleates, will form reverse micelles. Increasing the temperature will increase the critical micelle concentration of most surfactants, suggesting that the primary state function driving reverse micelle formation in apolar solvents is enthalpy. Only one surfactant showed an entropically driven micellization process, where increasing the temperature resulted in a decrease in CMC [141]. This sort of investigation is most pertinent, primarily when intended to employ biosurfactants in the petroleum industry or for bioremediation of oil spilled areas since petroleum is an admixture mostly of hydrophobic compounds.
Other important physicochemical parameters of biosurfactants that should be looked at more in-depth are surface and interfacial tensions [34]. As mentioned before, solvents present a cohesion related to their intermolecular interactions, which results in the solvent’s propensity to resist rupture when exposed to an external force. In this sense, biosurfactants’ properties can lower this cohesive force, increasing the solubilization of poorly soluble substances. The lowest surface/interfacial tension is achieved when the biosurfactant concentration reaches CMC [142]. Thus, surface and interfacial tensions are related to CMC. The lower the concentration required for a given biosurfactant to assemble micelles is the concentration where surface tension is minimum. More practically, when adding biosurfactant molecules to a pure water system, the solution’s surface contains a much higher concentration of biosurfactant molecules than the bulk, even when CMC is reached. Since the water surface spontaneously presents a higher biosurfactant concentration than the bulk, increasing the water surface area would result in a withdrawal of biosurfactant molecules from the bulk to compensate for the new surface. In this scenario, if the biosurfactant concentration is increased, the bulk will present a higher concentration, increasing the chemical potential [143]. Consequently, it will be easier to withdraw molecules from the bulk when the surface area increases, lowering the surface tension.
One concept that comes to be important is the distinction between biosurfactant and bioemulsifier compounds. Even though they present similar structural features and some chemical properties, they have enough remarks to categorize them, as reviewed by Uzoigwe et al. [36]. The first difference between these two molecule classes is their size. Bioemulsifiers are higher in molecular weight compared to biosurfactants. Chemically, they are biopolymers or exopolysaccharides, constituted of a complex mixture of heteropolysaccharides, lipopolysaccharides, lipoproteins, and proteins.
On the other hand, as mentioned before, biosurfactants are oligosaccharides, amino acids, and fatty acids derived compounds, presenting a considerably lower molecular weight than bioemulsifiers [36]. Another difference regards the ability to lower the surface and interfacial tensions. Whereas both compounds can efficiently emulsify two immiscible liquids or poorly soluble compounds, biosurfactants are more effective at reducing the surface tension [144]. The chemical aspect of these classes of compounds might rest some clues about the evolutionary traits of organisms adapted to produce them. To further discuss the biological implications, first, we must take an additional glimpse into the thermodynamics of surface tension, especially its relationship with temperature and concentration, along with the biosurfactant solubility nature.
As for the CMC, surface tension is also dependent on temperature. Generally, interfacial and surface tensions decrease with temperature increase [145]. This observation is due to a system’s energy compensation. When molecules are transferred from the bulk to the surface, there is an increase in the system’s energy. That is due to the balance between intermolecular attractive and repulsive forces of neighboring molecules and the energy necessary to overcome those forces for motion. Given that this is a spontaneous process, the gain of energy is compensated by reducing the surface area and, consequently, surface tension. At higher temperatures, the greater kinetic energy of the molecules results in a decrease of the attractive forces, exacerbating the reduction of surface tension. Experimentally, the relationship between temperature and surface tension was modeled by different scientists throughout history. Eötvös found a correlation between temperature and volume-based surface area, and Guggenheim found a formula based on the property of each liquid [145,146], observing the same temperature-surface tension relationship.
6.1. Surfactin and Hydrophobin—Case Studies
Surfactin is one of the most well-studied biosurfactants, and surfactin-like molecules have recently been extensively reviewed by Théatre et al. [147]. Its structure is a cyclic lipoheptapeptide composed of L-Glu1-L-Leu2-D-Leu3-L-Val4-L-Asp5-D-Leu6-L-Leu7, which assembles into a lactone ring structure with a β-hydroxy fatty acid chain [148,149]. Multimodular mega-enzymes synthesize the peptide moiety denominated non-ribosomal peptide synthetases [43]. On the other hand, the fatty chain moiety is synthesized through the classical fatty acid biosynthesis pathway [150], followed by the production of 3-hydroxy-acyl-coenzyme A [151]. Later, this substrate is connected to the peptide moiety through the action of the surfactin synthetase [152]. Several structural variations are observed in surfactin molecules produced by different Bacillus species. Amidst the diversity of structural features documented, there are Val4Leu, and Leu7Ile/Val (pumilacidin) [153]; Glu1Gln (lichenysin) [154]; fatty acid chains varying from 12 to 17 carbons, whereas the most common length is 14-15 carbons [147,155]; isomers in the lipid chain, where it can be linear or branched, iso and anteiso; linearization of the peptide moiety (not frequent) [156]. Additionally, a modified C15-surfactin-O-methyl was seen in B. subtilis HSO121 [157] and C14-surfactin methyl ester in Bacillus pumilus KMM 456 [158]. There are also synthetic structural modifications in surfactin that alter its natural properties, such as esterification [159], linearization of the cyclic surfactin [160], epimerization of L-Leu2 to D-Leu2, charge modification when Asp5Gln, and the switch of residues Asp4-Leu5 [161].
As discussed before, there are various physicochemical properties of biosurfactants, and their understanding is pivotal for industrial or bioremediation applications and for understanding the biosurfactant-producing organisms’ evolutionary process. For surfactin, the CMC for a C15 fatty acid moiety is 20 µM in Tris-HCl pH 8, and the CMC increases as the hydrophobic chain length decrease [162]. This is a result of the packing of surfactin molecules during the micellization. Longer fatty acid chains allow an improved hydrophobic core assembly once the peptide ring is large and hinders the linear chain contacts. Therefore, branched chains are expected to decrease CMC since it is unfavorable to van der Waals interactions. Moreover, linearization of the peptide moiety increases the CMC significantly [163]. Another aspect, the micelle geometry can vary considerably according to the system’s parameters, such as temperature, pH, ionic strength, and surfactin concentration [164,165]. In this sense, surfactin micelles’ shape can be sphere-like, worm-like, and unilamellar bilayers [1].
Another property of biosurfactants previously discussed is surface tension. Surfactin reduces surface tension when in CMC and Tris pH 9.4 to 37 mN/m. The lichensyin variant reaches 35 mN/m under the same conditions [166]. Additionally, lichensyin CMC is tenfold lower than surfactin in the absence of cations [166]. The fatty acid chain influences the surface tension reduction of surfactin, whereas the linear, long-chain is more efficient.
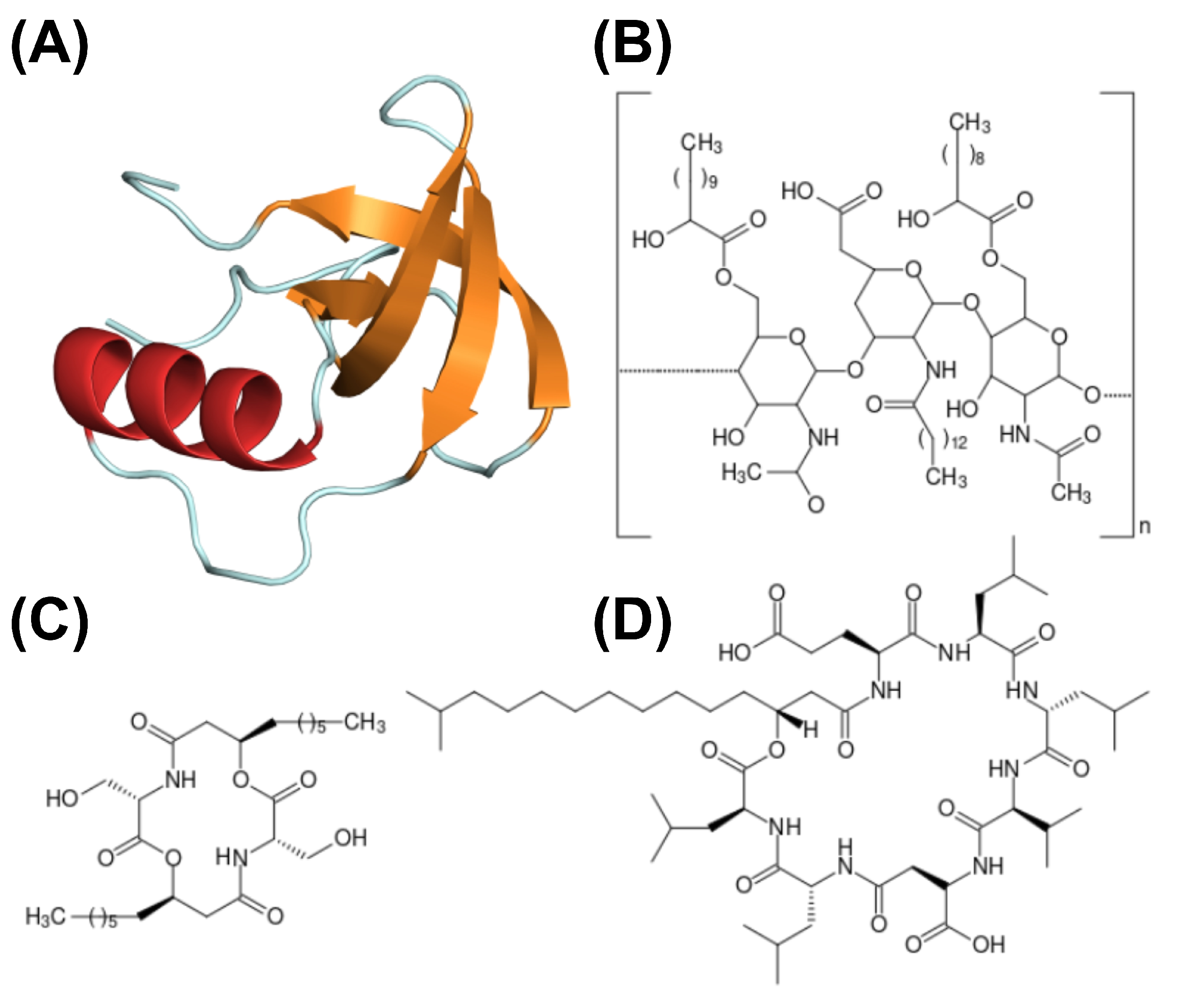
Another type of biosurfactant is hydrophobin, which has also been reviewed over the last years [42,167,168]. Hydrophobins are small, surface-active proteins produced by filamentous fungi. This protein is structurally characterized by a -sheet core with four anti-parallel -strands, containing eight conserved cysteine residues, which form four disulfide bridges, and an external -helix. Hydrophobins have been proven helpful in various industrial applications, such as protein purification processes, biosensors, drug solubility, food dispersion, and tissue engineering. Furthermore, hydrophobins are classified into two groups based on their solubility: class I and II.
Molecular dynamics elucidate hydrophobins’ molecular behavior and physical-chemical properties. Euston [169] investigated the adsorption property of the class II hydrophobin HFBI at different interfaces: vacuum-water, di-palmitoyl-phosphatidylcholine (DPPC)-water, and decane-water. HFBI showed, generally, an anisotropic structure characteristic, acting mainly as a rigid body when adsorbed and presenting high structural stability. However, the protein’s behavior is dependent on the type of interface. The biosurfactant displays spontaneous dimerization conduct in water, aggregating by their hydrophobic patch. On the other hand, HFBI showed a propensity to denaturate its surface and lose secondary structure, reducing its -structure when at the decane-water interface. Moreover, HFBI change in its tertiary structure is slightly more significant at the DPPC-water interface when compared to the vacuum-water interface.
Next, Raffaini et al. [170] investigated the molecular behavior of HFBII in water, fluorinated solution, and water-vacuum interface at an atom level. HFBII showed only minor local changes in the hydrophobic patch when in the fluorinated solution; As expected, when in water, HFBII relocates to the water-vacuum interface, keeping the hydrophilic residues in contact with water and exposing its hydrophobic side to the vacuum. These results are essential for future investigation of the HFBII self-assembly process.
6.2. Physical-Chemical Selective Pressure of Biosurfactant Molecules
What do the physical-chemical aspects of surfactants reveal about evolution of the biosurfactant-producing organisms? Temperature, pH, and ionic strength are the parameters that vary the most in different environments. Organisms from fresh, warm water are subjected to different evolutionary pressures than organisms from brine, cold water. Higher temperatures induce biosurfactants to quickly lower surface tension, while low-temperature environments are more challenging to reduce surface tension. Thus, from a physical-chemical perspective, it can be expected that organisms from cold environments present a more extended, less-branched hydrophobic chain to compensate for the temperature barrier for lowering surface tension.
Biosurfactant properties are intrinsically connected to the molecule structure. Small proteins, like hydrophobin, will display a different molecular behavior compared to small, peptide-derived biosurfactants. Peptide and protein biosurfactants may present different net charges, molecular polarity distribution, alkyl chain length, molecular mass, and other characteristics that influence their biosurfactant properties. Moreover, biosurfactants may present as polymers, such as emulsan. Figure 3 depicts the structure of different biosurfactants.
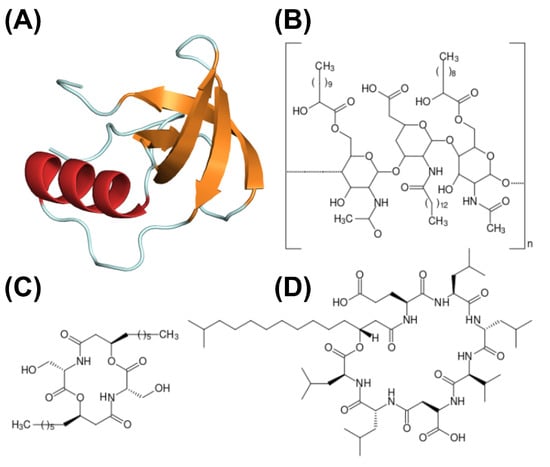
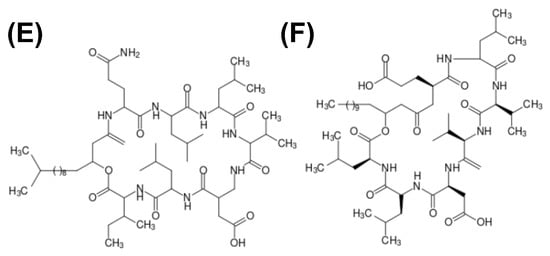
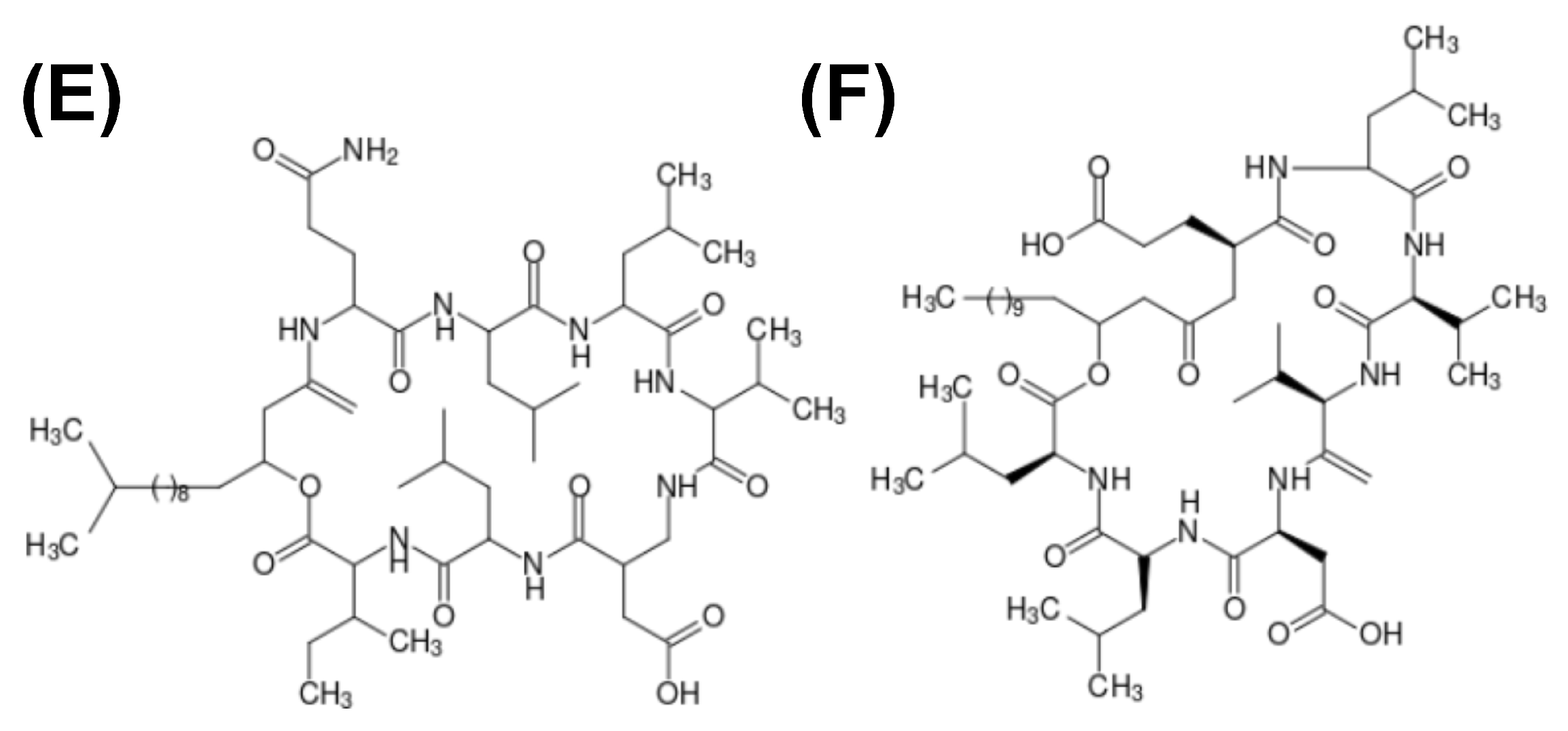
Figure 3.
Structure schematics of biosurfactants. (A) Hydrophobin (PDB ID: 2PL6), structure of -sheet core represented in orange, and external -helix colored in red; (B) emulsan, a polymeric biosurfactant; (C) serrawerttin; (D) surfactin; (E) lichenysin; (F) iturin.
7. Omics Technology and Bioinformatic Analysis as a Tool for Biosurfactant Identification
The appropriate use of biosurfactants, in general, is only possible when metabolic and bacterial diversity is known. Understanding how interaction occurs among microbial communities is essential to elucidate biosurfactant production. The richest source for biosurfactant-producing microorganisms is seawater, as the conditions of this environment allow for the survival of only the most tolerant organisms [171]. Nevertheless, even though around bacteria are present in 1 mL of seawater, only about 0.0001–0.1% of these are culturable [171,172]. The same goes for soil microorganisms, where less than 1% of those are cultivable [173].
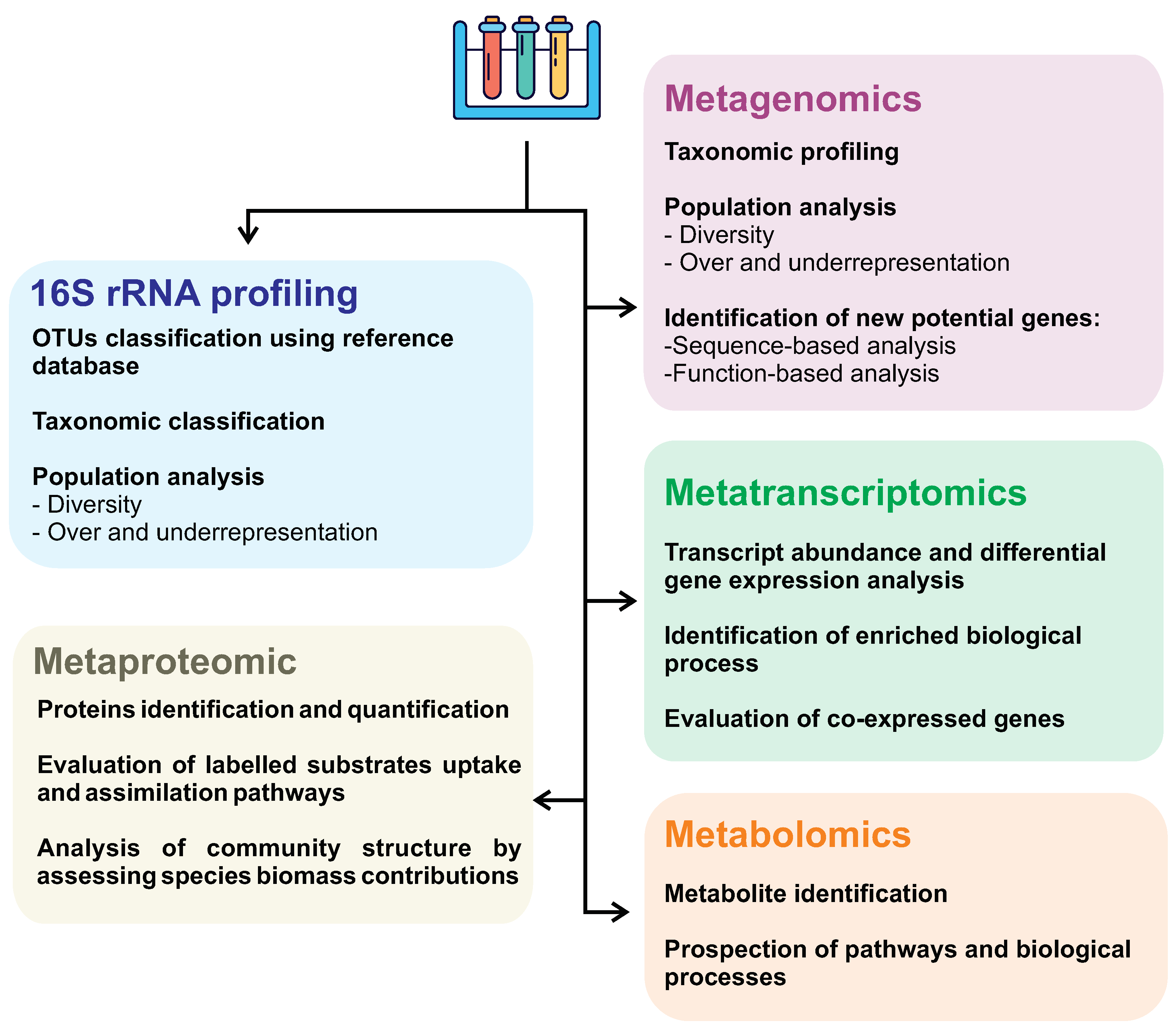
First described in 1998 as the total of genomes found in a sample [174], metagenomics is the analysis of genomes contained within an environmental sample with no need for prior cultivation or classification of the species [175]. Therefore, this opens a wide array of opportunities to discover novel species and compounds, as said before, because most of the microorganisms are uncultivable in lab conditions. Not only that, but the natural conditions in situ that these life forms exist are so complex that reproducing them artificially is practically impossible. On top of that, metagenomic and genetic data, in general, are widely available for use in public databases over the internet, making the information much more accessible for use. In addition, other large-scale approaches can also be employed, as shown in Figure 4.
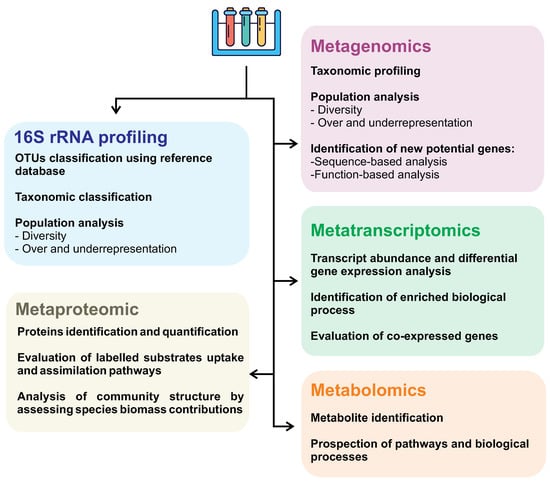
Figure 4.
Omics approaches and their applications as strategies in biosurfactant research.
Metagenomics allows us to access the genetic information of microorganisms that cannot be cultured in laboratory conditions, allowing environmental DNA extraction and the evaluation of microbial consortium, which can significantly contribute to the discovery of new biosurfactant producers and molecules. In contrast, 16S rRNA gene sequencing can’t retrieve the microorganisms’ genome being able to give an accurate microbial composition profiling sampled environment. Metagenomic studies consist of main approaches: (i) sequence-based, and (ii) function-based strategies. Sequence-based analyses have steadily advanced over the years alongside next-generation sequencing platforms. This technique focuses on discovering protein-coding sequences based on homology using curated data from databases. This method has been used to identify genes that regulate biosurfactant production in the genome of S. marcescens strain Db10 [171,176]. Despite its success, sequence-based screening is widely believed to be limited. It arises from the fact that existing homologous genes are being targeted, and very few novel genes are detected [171,177].
The function-based analysis involves screening the metagenomic library for functional activities from the expression of genes in the bacterial metagenomic DNA [178]. This approach is much more time-consuming and laborious than homology-based approaches but yields better results in identifying novel genes [177]. Even so, this technique is ever-evolving, and with time several different methods were elaborated and put into action to make it even more efficient [179].
The metagenomic approach has been growing for the past few years as an efficient way to elucidate biosurfactant production and bioremediation efficiency [180,181,182,183]. Some key biosurfactant producers identified in marine samples are the Bacillus and Rhodococcus genera [184]. Examples of novel biosurfactants discovered in saltwater environments are Aneurinifactin [185], a laccase-like enzyme [186], and an esterase called EstATII found in the red sea [187]. Although described in soil, the archaeal biosurfactant MBSP1 is thought to have great efficiency in marine environments [188].
Moreover, soil contamination has also been in the spotlight, as it is an environment considered of vital importance and one of earth’s “system life support” [189]. Metagenomics is a pivotal component in understanding soil contamination and has been used in soil-oriented biosurfactant research. One example is hydrocarbon-contaminated arctic soil in Canada, where the soil metagenome was sequenced over time. As results, Pseudomonas was the most active microorganism during peak contamination, followed by Rhodococcus [190]. Experiments followed over time allow monitoring the microbial population dynamics and its variation as a response to the interaction of organisms with each other and with the environment. Furthermore, evaluating multiple interacting organisms better reproduces the biological reality that will be faced in most biosurfactant applications.
Different omic approaches (such as metagenomic, metatranscriptomic, metabolomic, and metaproteomic) can evaluate different levels of knowledge of a system and, therefore, should be chosen and combined to address the research aim better. Despite metagenomics being more widely used in the investigation of new biosurfactants, the combination of multi-omic approaches contributes to greater accuracy in investigating surfactant biosynthesis pathways, providing a more detailed description of interacting microorganisms, especially in taxonomically very diverse communities. Metagenomic analyses give an insight into the present microorganisms, their abundance, and the functional aspects of their genome. On the other hand, metatranscriptomics can better understand the whole gene expression profile, shedding light on microbial communities’ ecological response subjected to specific environmental conditions, such as oil spills. In other words, metatranscriptome allows us to assess which molecules are active at a given time and how abundant they are, discriminating members actively contributing to a specific response or behavior from those quiescent members.
Other omics approaches, such as metabolomics and metaproteomic, are barely discussed in the literature searching for new biosurfactants. One possible reason is the high cost of these analyses associated with numerous challenges related to the analysis workflow. Metaproteomic generally shows lower measurements depth than metagenomic and metatranscriptomic, and protein inference may be challenging due to redundant identification caused by homologous proteins [191,192]. However, with a highly curated and comprehensive species protein database, it is possible to precisely identify and assign these proteins to their respective taxa, providing a picture of microbial activity and the exploration of novel functional genes and biochemical pathways in response to specific conditions [193]. Despite this, proteomics can generate exciting results, as demonstrated by an analysis conducted by Pitocchi et al. from two fungi species isolated from the spill-oil polluted marine site [194]. In this research, it was identified a potent biosurfactant secreted by Aspergillus terreus MUT 271 and Trichoderma harzianum MUT 290 belonging to cerato-platanins protein family with similar properties of hydrophobins [194].
Metabolomics provides a global picture of metabolites, reflecting the biochemical activity of microorganisms. In microbiomes analysis, it is challenging to relate metabolites to specific taxa; being possible to solve this issue by identification of covariates metabolite integrated with a species composition analysis, which may indicate species-specific metabolite production [191]. Interestingly, an untargeted metabolomics study from one thousand marine isolates microorganisms identified several molecule classes and annotated 76 molecular families, among them, the surfactin biosurfactant [195].
Studies at the biological system level open the opportunity to explore new molecules and identify new organisms that produce biosurfactants, generating knowledge about the ecological systems where these organisms are found. Knowledge bases on biological pathways and functional classification are constantly expanding. A specific repository to assist in biosurfactant research has already been launched, BioSurfDB, which contains information on metagenomes, organisms, cured biosurfactants, bioremediation experiments results, and biodegradation relevant genes Oliveira et al. [196].
Collecting samples and sequencing are only some essential steps in omics studies. After gathering data, all that information needs to be analyzed through bioinformatics methods. In addition to the well-established workflows appropriate for different omic data, the integration of omic analyzes could lead to more consistent and informative results.
7.1. Network Analysis
Understanding the relationship between molecules and the organization of possible molecular pathways associated with biosurfactant production is crucial. Interaction networks are valuable tools to deal with a massive amount of information, whether from a single methodological approach, such as protein-protein interaction networks or a combination of data from multiple levels of knowledge. These networks represent direct and indirect interactions between the components of that system, which can describe genes, proteins, and any other target molecule. It is possible to build networks from multiple omics approaches, linking the results of genomics, transcriptomics, proteomics, and metabolomics within a single system through multilayer networks. Numerous works have used interaction networks to understand the relationship between genes/proteins and different compounds [195,197,198,199]. Also, metabolomic-based networks were used to elucidate metabolic profiles and chemical structures from secondary metabolites produced by S. marcescens strains, resulting in the identification of serrawettin W1 [200].
However, the integration of multi-omic data is a challenge on its own. Compiling different data layers requires high computational costs and robust pipelines that ensure the quality of the combined information. Additionally, creating consistent workflows requires substantial knowledge of the analyzed systems. Identifying modules capable of participating in a common biological function during network analysis is essential for understanding a complex network’s structural and functional aspects. Many modules are conserved throughout evolution and may represent core functions or processes for biological regulation [201]. Clustering and topological analysis are powerful methods for elucidating molecular machinery. It is not unusual in omics analyses to identify new molecules of unknown function. Although sequence-based analysis is usually used, module analysis can be very useful for applying a principle called guilt-by-association, especially in co-expression networks [202]. In this sense, poorly annotated genes within the same module are more likely to be associated with similar processes. However, this approach must be undertaken rigorously to avoid misinterpreting these potential genes [202].
Significant advances also have been made in the area of flux balance analysis (FBA). This research area estimates metabolic flux in genome-scale metabolic network reconstructions using the constraints imposed by stoichiometric coefficients of each reaction [203]. FBA can be used for a wide variety of applications, being able to incorporate data from different omics. Thus, it is possible to create FBA models to study the tolerance of an organism to mutations and physiological stress, the organism’s growth rate, or even the production rate of a specific metabolite. FBA analysis can be of great importance for metabolic engineering, whose knowledge can provide insight into the large-scale production of biosurfactants. Occhipinti et al. [204] used in silico engineering to introduce the RhlA and RhlB genes and reactions responsible for rhamnolipids biosynthesis from Pseudomonas aeruginosa to an existing genome-scale model of P. putida. In silico models can be useful to predict the metabolic and genetic engineering steps needed for maximizing the production of a target compound.
Systems biology becomes a great ally in understanding microbial diversity and composition and in analyses related to cellular processes and responses to certain conditions. Furthermore, understanding natural or endemic bacterial communities in regions that require attention, such as crude oil spill-affected regions, can aid prospecting for biosurfactant-producing microorganisms. Different network analysis methods for studying microbial communities have been proposed and are explored in detail in [205,206]. A study conducted in a microcosm showed that microbial biofilm diversity decreased after exposition to the biosurfactant rhamnolipid. Furthermore, exposure of the microbial community to rhamnolipid considerably changed extracellular enzyme activity, reducing phosphatase activity and increasing beta-glucosidase levels. Network analysis can also be applied to assess the impact of surfactant additions on natural microbial communities to support (or not) their applications once their impact on the environment is proven. In this sense, a study in a northeast Atlantic marine community exposed to crude oil spills investigated how the biosurfactant rhamnolipids or synthetic dispersants could impact community dynamics and functional diversity. The synthetic surfactant negatively affected taxa diversity and impacted the microbial community’s functional robustness more significantly than biosurfactant use.
7.2. Machine Learning and Data Integration
Machine Learning (ML) is a field derived from studies of artificial intelligence, pattern recognition, statistics, and optimization, which aims to develop algorithms capable of performing functions without being explicitly controlled by a user. ML techniques “learn” how to make predictions and decisions through a given data or by integrating more than one [207,208], which could be genomic, transcriptomic, proteomic, or metabolomic. The main advantage of using ML is its ability to extract information from a massive amount of large-scale data, which can be used to create predictive models for a given situation quickly and with high precision, in addition to finding essential patterns for the description of the biological phenomenon [209].
Analyzing the omics datasets in an integrative way using ML techniques will return a complete picture of the whole system than separately analyzing them. Data integration can be defined as the integrative study of data from multiple sources to improve knowledge discovery [210,211,212]. Tarazona et al. [213] categorizes integration methods based on incorporating additional biological and using supervised or unsupervised approaches. Supervised methods can be used in two principal ways—the first is for predicting a response variable. The second way is to understand the communication between the omics layers or, more specifically, to model the potential regulations to build the regulatory network of the biological system studied [214]. Unsupervised methods are mainly applied for a preliminary exploration of datasets and sometimes also for clustering observations. In the case of multi-omic experiments, an unsupervised strategy can help us to understand the relationship among omics and find common and different patterns among them. Unsupervised strategies comprise clustering, dimension reduction methods (e.g., principal component analysis, T-distributed stochastic neighbor embedding), or machine-learning strategies (e.g., artificial neural networks).
Machine learning approaches have been increasingly applied to predict suitable parameters to improve biosurfactant production yield for scale-up toward industrial production. Bioprocess engineering involving biosurfactant production faces several challenges since different factors affect its production, such as carbon and nitrogen sources, temperature, pH, oxygen requirements, metal ions, and agitation speed [215]. The incorrect adjustment of these factors could affect the system’s efficiency, influencing the microorganisms’ growth and metabolic activity [215]. In this sense, different machine learning algorithms have been applied to optimize biosurfactant production, such as artificial neural network (ANN) [216,217], support vector regression (SVR) and support vector regression coupled with firefly algorithm (SVR-FFA) [218], and support vector machine (SVM) [219].
Interspecific interactions within a population are paramount to understanding the relationships in a synthetic consortium or between the microbial community and their environment. In this sense, Chang et al. applied ML using the Random Forest method on metagenome data to assess whether the composition of microorganisms is related to crop productivity [220]. Interestingly, using ML approaches, it was possible to identify a subset of taxa that could be responsible for functional aspects of the community [221]. Also, a study showed that support vector regression-fruit fly optimization algorithm (SVR-FOA) was able to accurately estimate the simultaneous biodegradation of azo dyes and hexavalent chromium by a biosurfactant-producing bacterial strain Klebsiella sp. KOD36 [222]. ML applications in microbial analysis focusing on data integration and understanding cooperative and competitive relationships within a microbial ecosystem are an important direction for future research.
7.3. Molecular Dynamics (MD) Simulations
The principle behind MD is to represent computationally molecules and ions, which can be at an atomistic level or coarse-grained; define a volume for the experiment to take place, which will be represented as a box, and apply Newtonian physics principles in the system to calculate all bonded and non-bonded forces in motion. In other words, MD allows us to use computer processing to simulate and visualize how a set of molecules behave under specific conditions in a given time. From one typical simulation, one can obtain a myriad of information about structural features and physical-chemical properties. Moreover, a molecule, or thousands of the same molecule, such as a specific biosurfactant, can be simulated in different solvents. This way, the properties in different interfaces can be investigated [223,224,225].
Furthermore, MD can be performed at varying temperature, pressure, ionic force, solvent type, the number of particles (concentration), and other parameters that might be relevant to a specific experiment. Therefore, one can investigate structural features, such as hydrogen bonds through time, atoms position and fluctuation through time, as well as physical-chemical properties like density and free energy of solvation. A handful of free software programs for MD, such as GROMACS [226], NAMD [227] and OpenMM [228]. An easy, straightforward tutorial to learn how to simulate from scratch using GROMACS can be found in [229].
Moreover, surfactants properties, such as CMC, interfacial/surface tension, Winsor I-II-III-IV transition, Hydrophile-Lipophile Balance (HLB), and structure-property relationships can all be assessed through MD techniques [230]. Furthermore, variations of MD have been developed to investigate free energy-based processes to overcome limitations from classical, atomistic MD simulations. Since MD keeps
Assessing CMC through computational methods depends on the calculation of the aggregation probability number (P(N)), which is the likelihood of N molecules aggregating [230]. However, the minimum of biosurfactants molecules needed to produce a micelle varies depending on the system. In this sense, MD can predict biosurfactants’ aggregation number, structure, and CMC. It has been employed before to study a variety of surfactants with different properties, such as ionic [231], and nonionic [232].
Next, MD simulations have been employed to investigate interfacial/surface tension, which is accomplished based on mechanical laws through pressure tensor calculation, or the system’s free energy derivative concerning the surface area [230]. For surfactants in general, experiments at the wet lab and MD simulation results correspond well, especially regarding the density distribution of alkyl chains and surfactant headgroups. However, MD cannot forecast the counterion distribution effectively, leading to the opposite results in experiments and modeling for the counterion affinity with the surfactant headgroup [233,234,235]. The core reason for this limitation is that by utilizing default Lennard-Jones values from classical force fields, the attraction between the surfactant headgroup and the counterions may be exaggerated. By lessening this attraction, it is possible to achieve a surfactant/counterion interaction that is realistic and consistent with the experimental findings [236].
Furthermore, it is possible to employ MD to investigate Winsor transitions [230], which are related to the different types of microemulsions. Four different types of microemulsions, known as Winsor-type microemulsions, exist [237]. Type I microemulsions are defined by the solubilization of oil in spherical, normal micelles within the water-continuous phase; type II presents solubilization in reverse micelles within the oil-continuous phase; type III exists in three-phase systems in which the middle phase microemulsions remain in equilibrium with both aqueous and organic phases and; type IV, characterized when the middle phase microemulsions are expanded at a high surfactant concentration, resulting in the formation of a single phase from the combination of aqueous and organic phases [238]. Moreover, parameters such as the temperature, the surfactant concentration, and the salinity all affect how many phases a system consists of (bio)surfactants, an organic phase, and a brine, aqueous phase. Changing the parameters will result in a phase transition. Thus, performing simulations with systems at different temperatures, types of biosurfactant (i.e., different size, shape, and hydrophile/hydrophobic moieties proportion), and nature of the organic phase will influence Winsor I-II-III-IV transitions.
Another important property of amphiphilic biosurfactants, such as lipopeptides, is the HLB. A biosurfactant’s HLB is the balance between the size and the tensile strength of its hydrophilic and lipophilic moieties [239]. Among the structural features that influence lipopeptides self-assembly, HLB is one important parameter to consider. Studies have indicated that the minimum lipid moiety size is crucial for self-assembly. However, there is no universal rule to predict such property since the sequence and length of the peptide moiety also influence amphiphilicity. Many other factors, including electrostatic interactions, hydrogen bonding, van der Waals interactions - stacking interactions [240]. In this sense, MD simulations provide an in-depth analysis of all these factors, and investigation of systems varying temperature, molecule type, and salt concentration is easily achieved. Additionally, non-bonded interactions are routinely discrete by MD analyses, thoroughly scrutinizing how much each factor contributes to the overall net result.
Structural analyses of biomolecules are, perhaps, the most common application of MD simulations. Understanding the molecular behavior of biosurfactants through time, such as the influence of specific intermolecular interactions, the impact of angle and dihedral conformations, the variation of position and fluctuation of atoms, solvent accessible area, and energy profile, are typical MD analyses. Moreover, these analyses are conveniently applied to evaluate the effect of biosurfactants on other proteins [241]. This approach has been employed for decades [223], and it is not only pertinent nowadays, but its use is compelling since computational resources become better over time and force field parameters are more accurate. A myriad of studies regarding MD simulations and peptide biosurfactants are available in the literature [223,225,241,242,243,244,245,246,247,248,249,250,251,252,253,254,255]. Moreover, specific reviews about MD and biosurfactants are available [256,257,258].
Although MD simulations have been consistently employed to investigate surfactants’ properties, only a few types of peptide biosurfactants were regularly assessed through the lens of MD. This way, there is a sea of opportunities for different works for MD and biosurfactants. Furthermore, MD simulations present as the nexus between physical-chemical theory and wet lab experiments. Biosurfactant studies employing MD techniques have been carried out consistently for a long time, and the results have shown to agree with the literature. Thus, incorporating computational methods in the experimental workflow will provide in-depth, comprehensive results to conduct global research at lower costs.
8. Conclusions and Perspectives
Lipopeptides are a class of compounds with a large diversity of structures and functions, resulting in a variety of applications as ecological roles. And, as we can see, the study of biosurfactants has been exponentially growing during the past years. While, as a subject of study, they are rather old, new research tools have enabled a much better understanding of biosurfactant production, composition, and function. More efficient ways of cultivating microorganisms and boosting biosurfactant yields and the combined production alongside other compounds of interest during co-production are here to ease the market for their use. While research using bioinformatics tools is giving us new ways to understand biosurfactants and discover new natural surfactants, as well as insight into how microbial populations function during contamination and how the composition of these populations can affect the biosurfactant yield. All the information and research tools discussed here are essential going forward on biosurfactant research. Although it seems like much is known in this subject, around 30% of bacterial lipoproteins are still not functionally characterized. And this is were powerful means of complex data analysis in molecular dynamics and meta-mics research come into play.
By using the technologial assets presented here, thorough studies on biosurfactants can be envisioned. For example, in a contaminated site the whole genomic and transcriptomic profiles of microbial populations can be sampled, analyzed, and these results compared in multi-field research. We can detect biosurfactant producers via metagenomics and metatranscriptomics, test biosurfactant yield and degradation capabilities in lab trials, and elucidate the physical and chemical properties of biosurfactant molecules, while understanding the environmental influence on their molecular behavior. Furthermore, machine learning methodologies can be used to predict output parameters of biosurfactants to optimize the conceptual phase of studies. Predicting the total biosurfactant production is also a great asset to understand total costs of practical applications and use of more cost-effective substrates.
Biosurfactant-producing microorganisms, especially from extreme environments, show great promise for the discovery of novel lipopeptides. As more powerful research tools emerge, such as metagenomics, metatranscriptomics, MD simulations and machine learning are coupled with traditional laboratory research methods, the discovery of diverse lipopetides structures and functions becomes possible.
Author Contributions
Conceptualization, M.D. and J.d.F.P.; methodology, R.A.J.; investigation, R.A.J., J.d.F.P. and É.S.M.P.; resources, M.D.; writing—original draft preparation, R.A.J., J.d.F.P. and É.S.M.P.; writing—review and editing, M.D., J.d.F.P. and É.S.M.P.; visualization, R.A.J.; supervision, M.D.; project administration, M.D.; funding acquisition, M.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul—FAPERGS [19/2551-0001906-8; 17/2551-0000520-1], Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq [314082/2021-2; 465450/2014-8; 440279/2022-4], and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES/STICAMSUD [88881.522073/2020-01] and DAAD/CAPES PROBRAL 88881.198766/2018-01]. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jahan, R.; Bodratti, A.M.; Tsianou, M.; Alexandridis, P. Biosurfactants, natural alternatives to synthetic surfactants: Physicochemical properties and applications. Adv. Colloid Interface Sci. 2020, 275, 102061. [Google Scholar] [CrossRef] [PubMed]
- Hamme, J.D.V.; Singh, A.; Ward, O.P. Physiological aspects: Part 1 in a series of papers devoted to surfactants in microbiology and biotechnology. Biotechnol. Adv. 2006, 24, 604–620. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Patil, Y.; Rale, V. Biosurfactant production: Emerging trends and promising strategies. J. Appl. Microbiol. 2019, 126, 2–13. [Google Scholar] [CrossRef]
- Desai, J.D.; Banat, I.M. Microbial production of surfactants and their commercial potential. Microbiol. Mol. Biol. Rev. 1997, 61, 47–64. [Google Scholar] [PubMed]
- Machlin, E.S. CHAPTER XIV—Thermodynamics of Micelles. In An Introduction to Aspects of Thermodynamics and Kinetics Relevant to Materials Science, 3rd ed.; Machlin, E.S., Ed.; Elsevier Science Ltd.: Oxford, UK, 2007; pp. 425–454. [Google Scholar]
- Banat, I.M.; Makkar, R.S.; Cameotra, S.S. Potential commercial applications of microbial surfactants. Appl. Microbiol. Biotechnol. 2000, 53, 495–508. [Google Scholar] [CrossRef]
- Akbari, S.; Abdurahman, N.H.; Yunus, R.M.; Fayaz, F.; Alara, O.R. Biosurfactants—A new frontier for social and environmental safety: A mini review. Biotechnol. Res. Innov. 2018, 2, 81–90. [Google Scholar] [CrossRef]
- Hazra, C.; Kundu, D.; Chaudhari, A. Biosurfactant-Assisted Bioaugmentation in Bioremediation; Springer: Dordrecht, The Netherlands, 2012; pp. 631–664. [Google Scholar]
- Wise, J.; Wise, J.P. A review of the toxicity of chemical dispersants. Rev. Environ. Health 2011, 26, 281–300. [Google Scholar] [CrossRef]
- Bejarano, A.C. Critical review and analysis of aquatic toxicity data on oil spill dispersants. Environ. Toxicol. Chem. 2018, 37, 2989–3001. [Google Scholar] [CrossRef]
- Hamdan, L.J.; Fulmer, P.A. Effects of COREXIT® EC9500A on bacteria from a beach oiled by the Deepwater Horizon spill. Aquat. Microb. Ecol. 2011, 63, 101–109. [Google Scholar] [CrossRef]
- Bookstaver, M.; Bose, A.; Tripathi, A. Interaction of Alcanivorax borkumensis with a surfactant decorated oil–water Interface. Langmuir 2015, 31, 5875–5881. [Google Scholar] [CrossRef]
- Souza, K.S.T.; Gudiña, E.J.; Schwan, R.F.; Rodrigues, L.R.; Dias, D.R.; Teixeira, J.A. Improvement of biosurfactant production by Wickerhamomyces anomalus CCMA 0358 and its potential application in bioremediation. J. Hazard. Mater. 2018, 346, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Cameotra, S.S.; Makkar, R.S.; Kaur, J.; Mehta, S.K. Synthesis of Biosurfactants and Their Advantages to Microorganisms and Mankind. In Biosurfactants; Sen, R., Ed.; Springer: New York, NY, USA, 2010; pp. 261–280. [Google Scholar]
- Kiran, G.S.; Sabarathnam, B.; Selvin, J. Biofilm disruption potential of a glycolipid biosurfactant from marine Brevibacterium casei. FEMS Immunol. Med. Microbiol. 2010, 59, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Franzetti, A.; Gandolfi, I.; Fracchia, L.; Van Hamme, J.; Gkorezis, P.; Marchant, R.; Banat, I.M. Biosurfactant use in heavy metal removal from industrial effluents and contaminated sites. Biosurfactants Prod. Util. Technol. Econ. 2014, 159, 361. [Google Scholar]
- Reis, R.; Pacheco, G.; Pereira, A.; Freire, D. Biosurfactants: Production and Applications. In Biodegradation; Chamy, R., Rosenkranz, F., Eds.; IntechOpen: Rijeka, Croatia, 2013; Chapter 2. [Google Scholar]
- Andrade, R.; Silva, T.; Rubio, D.; Montero Rodríguez, D.; Souza, A.; Lima, M.; Lima, R.; Silva, C.; Campos Takaki, G. Promising Biosurfactant Produced by Cunninghamella echinulata UCP 1299 Using Renewable Resources and Its Application in Cotton Fabric Cleaning Process. Adv. Mater. Sci. Eng. 2018, 2018, 1624573. [Google Scholar] [CrossRef]
- Soares da Silva, R.; de Almeida, D.; Brasileiro, P.E.A. Production, formulation and cost estimation of a commercial biosurfactant. Biodegradation 2019, 159, 191–201. [Google Scholar] [CrossRef]
- Soares Dos Santos, A.; Pereira, N.J.; Freire, D.M. Strategies for improved rhamnolipid production by Pseudomonas aeruginosa PA1. PeerJ 2016, 4, e2078. [Google Scholar] [CrossRef]
- Sahebnazar, Z.; Mowla, D.; Karimi, G. Enhancement of Pseudomonas aeruginosa Growth and Rhamnolipid Production Using Iron-Silica Nanoparticles in Low-Cost Medium. J. Nanostruct. 2018, 8, 1–10. [Google Scholar]
- Vijayakumar, S.; Saravanan, V. Biosurfactants-Types, Sources and Applications. Res. J. Microbiol. 2015, 10, 181–192. [Google Scholar]
- Roy, A. A Review on the Biosurfactants: Properties, Types and its Applications. J. Fundam. Renew. Energy Appl. 2017, 8, 1–14. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour. Technol. 2017, 232, 389–397. [Google Scholar] [CrossRef]
- Henkel, M.; Hausmann, R. Chapter 2—Diversity and Classification of Microbial Surfactants. In Biobased Surfactants, 3rd ed.; Hayes, D.G., Solaiman, D.K., Ashby, R.D., Eds.; AOCS Press: London, UK, 2019; pp. 41–63. [Google Scholar]
- Kashif, A.; Rehman, R.; Fuwad, A.; Shahid, M.K.; Dayarathne, H.; Jamal, A.; Aftab, M.N.; Mainali, B.; Choi, Y. Current advances in the classification, production, properties and applications of microbial biosurfactants—A critical review. Adv. Colloid Interface Sci. 2022, 2022, 102718. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.K.F.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional Biomolecules of the 21st Century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Campos, J.M.; Montenegro Stamford, T.L.; Sarubbo, L.A.; de Luna, J.M.; Rufino, R.D.; Banat, I.M. Microbial biosurfactants as additives for food industries. Biotechnol. Prog. 2013, 29, 1097–1108. [Google Scholar] [CrossRef]
- Kaur, P.; Singh, S.; Kumar, V.; Singh, N.; Singh, J. Effect of rhizobacteria on arsenic uptake by macrophyte Eichhornia crassipes (Mart.) Solms. Int. J. Phytoremediat. 2017, 20, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Ibrar, M.; Zhang, H. Construction of a hydrocarbon-degrading consortium and characterization of two new lipopeptides biosurfactants. Sci. Total. Environ. 2020, 714, 136400. [Google Scholar] [CrossRef] [PubMed]
- Ibrar, M.; Yang, X. Reconstructing polyaromatic hydrocarbons degrading pathways in the enriched bacterial consortium and their biosurfactants characterization. J. Environ. Chem. Eng. 2022, 10, 107219. [Google Scholar] [CrossRef]
- Bezza, F.A.; Chirwa, E.M.N. Production and applications of lipopeptide biosurfactant for bioremediation and oil recovery by B. subtilis CN2. Biochem. Eng. J. 2015, 101, 168–178. [Google Scholar] [CrossRef]
- Habib, S.; Ahmad, S.A.; Wan Johari, W.L.; Abd Shukor, M.Y.; Alias, S.A.; Smykla, J.; Saruni, N.H.; Abdul Razak, N.S.; Yasid, N.A. Production of lipopeptide biosurfactant by a hydrocarbon-degrading Antarctic Rhodococcus. Int. J. Mol. Sci. 2020, 21, 6138. [Google Scholar] [CrossRef] [PubMed]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef]
- Calvo, C.; Manzanera, M.; Silva-Castro, G.; Uad, I.; González-López, J. Application of bioemulsifiers in soil oil bioremediation processes. Future prospects. Sci. Total. Environ. 2009, 407, 3634–3640. [Google Scholar] [CrossRef]
- Uzoigwe, C.; Burgess, J.G.; Ennis, C.J.; Rahman, P.K. Bioemulsifiers are not biosurfactants and require different screening approaches. Front. Microbiol. 2015, 6, 245. [Google Scholar] [CrossRef] [PubMed]
- Smyth, T.; Perfumo, A.; McClean, S.; Marchant, R.; Banat, I. Isolation and Analysis of Lipopeptides and High Molecular Weight Biosurfactants. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3688–3704. [Google Scholar]
- Navon-Venezia, S.; Zosim, Z.; Gottlieb, A.; Legmann, R.; Carmeli, S.; Ron, E.; Rosenberg, E. Alasan, a new bioemulsifier from Acinetobacter radioresistens. Appl. Environ. Microbiol. 1995, 61, 3240–3244. [Google Scholar] [CrossRef] [PubMed]
- Barkay, T.; Navon-Venezia, S.; Ron, E.; Rosenberg, E. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Appl. Environ. Microbiol. 1999, 65, 2697–2702. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Sundar, D.; Srivastava, P. Biosurfactants: Potential Agents for Controlling Cellular Communication, Motility, and Antagonism. Front. Mol. Biosci. 2021, 2021, 893. [Google Scholar] [CrossRef]
- Linder, M.B. Hydrophobins: Proteins that self assemble at interfaces. Curr. Opin. Colloid Interface Sci. 2009, 5, 356–363. [Google Scholar] [CrossRef]
- Berger, B.W.; Sallada, N.D. Hydrophobins: Multifunctional biosurfactants for interface engineering. J. Biol. Eng. 2019, 13, 10. [Google Scholar] [CrossRef]
- Roongsawang, N.; Washio, K.; Morikawa, M. Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int. J. Mol. Sci. 2010, 12, 141–172. [Google Scholar] [CrossRef]
- National Library of Medicine. PubChem Compound Summary for CID 23724538, Arthrofactin; NCBI: Bethesda, MD, USA, 2004.
- Zhang, L.; Sun, C. Fengycins, cyclic lipopeptides from marine B. subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef]
- Sur, S.; Romo, T.D.; Grossfield, A. Selectivity and mechanism of fengycin, an antimicrobial lipopeptide, from molecular dynamics. J. Phys. Chem. B 2018, 122, 2219–2226. [Google Scholar] [CrossRef]
- National Library of Medicine. PubChem Compound Summary for CID 443591, Fengycin; NCBI: Bethesda, MD, USA, 2004.
- Deleu, M.; Paquot, M.; Nylander, T. Effect of fengycin, a lipopeptide produced by B. subtilis, on model biomembranes. Biophys. J. 2008, 94, 2667–2679. [Google Scholar] [CrossRef]
- Morikawa, M.; Daido, H.; Takao, T.; Murata, S.; Shimonishi, Y.; Imanaka, T. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J. Bacteriol. 1993, 175, 6459–6466. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhu, X.; Lu, Y.; Zhao, H.; Lu, F.; Lu, Z. Improving iturin A production of Bacillus amyloliquefaciens by genome shuffling and its inhibition against Saccharomyces cerevisiae in orange juice. Front. Microbiol. 2018, 9, 2683. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Du, Z.; Cui, Q.; Dong, H.; Wang, F.; He, P.; Tang, Y. Biosurfactant produced by novel Pseudomonas sp. WJ6 with biodegradation of n-alkanes and polycyclic aromatic hydrocarbons. J. Hazard. Mater. 2014, 276, 489–498. [Google Scholar] [CrossRef]
- Anuradha, S.N. Structural and molecular characteristics of lichenysin and its relationship with surface activity. Adv. Exp. Med. Biol. 2010, 672, 304–315. [Google Scholar]
- Cameotra, S.S.; Makkar, R.S. Biosurfactant-enhanced bioremediation of hydrophobic pollutants. Pure Appl. Chem. 2010, 82, 97–116. [Google Scholar] [CrossRef]
- Matsuyama, T.; Tanikawa, T.; Nakagawa, Y. Serrawettins and other surfactants produced by Serratia. In Biosurfactants; Springer: Berlin/Heidelberg, Germany, 2011; pp. 93–120. [Google Scholar]
- Clements, T.; Ndlovu, T.; Khan, S.; Khan, W. Biosurfactants produced by Serratia species: Classification, biosynthesis, production and application. Appl. Microbiol. Biotechnol. 2019, 103, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Scholz-Schroeder, B.K.; Hutchison, M.L.; Grgurina, I.; Gross, D.C. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol.-Plant-Microbe Interact. 2001, 14, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Geudens, N.; Martins, J.C. Cyclic lipodepsipeptides from Pseudomonas spp.–biological swiss-army knives. Front. Microbiol. 2018, 9, 1867. [Google Scholar] [CrossRef] [PubMed]
- Dalla Serra, M.; Bernhart, I.; Nordera, P.; Di Giorgio, D.; Ballio, A.; Menestrina, G. Conductive properties and gating of channels formed by syringopeptin 25A, a bioactive lipodepsipeptide from Pseudomonas syringae pv. syringae, in planar lipid membranes. MPMI-Mol. Plant Microbe Interact. 1999, 12, 401–409. [Google Scholar] [CrossRef]
- Saini, H.S.; Barragán-Huerta, B.E.; Lebrón-Paler, A.; Pemberton, J.E.; Vázquez, R.R.; Burns, A.M.; Marron, M.T.; Seliga, C.J.; Gunatilaka, A.L.; Maier, R.M. Efficient purification of the biosurfactant viscosin from Pseudomonas libanensis strain M9-3 and its physicochemical and biological properties. J. Nat. Prod. 2008, 71, 1011–1015. [Google Scholar] [CrossRef]
- National Library of Medicine. PubChem Compound Summary for CID 72937, Calamenene; NCBI: Bethesda, MD, USA, 2004.
- De Bruijn, I.; Raaijmakers, J. Diversity and functional analysis of LuxR-type transcriptional regulators of cyclic lipopeptide biosynthesis in Pseudomonas fluorescens. Appl. Environ. Microbiol. 2009, 75, 4753–4761. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [PubMed]
- Marchant, R.; Funston, S.; Uzoigwe, C.; Rahman, P.; Banat, I.M. Production of Biosurfactants from Nonpathogenic Bacteria. In Biosurfactants: Production and Utilization-Processes, Technologies, and Economics; Kosaric, N., Sukan, F.V., Eds.; CRC Press: Boca Raton, FL, USA, 2014; pp. 73–81. [Google Scholar]
- Das, P.; Mukherjee, S.; Sen, R. Genetic regulations of the biosynthesis of microbial surfactants: An overview. Biotechnol. Genet. Eng. Rev. 2008, 25, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Carolin C, F.; Kumar, P.S.; Ngueagni, P.T. A review on new aspects of lipopeptide biosurfactant: Types, production, properties and its application in the bioremediation process. J. Hazard. Mater. 2021, 407, 124827. [Google Scholar] [CrossRef]
- Long, X.; He, N.; He, Y.; Jiang, J.; Wu, T. Biosurfactant surfactin with pH-regulated emulsification activity for efficient oil separation when used as emulsifier. Bioresour. Technol. 2017, 241, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Pradel, E.; Zhang, Y.; Pujol, N.; Matsuyama, T.; Bargmann, C.I.; Ewbank, J.J. Detection and avoidance of a natural product from the pathogenic bacterium S. marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2007, 104, 2295–2300. [Google Scholar] [CrossRef]
- Huang, X.; Lu, Z.; Zhao, H.; Bie, X.; Lü, F.; Yang, S. Antiviral activity of antimicrobial lipopeptide from B. subtilis fmbj against pseudorabies virus, porcine parvovirus, newcastle disease virus and infectious bursal disease virus in vitro. Int. J. Pept. Res. Ther. 2006, 12, 373–377. [Google Scholar] [CrossRef]
- Vollenbroich, D.; Pauli, G.; Ozel, M.; Vater, J. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from B. subtilis. Appl. Environ. Microbiol. 1997, 63, 44–49. [Google Scholar] [CrossRef]
- Yeh, M.S.; Wei, Y.H.; Chang, J.S. Enhanced Production of Surfactin from B. subtilis by addition of solid carriers. Biotechnol. Prog. 2005, 21, 1329–1334. [Google Scholar] [CrossRef]
- Whang, L.M.; Liu, P.W.G.; Ma, C.C.; Cheng, S.S. Application of biosurfactants, rhamnolipid, and surfactin, for enhanced biodegradation of diesel-contaminated water and soil. J. Hazard. Mater. 2008, 151, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Schor, M.; Reid, J.L.; MacPhee, C.E.; Stanley-Wall, N.R. The Diverse Structures and Functions of Surfactant Proteins. Trends Biochem. Sci. 2016, 41, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Whitsett, J.A.; Weaver, T.E. Hydrophobic Surfactant Proteins in Lung Function and Disease. N. Engl. J. Med. 2002, 347, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Cirigliano, M.C.; Carman, G.M. Purification and characterization of liposan, a bioemulsifier from Candida lipolytica. Appl. Environ. Microbiol. 1985, 50, 846–850. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Iwano, M. BslA (YuaB) forms a hydrophobic layer on the surface of B. subtilis biofilms. Mol. Microbiol. 2012, 85, 51–66. [Google Scholar] [CrossRef]
- Ostrowski, A.; Mehert, A.; Prescott, A.; Kiley, T.B.; Stanley-Wall, N.R. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by B. subtilis. J. Bacteriol. 2011, 193, 4821–4831. [Google Scholar] [CrossRef]
- Ahn, S.O.; Lim, H.D.; You, S.H.; Cheong, D.E.; Kim, G.J. Soluble Expression and Efficient Purification of Recombinant Class I Hydrophobin DewA. Int. J. Mol. Sci. 2021, 22, 7843. [Google Scholar] [CrossRef]
- Claessen, D.; Wösten, H.A.; Keulen, G.v.; Faber, O.G.; Alves, A.M.; Meijer, W.G.; Dijkhuizen, L. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 2002, 44, 1483–1492. [Google Scholar] [CrossRef]
- Morris, V.K.; Kwan, A.H.; Sunde, M. Analysis of the structure and conformational states of DewA gives insight into the assembly of the fungal hydrophobins. J. Mol. Biol. 2013, 425, 244–256. [Google Scholar] [CrossRef]
- Wösten, H.A.; de Vocht, M.L. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 2000, 1469, 79–86. [Google Scholar] [CrossRef]
- Wessels, J.G. Hydrophobins: Proteins that change the nature of the fungal surface. Adv. Microb. Physiol. 1996, 38, 1–45. [Google Scholar]
- Ren, Q.; Kwan, A.H.; Sunde, M. Two forms and two faces, multiple states and multiple uses: Properties and applications of the self-assembling fungal hydrophobins. Pept. Sci. 2013, 100, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Soltani, J. Chapter 22—Secondary Metabolite Diversity of the Genus Aspergillus: Recent Advances. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 275–292. [Google Scholar]
- Roongsawang, N.; Lim, S.P.; Washio, K.; Takano, K.; Kanaya, S.; Morikawa, M. Phylogenetic analysis of condensation domains in the nonribosomal peptide synthetases. FEMS Microbiol. Lett. 2005, 252, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kowall, M.; Vater, J.; Kluge, B.; Stein, T.; Franke, P.; Ziessow, D. Separation and Characterization of Surfactin Isoforms Produced by B. subtilis OKB 105. J. Colloid Interface Sci. 1998, 204, 1–8. [Google Scholar] [CrossRef]
- de Bruijn, I.; de Kock, M.J.D.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Massetolide A Biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 2777–2789. [Google Scholar] [CrossRef] [PubMed]
- Kitten, T.; Kinscherf, T.G.; McEvoy, J.L.; Willis, D.K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol. Microbiol. 1998, 28, 917–929. [Google Scholar] [CrossRef]
- Reimmann, C.; Beyeler, M.; Latifi, A.; Winteler, H.; Foglino, M.; Lazdunski, A.; Haas, D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 1997, 24, 309–319. [Google Scholar] [CrossRef]
- de Bruijn, I.; de Kock, M.J.D.; Yang, M.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol. Microbiol. 2007, 63, 417–428. [Google Scholar] [CrossRef]
- Moran, S.; Rai, D.K.; Clark, B.R.; Murphy, C.D. Precursor-directed biosynthesis of fluorinated iturin A in Bacillus spp. Org. Biomol. Chem. 2009, 7, 644–646. [Google Scholar] [CrossRef]
- Yang, R.; Lei, S.; Xu, X.; Jin, H.; Sun, H.; Zhao, X.; Pang, B.; Shi, J. Key elements and regulation strategies of NRPSs for biosynthesis of lipopeptides by Bacillus. Appl. Microbiol. Biotechnol. 2020, 104, 8077–8087. [Google Scholar] [CrossRef]
- Zhang, Z.; Ding, Z.; Zhong, J.; Zhou, J.; Shu, D.; Luo, D.; Yang, J.; Tan, H. Improvement of iturin A production in B. subtilis ZK 0 by overexpression of the comA and sigA genes. Lett. Appl. Microbiol. 2017, 64, 452–458. [Google Scholar] [PubMed]
- Cosmina, P.; Rodriguez, F.; de Ferra, F.; Grandi, G.; Perego, M.; Venema, G.; van Sinderen, D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in B. subtilis. Mol. Microbiol. 1993, 8, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Liu, Y.; Li, S. Rational strain improvement for surfactin production: Enhancing the yield and generating novel structures. Microb. Cell Factories 2019, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Williams, P.; Cámara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Lindum, P.W.; Anthoni, U.; Christophersen, C.; Eberl, L.; Molin, S.; Givskov, M. N -Acyl-l-Homoserine Lactone Autoinducers Control Production of an Extracellular Lipopeptide Biosurfactant Required for Swarming Motility of Serratia liquefaciens MG1. J. Bacteriol. 1998, 180, 6384–6388. [Google Scholar] [CrossRef]
- Solomon, J.M.; Lazazzera, B.A.; Grossman, A.D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in B. subtilis. Genes Dev. 1996, 10, 2014–2024. [Google Scholar] [CrossRef]
- Liu, H.; Qu, X.; Gao, L.; Zhao, S.; Lu, Z.; Zhang, C.; Bie, X. Characterization of a B. subtilis surfactin synthetase knockout and antimicrobial activity analysis. J. Biotechnol. 2016, 237, 1–12. [Google Scholar] [CrossRef]
- Coutte, F.; Leclère, V.; Béchet, M.; Guez, J.S.; Lecouturier, D.; Chollet-Imbert, M.; Dhulster, P.; Jacques, P. Effect of pps disruption and constitutive expression of srfA on surfactin productivity, spreading and antagonistic properties of B. subtilis 168 derivatives. J. Appl. Microbiol. 2010, 109, 480–491. [Google Scholar] [CrossRef]
- Kim, Y.T.; Park, B.K.; Kim, S.E.; Lee, W.J.; Moon, J.S.; Cho, M.S.; Park, H.Y.; Hwang, I.; Kim, S.U. Organization and characterization of genetic regions in B. subtilis subsp. krictiensis ATCC55079 associated with the biosynthesis of iturin and surfactin compounds. PLoS ONE 2017, 12, e0188179. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 4360. [Google Scholar] [CrossRef] [PubMed]
- Poremba, K.; Gunkel, W.; Lang, S.; Wagner, F. Marine Biosurfactants, III. Toxicity Testing with Marine Microorganisms and Comparison with Synthetic Surfactants. Z. Naturforschung C 1991, 46, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, B.G.; Guerra, J.M.C.; Sarubbo, L.A. Biosurfactants: Production and application prospects in the food industry. Biotechnol. Prog. 2020, 36, e3030. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.R.; Lepo, J.E.; Lewis, M.A. Toxicity comparison of biosurfactants and synthetic surfactants used in oil spill remediation to two estuarine species. Mar. Pollut. Bull. 2003, 46, 1309–1316. [Google Scholar] [CrossRef]
- Thies, S.; Santiago-Schübel, B.; Kovacic, F.; Rosenau, F.; Hausmann, R.; Jaeger, K.E. Heterologous production of the lipopeptide biosurfactant serrawettin W1 in Escherichia coli. J. Biotechnol. 2014, 181, 27–30. [Google Scholar] [CrossRef]
- Syldatk, C.; Hausmann, R. Microbial biosurfactants. Eur. J. Lipid Sci. Technol. 2010, 112, 615–616. [Google Scholar] [CrossRef]
- Chan, X.Y.; Chang, C.Y.; Hong, K.W.; Tee, K.K.; Yin, W.F.; Chan, K.G. Insights of biosurfactant producing S. marcescens strain W2.3 isolated from diseased tilapia fish: A draft genome analysis. Gut Pathog. 2013, 5, 29. [Google Scholar] [CrossRef]
- Uzoigwe, C.; Ennis, C.; Rahman, P. Production of Biosurfactants Using Eco-friendly Microorganisms. In Environmental Sustainability; Springer: New Delhi, India, 2015; pp. 185–204. [Google Scholar]
- Velioğlu, Z.; Ozturk Urek, R. Biosurfactant production by Pleurotus ostreatus in submerged and solid-state fermentation systems. Turk. J. Biol. 2015, 39, 160–166. [Google Scholar] [CrossRef]
- Magro, A.E.A.; Silva, L.C.; Rasera, G.B.; de Castro, R.J.S. Solid-state fermentation as an efficient strategy for the biotransformation of lentils: Enhancing their antioxidant and antidiabetic potentials. Bioresour. Bioprocess. 2019, 6, 38. [Google Scholar] [CrossRef]
- Cerda, A.; Artola, A.; Barrena, R.; Font, X.; Gea, T.; Sánchez, A. Innovative Production of Bioproducts from Organic Waste through Solid-State Fermentation. Front. Sustain. Food Syst. 2019, 3, 63. [Google Scholar] [CrossRef]
- Das, K.; Mukherjee, A. Comparison of lipopeptide biosurfactants production by B. subtilis strains in submerged and solid state fermentation systems using a cheap carbon source: Some industrial applications of biosurfactants. Process. Biochem. 2007, 42, 1191–1199. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, G.; Luo, Y.; Ran, W.; Shen, Q. Production of lipopeptides by Bacillus amyloliquefaciens XZ-173 in solid state fermentation using soybean flour and rice straw as the substrate. Bioresour. Technol. 2012, 112, 254–260. [Google Scholar] [CrossRef]
- Zouari, R.; Ellouze-Chaabouni, S.; Ghribi-Aydi, D. Optimization of B. subtilis SPB1 Biosurfactant Production under Solid-state Fermentation Using By-Products of a Traditional Olive Mill Factory. Achieve Life Sci. 2014, 8, 162–169. [Google Scholar] [CrossRef]
- Jajor, P.; Piłakowska-Pietras, D.; Krasowska, A.; Łukaszewicz, M. Surfactin analogues produced by B. subtilis strains grown on rapeseed cake. J. Mol. Struct. 2016, 1126, 141–146. [Google Scholar] [CrossRef]
- Das, S.; Das, P. Effects of cultivation media components on biosurfactant and pigment production from Pseudomonas aeruginosa PAO1. Braz. J. Chem. Eng. 2015, 32, 317–324. [Google Scholar] [CrossRef][Green Version]
- Sharma, R.; Singh, J.; Verma, N. Optimization of rhamnolipid production from Pseudomonas aeruginosa PBS towards application for microbial enhanced oil recovery. 3Biotech 2018, 8, 20. [Google Scholar] [CrossRef]
- Ghasemi, A.; Moosavi-Nasab, M.; Setoodeh, P.; Mesbahi, G.; Yousefi, G. Biosurfactant Production by Lactic Acid Bacterium Pediococcus dextrinicus SHU1593 Grown on Different Carbon Sources: Strain Screening Followed by Product Characterization. Sci. Rep. 2019, 9, 5287. [Google Scholar] [CrossRef]
- Andualem Bezza, F.; Chirwa, E.M. Improvement of Biosurfactant Production by Microbial Strains through Supplementation of Hydrophobic Substrates. Chem. Eng. Trans. 2020, 79, 91–96. [Google Scholar]
- Kavuthodi, B.; Thomas, S.K.; Sebastian, D. Co-production of Pectinase and Biosurfactant by the Newly Isolated Strain B. subtilis BKDS1. Microbiol. Res. J. Int. 2015, 10, 1–12. [Google Scholar] [CrossRef]
- Li, H.; Tanikawa, T.; Sato, Y.; Nakagawa, Y.; Matsuyama, T. S. marcescens Gene Required for Surfactant Serrawettin W1 Production Encodes Putative Aminolipid Synthetase Belonging to Nonribosomal Peptide Synthetase Family. Microbiol. Immunol. 2005, 49, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Feng, S.; Huang, Y.; Li, S.; Xu, H.; Zhang, X.; Bai, Y.; Qiao, M. Expression and characterization of a Grifola frondosa hydrophobin in Pichia pastoris. Protein Expr. Purif. 2010, 72, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Dexter, A.F.; Middelberg, A.P. Peptides as functional surfactants. Ind. Eng. Chem. Res. 2008, 47, 6391–6398. [Google Scholar] [CrossRef]
- Marchant, R.; Banat, I.M. Microbial biosurfactants: Challenges and opportunities for future exploitation. Trends Biotechnol. 2012, 30, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Belhaj, A.F.; Elraies, K.A.; Mahmood, S.M.; Zulkifli, N.N.; Akbari, S.; Hussien, O.S. The effect of surfactant concentration, salinity, temperature, and pH on surfactant adsorption for chemical enhanced oil recovery: A review. J. Pet. Explor. Prod. Technol. 2020, 10, 125–137. [Google Scholar] [CrossRef]
- De Almeida, D.G.; Soares Da Silva, R.d.C.F.; Luna, J.M.; Rufino, R.D.; Santos, V.A.; Banat, I.M.; Sarubbo, L.A. Biosurfactants: Promising molecules for petroleum biotechnology advances. Front. Microbiol. 2016, 7, 1718. [Google Scholar] [CrossRef]
- Bognolo, G. Biosurfactants as emulsifying agents for hydrocarbons. Colloids Surfaces Physicochem. Eng. Asp. 1999, 152, 41–52. [Google Scholar] [CrossRef]
- Satpute, S.K.; Banpurkar, A.G.; Dhakephalkar, P.K.; Banat, I.M.; Chopade, B.A. Methods for investigating biosurfactants and bioemulsifiers: A review. Crit. Rev. Biotechnol. 2010, 30, 127–144. [Google Scholar] [CrossRef]
- Bouchemal, K.; Agnely, F.; Koffi, A.; Djabourov, M.; Ponchel, G. What can isothermal titration microcalorimetry experiments tell us about the self-organization of surfactants into micelles? J. Mol. Recognit. 2010, 23, 335–342. [Google Scholar] [CrossRef]
- Wu, S.; Liang, F.; Hu, D.; Li, H.; Yang, W.; Zhu, Q. Determining the critical micelle concentration of surfactants by a simple and fast titration method. Anal. Chem. 2019, 92, 4259–4265. [Google Scholar] [CrossRef]
- Scholz, N.; Behnke, T.; Resch-Genger, U. Determination of the critical micelle concentration of neutral and ionic surfactants with fluorometry, conductometry, and surface tension—A method comparison. J. Fluoresc. 2018, 28, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Gerola, A.P.; Costa, P.F.; Nome, F.; Quina, F. Micellization and adsorption of zwitterionic surfactants at the air/water interface. Curr. Opin. Colloid Interface Sci. 2017, 32, 48–56. [Google Scholar] [CrossRef]
- Abe, M.; Kato, K.; Ogino, K. Effects of inorganic electrolytes and of pH on micelle formation of amphoteric-anionic mixed surfactant systems. J. Colloid Interface Sci. 1989, 127, 328–335. [Google Scholar] [CrossRef]
- Puvvada, S.; Blankschtein, D. Molecular-thermodynamic approach to predict micellization, phase behavior and phase separation of micellar solutions. I. Application to nonionic surfactants. J. Chem. Phys. 1990, 92, 3710–3724. [Google Scholar] [CrossRef]
- Nagarajan, R.; Ruckenstein, E. Theory of surfactant self-assembly: A predictive molecular thermodynamic approach. Langmuir 1991, 7, 2934–2969. [Google Scholar] [CrossRef]
- Khoshnood, A.; Lukanov, B.; Firoozabadi, A. Temperature effect on micelle formation: Molecular thermodynamic model revisited. Langmuir 2016, 32, 2175–2183. [Google Scholar] [CrossRef]
- Mozrzymas, A. On the hydrophobic chains effect on critical micelle concentration of cationic gemini surfactants using molecular connectivity indices. Monatshefte Chem.—Chem. Mon. 2020, 151, 525–531. [Google Scholar] [CrossRef]
- Michor, E.L.; Berg, J.C. Temperature effects on micelle formation and particle charging with span surfactants in apolar media. Langmuir 2015, 31, 9602–9607. [Google Scholar] [CrossRef]
- De, S.; Malik, S.; Ghosh, A.; Saha, R.; Saha, B. A review on natural surfactants. RSC Adv. 2015, 5, 65757–65767. [Google Scholar] [CrossRef]
- Hui, C.Y.; Jagota, A. Surface tension, surface energy, and chemical potential due to their difference. Langmuir 2013, 29, 11310–11316. [Google Scholar] [CrossRef]
- Willumsen, P.A.; Karlson, U. Screening of bacteria, isolated from PAH-contaminated soils, for production of biosurfactants and bioemulsifiers. Biodegradation 1996, 7, 415–423. [Google Scholar] [CrossRef]
- Glasser, L. Volume-based thermodynamics of organic liquids: Surface tension and the Eötvös equation. J. Chem. Thermodyn. 2021, 157, 106391. [Google Scholar] [CrossRef]
- Guggenheim, E.A. The principle of corresponding states. J. Chem. Phys. 1945, 13, 253–261. [Google Scholar] [CrossRef]
- Théatre, A.; Cano-Prieto, C.; Bartolini, M.; Laurin, Y.; Deleu, M.; Niehren, J.; Fida, T.; Gerbinet, S.; Alanjary, M.; Medema, M.H.; et al. The surfactin-like lipopeptides from Bacillus spp.: Natural biodiversity and synthetic biology for a broader application range. Front. Bioeng. Biotechnol. 2021, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Kakinuma, A.; Hori, M.; Isono, M.; Tamura, G.; Arima, K. Determination of amino acid sequence in surfactin, a crystalline peptidelipid surfactant produced by B. subtilis. Agric. Biol. Chem. 1969, 33, 971–972. [Google Scholar] [CrossRef]
- Kakinuma, A.; Sugino, H.; Isono, M.; Tamura, G.; Arima, K. Determination of fatty acid in surfactin and elucidation of the total structure of surfactin. Agric. Biol. Chem. 1969, 33, 973–976. [Google Scholar] [CrossRef]
- Wakil, S.J.; Stoops, J.K.; Joshi, V.C. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 1983, 52, 537–579. [Google Scholar] [CrossRef]
- Youssef, N.H.; Wofford, N.; McInerney, M.J. Importance of the long-chain fatty acid beta-hydroxylating cytochrome P450 enzyme YbdT for lipopeptide biosynthesis in B. subtilis strain OKB105. Int. J. Mol. Sci. 2011, 12, 1767–1786. [Google Scholar] [CrossRef]
- Steller, S.; Sokoll, A.; Wilde, C.; Bernhard, F.; Franke, P.; Vater, J. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry 2004, 43, 11331–11343. [Google Scholar] [CrossRef]
- Naruse, N.; Tenmyo, O.; Kobaru, S.; Kamei, H.; Miyaki, T.; Konishi, M.; Oki, T. Pumilacidin, a complex of new antiviral antibiotics production, isolation, chemical properties, structure and biological activity. J. Antibiot. 1990, 43, 267–280. [Google Scholar] [CrossRef]
- Horowitz, S.; Gilbert, J.; Griffin, W.M. Isolation and characterization of a surfactant produced by Bacillus licheniformis 86. J. Ind. Microbiol. 1990, 6, 243–248. [Google Scholar] [CrossRef]
- Janek, T.; Gudiña, E.J.; Połomska, X.; Biniarz, P.; Jama, D.; Rodrigues, L.R.; Rymowicz, W.; Lazar, Z. Sustainable surfactin production by B. subtilis using crude glycerol from different wastes. Molecules 2021, 26, 3488. [Google Scholar] [CrossRef]
- Gao, L.; Han, J.; Liu, H.; Qu, X.; Lu, Z.; Bie, X. Plipastatin and surfactin coproduction by B. subtilis pB2-L and their effects on microorganisms. Antonie Van Leeuwenhoek 2017, 110, 1007–1018. [Google Scholar] [CrossRef]
- Liu, X.Y.; Yang, S.Z.; Mu, B.Z. Production and characterization of a C15-surfactin-O-methyl ester by a lipopeptide producing strain B. subtilis HSO121. Process. Biochem. 2009, 44, 1144–1151. [Google Scholar] [CrossRef]
- Zhuravleva, O.I.; Afiyatullov, S.S.; Ermakova, S.P.; Nedashkovskaya, O.I.; Dmitrenok, P.S.; Denisenko, V.A.; Kuznetsova, T.A. New C14-surfactin methyl ester from the marine bacterium Bacillus pumilus KMM 456. Russ. Chem. Bull. 2010, 59, 2137–2142. [Google Scholar] [CrossRef]
- Shao, C.; Liu, L.; Gang, H.; Yang, S.; Mu, B. Structural diversity of the microbial surfactin derivatives from selective esterification approach. Int. J. Mol. Sci. 2015, 16, 1855–1872. [Google Scholar] [CrossRef]
- Eeman, M.; Berquand, A.; Dufrêne, Y.; Paquot, M.; Dufour, S.; Deleu, M. Penetration of surfactin into phospholipid monolayers: Nanoscale interfacial organization. Langmuir 2006, 22, 11337–11345. [Google Scholar] [CrossRef]
- Pagadoy, M.; Peypoux, F.; Wallach, J. Solid-phase synthesis of surfactin, a powerful biosurfactant produced by B. subtilis, and of four analogues. Int. J. Pept. Res. Ther. 2005, 11, 195–202. [Google Scholar] [CrossRef]
- Liu, Q.; Lin, J.; Wang, W.; Huang, H.; Li, S. Production of surfactin isoforms by B. subtilis BS-37 and its applicability to enhanced oil recovery under laboratory conditions. Biochem. Eng. J. 2015, 93, 31–37. [Google Scholar] [CrossRef]
- Dufour, S.; Deleu, M.; Nott, K.; Wathelet, B.; Thonart, P.; Paquot, M. Hemolytic activity of new linear surfactin analogs in relation to their physico-chemical properties. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2005, 1726, 87–95. [Google Scholar] [CrossRef]
- Taira, T.; Yanagisawa, S.; Nagano, T.; Tsuji, T.; Endo, A.; Imura, T. pH-induced conformational change of natural cyclic lipopeptide surfactin and the effect on protease activity. Colloids Surfaces B Biointerfaces 2017, 156, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Zou, A.; Liu, J.; Garamus, V.M.; Yang, Y.; Willumeit, R.; Mu, B. Micellization activity of the natural lipopeptide [Glu1, Asp5] surfactin-C15 in aqueous solution. J. Phys. Chem. B 2010, 114, 2712–2718. [Google Scholar] [CrossRef] [PubMed]
- Grangemard, I.; Wallach, J.; Maget-Dana, R.; Peypoux, F. Lichenysin. Appl. Biochem. Biotechnol. 2001, 90, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Dokouhaki, M.; Hung, A.; Kasapis, S.; Gras, S.L. Hydrophobins and chaplins: Novel bio-surfactants for food dispersions a review. Trends Food Sci. Technol. 2021, 111, 378–387. [Google Scholar] [CrossRef]
- Khalesi, M.; Gebruers, K.; Derdelinckx, G. Recent advances in fungal hydrophobin towards using in industry. Protein J. 2015, 34, 243–255. [Google Scholar] [CrossRef]
- Euston, S.R. Molecular simulation of adsorption of hydrophobin HFBI to the air–water, DPPC–water and decane–water interfaces. Food Hydrocoll. 2014, 42, 66–74. [Google Scholar] [CrossRef]
- Raffaini, G.; Milani, R.; Ganazzoli, F.; Resnati, G.; Metrangolo, P. Atomistic simulation of hydrophobin HFBII conformation in aqueous and fluorous media and at the water/vacuum interface. J. Mol. Graph. Model. 2016, 63, 8–14. [Google Scholar] [CrossRef]
- Jackson, S.A.; Borchert, E.; O’Gara, F.; Dobson, A.D. Metagenomics for the discovery of novel biosurfactants of environmental interest from marine ecosystems. Curr. Opin. Biotechnol. 2015, 33, 176–182. [Google Scholar]
- Ghosh, A.; Mehta, A.; Khan, A.M. Metagenomic Analysis and its Applications. In Encyclopedia of Bioinformatics and Computational Biology; Ranganathan, S., Gribskov, M., Nakai, K., Schönbach, C., Eds.; Academic Press: Oxford, UK, 2019; pp. 184–193. [Google Scholar]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular Biological Access to the Chemistry of Unknown Soil Microbes: A New Frontier for Natural Products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- Oulas, A.; Pavloudi, C.; Polymenakou, P.; Pavlopoulos, G.A.; Papanikolaou, N.; Kotoulas, G.; Arvanitidis, C.; Iliopoulos, L. Metagenomics: Tools and Insights for Analyzing Next-Generation Sequencing Data Derived from Biodiversity Studies. Bioinform. Biol. Insights 2015, 9, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Gerc, A.J.; Stanley-Wall, N.R.; Coulthurst, S.J. Role of the phosphopantetheinyltransferase enzyme, PswP, in the biosynthesis of antimicrobial secondary metabolites by S. marcescens Db10. Microbiology 2014, 160, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Margassery, L.M.; Morrissey, J.P.; O’Gara, F.; Dobson, A.D. 5—Metagenomic strategies for the discovery of novel enzymes with biotechnological application from marine ecosystems. In Marine Enzymes for Biocatalysis; Trincone, A., Ed.; Woodhead Publishing Series in Biomedicine; Woodhead Publishing: Oxford, UK, 2013; pp. 109–130. [Google Scholar]
- Kennedy, J.; O’Leary, N.; Kiran, G.; Morrissey, J.; O’Gara, F.; Selvin, J.; Dobson, A. Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J. Appl. Microbiol. 2011, 111, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Ngara, T.R.; Zhang, H. Recent Advances in Function-based Metagenomic Screening. Genom. Proteom. Bioinform. 2018, 16, 405–415. [Google Scholar] [CrossRef]
- Bashir, Y.; Singh, S.P.; Konwar, B.K. Metagenomics: An Application Based Perspective. Chin. J. Biol. 2014, 2014, 146030. [Google Scholar] [CrossRef]
- Joshi, M.N.; Dhebar, S.V.; Bhargava, P.; Pandit, A.S.; P, P.R.; Saxena, A.K.; Bagatharia, S.B. Metagenomic Approach for Understanding Microbial Population from Petroleum Muck. Genome Announc. 2014, 2, e00533-14. [Google Scholar] [CrossRef]
- Kachienga, L.; Jitendra, K.; Momba, M. Metagenomic profiling for assessing microbial diversity and microbial adaptation to degradation of hydrocarbons in two South African petroleum-contaminated water aquifers. Sci. Rep. 2018, 8, 7564. [Google Scholar] [CrossRef]
- Datta, S.; Rajnish, K.N.; Samuel, M.S.; Pugazlendhi, A.; Selvarajan, E. Metagenomic applications in microbial diversity, bioremediation, pollution monitoring, enzyme and drug discovery. A review. Environ. Chem. Lett. 2020, 18, 1229–1241. [Google Scholar] [CrossRef]
- Cai, Q.; Zhang, B.; Chen, B.; Zhu, Z.; Lin, W.; Cao, T. Screening of biosurfactant producers from petroleum hydrocarbon contaminated sources in cold marine environments. Mar. Pollut. Bull. 2014, 86, 402–410. [Google Scholar] [CrossRef]
- Balan, S.S.; Kumar, C.G.; Jayalakshmi, S. Aneurinifactin, a new lipopeptide biosurfactant produced by a marine Aneurinibacillus aneurinilyticus SBP-11 isolated from Gulf of Mannar: Purification, characterization and its biological evaluation. Microbiol. Res. 2017, 194, 1–9. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, M.; Zhang, M.; Wang, C.; Liu, Y.; Fan, X.; Li, H. Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme with High Tolerance for Organic Solvents and Salt and Potent Dye Decolorization Ability, Derived from a Marine Metagenomic Library. Front. Microbiol. 2018, 9, 2998. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, Y.M.; Ghazy, M.A.; Sayed, A.; Ouf, A.; El-Dorry, H.; Siam, R. Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea brine pool. Sci. Rep. 2013, 3, 3358. [Google Scholar] [CrossRef] [PubMed]
- Araújo, S.C.S.; Silva-Portela, R.; Lima, D.; Fonsêca, M.; Araújo, W.; Silva, U.; Napp, A.; Pereira, E.; Vainstein, M.; Agnez-Lima, L.F. MBSP1: A biosurfactant protein derived from a metagenomic library with activity in oil degradation. Sci. Rep. 2020, 10, 1340. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.; Jackson, B.; Clothier, B.; Dominati, E.; Marchant, S.; Cooper, D.; Bristow, K. Advances in Soil Ecosystem Services: Concepts, Models, and Applications for Earth System Life Support. Vadose Zone J. 2013, 12, 1–13. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Beaumier, D.; Greer, C.W. Metagenomic Analysis of the Bioremediation of Diesel-Contaminated Canadian High Arctic Soils. PLoS ONE 2012, 7, e30058. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 2019, 7, 154. [Google Scholar] [CrossRef]
- Heyer, R.; Schallert, K.; Zoun, R.; Becher, B.; Saake, G.; Benndorf, D. Challenges and perspectives of metaproteomic data analysis. J. Biotechnol. 2017, 261, 24–36. [Google Scholar] [CrossRef]
- Gaur, V.K.; Sharma, P.; Gupta, S.; Varjani, S.; Srivastava, J.; Wong, J.W.; Ngo, H.H. Opportunities and challenges in omics approaches for biosurfactant production and feasibility of site remediation: Strategies and advancements. Environ. Technol. Innov. 2022, 25, 102132. [Google Scholar] [CrossRef]
- Pitocchi, R.; Cicatiello, P.; Birolo, L.; Piscitelli, A.; Bovio, E.; Varese, G.C.; Giardina, P. Cerato-platanins from marine fungi as effective protein biosurfactants and bioemulsifiers. Int. J. Mol. Sci. 2020, 21, 2913. [Google Scholar] [CrossRef]
- Floros, D.J.; Jensen, P.R.; Dorrestein, P.C.; Koyama, N. A metabolomics guided exploration of marine natural product chemical space. Metabolomics 2016, 12, 145. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Araujo, W.; Lopes Sales, A.I.; Brito Guerra, A.d.; Silva Araújo, S.C.d.; de Vasconcelos, A.T.R.; Agnez-Lima, L.F.; Freitas, A.T. BioSurfDB: Knowledge and algorithms to support biosurfactants and biodegradation studies. Database 2015, 2015, bav033. [Google Scholar] [CrossRef]
- de Faria Poloni, J.; Bonatto, D. Systems Chemo-Biology and Transcriptomic Meta-Analysis Reveal the Molecular Roles of Bioactive Lipids in Cardiomyocyte Differentiation. J. Cell. Biochem. 2015, 116, 2018–2031. [Google Scholar] [CrossRef]
- Feltes, B.C.; Poloni, J.d.F.; Notari, D.L.; Bonatto, D. Toxicological effects of the different substances in tobacco smoke on human embryonic development by a systems chemo-biology approach. PLoS ONE 2013, 8, e61743. [Google Scholar] [CrossRef]
- Feltes, B.C.; de Faria Poloni, J.; Nunes, I.J.G.; Bonatto, D. Fetal alcohol syndrome, chemo-biology and OMICS: Ethanol effects on vitamin metabolism during neurodevelopment as measured by systems biology analysis. Omics J. Integr. Biol. 2014, 18, 344–363. [Google Scholar] [CrossRef]
- Clements, T.; Rautenbach, M.; Ndlovu, T.; Khan, S.; Khan, W. A metabolomics and molecular networking approach to elucidate the structures of secondary metabolites produced by S. marcescens strains. Front. Chem. 2021, 9, 72. [Google Scholar] [CrossRef]
- Mitra, K.; Carvunis, A.R.; Ramesh, S.K.; Ideker, T. Integrative approaches for finding modular structure in biological networks. Nat. Rev. Genet. 2013, 14, 719–732. [Google Scholar] [CrossRef]
- Van Dam, S.; Vosa, U.; van der Graaf, A.; Franke, L.; de Magalhaes, J.P. Gene co-expression analysis for functional classification and gene–disease predictions. Briefings Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef]
- Dusad, V.; Thiel, D.; Barahona, M.; Keun, H.C.; Oyarzún, D.A. Opportunities at the interface of network science and metabolic modeling. Front. Bioeng. Biotechnol. 2021, 8, 591049. [Google Scholar] [CrossRef]
- Occhipinti, A.; Eyassu, F.; Rahman, T.J.; Rahman, P.K.; Angione, C. In silico engineering of Pseudomonas metabolism reveals new biomarkers for increased biosurfactant production. PeerJ 2018, 6, e6046. [Google Scholar] [CrossRef]
- Matchado, M.S.; Lauber, M.; Reitmeier, S.; Kacprowski, T.; Baumbach, J.; Haller, D.; List, M. Network analysis methods for studying microbial communities: A mini review. Comput. Struct. Biotechnol. J. 2021, 19, 2687–2698. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, A.; Mathé, E.; Merling, M.; Ma, Q.; Liu, B. Network analyses in microbiome based on high-throughput multi-omics data. Briefings Bioinform. 2021, 22, 1639–1655. [Google Scholar] [CrossRef]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion 2019, 50, 71–91. [Google Scholar] [CrossRef]
- Hudson, I.L. Data Integration Using Advances in Machine Learning in Drug Discovery and Molecular Biology. In Artificial Neural Networks; Cartwright, H., Ed.; Springer Science+Business Media: Oxford, UK, 2021; pp. 167–184. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J.; Data, M. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann: Elsevier: Cambridge, MA, USA, 2016. [Google Scholar]
- Next-generation genomics: An integrative approach. Nat. Rev. Genet. 2010, 11, 476–486. [CrossRef]
- Gomez-Cabrero, D.; Abugessaisa, I.; Maier, D.; Teschendorff, A.; Merkenschlager, M.; Gisel, A.; Ballestar, E.; Bongcam-Rudloff, E.; Conesa, A.; Tegnér, J. Data integration in the era of omics: Current and future challenges. BMC Syst. Biol. 2014, 8, I1. [Google Scholar] [CrossRef]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Briefings Bioinform. 2015, 17, 891–901. [Google Scholar] [CrossRef]
- Tarazona, S.; Balzano-Nogueira, L.; Conesa, A. Chapter Eighteen—Multiomics Data Integration in Time Series Experiments. In Data Analysis for Omic Sciences: Methods and Applications; Comprehensive Analytical Chemistry; Jaumot, J., Bedia, C., Tauler, R., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 82, pp. 505–532. [Google Scholar]
- Padayachee, T.; Khamiakova, T.; Shkedy, Z.; Perola, M.; Salo, P.; Burzykowski, T. The Detection of Metabolite-Mediated Gene Module Co-Expression Using Multivariate Linear Models. PLoS ONE 2016, 11, e0150257. [Google Scholar] [CrossRef]
- Adetunji, A.I.; Olaniran, A.O. Production and potential biotechnological applications of microbial surfactants: An overview. Saudi J. Biol. Sci. 2021, 28, 669–679. [Google Scholar] [CrossRef]
- Ekpenyong, M.; Asitok, A.; Antai, S.; Ekpo, B.; Antigha, R.; Ogarekpe, N.; Antai, A.; Ogbuagu, U.; Ayara, N. Kinetic modeling and quasi-economic analysis of fermentative glycolipopeptide biosurfactant production in a medium co-optimized by statistical and neural network approaches. Prep. Biochem. Biotechnol. 2021, 51, 450–466. [Google Scholar] [CrossRef]
- Tayyebi, S.; Bagheri Lotfabad, T.; Roostaazad, R. Applying neural network to dynamic modeling of biosurfactant production using soybean oil refinery wastes. Iran. J. Energy Environ. 2013, 4, 161–170. [Google Scholar] [CrossRef]
- Jovic, S.; Guresic, D.; Babincev, L.; Draskovic, N.; Dekic, V. Comparative efficacy of machine-learning models in prediction of reducing uncertainties in biosurfactant production. Bioprocess Biosyst. Eng. 2019, 42, 1695–1699. [Google Scholar] [CrossRef]
- Kaur, J.; Kaur, S.; Kumar, M.; Krishnan, P.; Priya Minhas, A. Studies on production, optimization and machine learning-based prediction of biosurfactant from Debaryomyces hansenii CBS767. Int. J. Environ. Sci. Technol. 2022, 19, 8465–8478. [Google Scholar] [CrossRef]
- Chang, H.X.; Haudenshield, J.S.; Bowen, C.R.; Hartman, G.L. Metagenome-wide association study and machine learning prediction of bulk soil microbiome and crop productivity. Front. Microbiol. 2017, 8, 519. [Google Scholar] [CrossRef]
- Thompson, J.; Johansen, R.; Dunbar, J.; Munsky, B. Machine learning to predict microbial community functions: An analysis of dissolved organic carbon from litter decomposition. PLoS ONE 2019, 14, e0215502. [Google Scholar] [CrossRef]
- Ahmad, Z.; Zhong, H.; Mosavi, A.; Sadiq, M.; Saleem, H.; Khalid, A.; Mahmood, S.; Nabipour, N. Machine Learning Modeling of Aerobic Biodegradation for Azo Dyes and Hexavalent Chromium. Mathematics 2020, 8, 913. [Google Scholar] [CrossRef]
- Nicolas, J. Molecular dynamics simulation of surfactin molecules at the water-hexane interface. Biophys. J. 2003, 85, 1377–1391. [Google Scholar] [CrossRef]
- Gang, H.Z.; Liu, J.F.; Mu, B.Z. Molecular dynamics study of surfactin monolayer at the air/water interface. J. Phys. Chem. B 2011, 115, 12770–12777. [Google Scholar] [CrossRef]
- Asadi, A.; Abdolmaleki, A.; Azizi-Shalbaf, S.; Gurushankar, K. Molecular Dynamics Study of Surfactin Interaction with Lipid Bilayer Membranes. Gene Cell Tissue 2021, 8, e112646. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Eastman, P.; Swails, J.; Chodera, J.D.; McGibbon, R.T.; Zhao, Y.; Beauchamp, K.A.; Wang, L.P.; Simmonett, A.C.; Harrigan, M.P.; Stern, C.D.; et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017, 13, e1005659. [Google Scholar] [CrossRef]
- Lemkul, J. From proteins to perturbed hamiltonians: A suite of tutorials for the gromacs-2018 molecular simulation package [article v1.0]. Living J. Comput. Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
- Creton, B.; Nieto-Draghi, C.; Pannacci, N. Prediction of surfactants’ properties using multiscale molecular modeling tools: A review. Oil Gas Sci. Technol. Rev. D’Ifp Energies Nouv. 2012, 67, 969–982. [Google Scholar] [CrossRef]
- Lbadaoui-Darvas, M.; Idrissi, A.; Jedlovszky, P. Computer Simulation of the Surface of Aqueous Ionic and Surfactant Solutions. J. Phys. Chem. B 2021, 126, 751–765. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, S.; Wang, X.; Yuan, S.; Yuan, S. Molecular dynamics study on emulsified oil droplets with nonionic surfactants. J. Mol. Liq. 2022, 346, 117102. [Google Scholar] [CrossRef]
- Peng, M.; Duignan, T.T.; Nguyen, C.V.; Nguyen, A.V. From Surface Tension to Molecular Distribution: Modeling Surfactant Adsorption at the Air–Water Interface. Langmuir 2021, 37, 2237–2255. [Google Scholar] [CrossRef]
- Wang, C.; Jiang, Z.; Xu, L.; Kan, A.; Fu, H.; Lin, X. Surface structure of aqueous ionic surfactant solutions and effects of solvent therein—A computer simulation study. Colloid Polym. Sci. 2016, 294, 575–581. [Google Scholar] [CrossRef]
- Hu, S.Q.; Ji, X.J.; Fan, Z.Y.; Zhang, T.T.; Sun, S.Q. Counterion effects on the properties of sulfate surfactants at the air/liquid interface by molecular dynamics simulation. Acta-Phys.-Chim. Sin. 2015, 31, 83–89. [Google Scholar]
- Kirby, B.J.; Jungwirth, P. Charge scaling manifesto: A way of reconciling the inherently macroscopic and microscopic natures of molecular simulations. J. Phys. Chem. Lett. 2019, 10, 7531–7536. [Google Scholar] [CrossRef]
- Abbasi, S.; Scanlon, M.G. Microemulsion: A novel alternative technique for edible oil extraction_a mechanistic viewpoint. Crit. Rev. Food Sci. Nutr. 2022. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Sabatini, D.A. Characterization and emulsification properties of rhamnolipid and sophorolipid biosurfactants and their applications. Int. J. Mol. Sci. 2011, 12, 1232–1244. [Google Scholar] [CrossRef]
- Zheng, Y.; Zheng, M.; Ma, Z.; Xin, B.; Guo, R.; Xu, X. 8-Sugar Fatty Acid Esters. In Polar Lipids; Ahmad, M.U., Xu, X., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 215–243. [Google Scholar]
- Edwards-Gayle, C.J.; Hamley, I.W. Self-assembly of bioactive peptides, peptide conjugates, and peptide mimetic materials. Org. Biomol. Chem. 2017, 15, 5867–5876. [Google Scholar] [CrossRef]
- Cob-Calan, N.N.; Chi-Uluac, L.A.; Ortiz-Chi, F.; Cerqueda-García, D.; Navarrete-Vázquez, G.; Ruiz-Sánchez, E.; Hernández-Núñez, E. Molecular docking and dynamics simulation of protein β-tubulin and antifungal cyclic lipopeptides. Molecules 2019, 24, 3387. [Google Scholar] [CrossRef]
- Schaller, A.; Connors, N.K.; Dwyer, M.D.; Oelmeier, S.A.; Hubbuch, J.; Middelberg, A.P. Computational study of elements of stability of a four-helix bundle protein biosurfactant. J. -Comput.-Aided Mol. Des. 2015, 29, 47–58. [Google Scholar] [CrossRef]
- Cheung, D.L. Molecular simulation of hydrophobin adsorption at an oil–water interface. Langmuir 2012, 28, 8730–8736. [Google Scholar] [CrossRef]
- Segura, S.M.A.; Macías, A.P.; Pinto, D.C.; Vargas, W.L.; Vives-Florez, M.J.; Barrera, H.E.C.; Álvarez, O.A.; Barrios, A.F.G. Escherichia coli’s OmpA as biosurfactant for cosmetic industry: Stability analysis and experimental validation based on molecular simulations. In Advances in Computational Biology; Springer: Berlin/Heidelberg, Germany, 2014; pp. 265–271. [Google Scholar]
- Aguilera-Segura, S.M.; Vélez, V.N.; Achenie, L.; Solano, O.Á.; Torres, R.; Barrios, A.F.G. Peptides design based on transmembrane Escherichia coli’s OmpA protein through molecular dynamics simulations in water–dodecane interfaces. J. Mol. Graph. Model. 2016, 68, 216–223. [Google Scholar] [CrossRef]
- Kodani, S.; Lodato, M.A.; Durrant, M.C.; Picart, F.; Willey, J.M. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol. Microbiol. 2005, 58, 1368–1380. [Google Scholar] [CrossRef]
- Fan, H.; Wang, X.; Zhu, J.; Robillard, G.T.; Mark, A.E. Molecular dynamics simulations of the hydrophobin SC3 at a hydrophobic/hydrophilic interface. Proteins Struct. Funct. Bioinform. 2006, 64, 863–873. [Google Scholar] [CrossRef]
- Lebecque, S.; Crowet, J.M.; Nasir, M.N.; Deleu, M.; Lins, L. Molecular dynamics study of micelles properties according to their size. J. Mol. Graph. Model. 2017, 72, 6–15. [Google Scholar] [CrossRef]
- Gang, H.; He, H.; Yu, Z.; Wang, Z.; Liu, J.; He, X.; Bao, X.; Li, Y.; Mu, B.Z. A coarse-grained model for microbial lipopeptide surfactin and its application in self-assembly. J. Phys. Chem. B 2020, 124, 1839–1846. [Google Scholar] [CrossRef]
- Gang, H.Z.; Bian, P.C.; He, X.; He, X.; Bao, X.; Mu, B.Z.; Li, Y.; Yang, S.Z. Mixing of surfactin, an anionic biosurfactant, with alkylbenzene sulfonate, a chemically synthesized anionic surfactant, at the n-decane/water interface. J. Surfactants Deterg. 2021, 24, 445–457. [Google Scholar] [CrossRef]
- Yu, H.; Yang, S.; Chen, Z.; Xu, Z.; Quan, X.; Zhou, J. Orientation and Conformation of Hydrophobin at the Oil–Water Interface: Insights from Molecular Dynamics Simulations. Langmuir 2022, 38, 6191–6200. [Google Scholar] [CrossRef]
- Ashokkumar, M.; Irudayaraj, G.; Yellapu, N.; Manonmani, A.M. Molecular characterization of bmyC gene of the mosquito pupicidal bacteria, Bacillus amyloliquefaciens (VCRC B483) and in silico analysis of bacillomycin D synthetase C protein. World J. Microbiol. Biotechnol. 2018, 34, 1–11. [Google Scholar] [CrossRef]
- Coronel, J.R.; Marqués, A.; Manresa, A.; Aranda, F.J.; Teruel, J.A.; Ortiz, A. Interaction of the lipopeptide biosurfactant lichenysin with phosphatidylcholine model membranes. Langmuir 2017, 33, 9997–10005. [Google Scholar] [CrossRef]
- De Roo, V.; Geudens, N.; Verleysen, Y.; Lee, T.H.; Madder, A.; Aguilar, M.; Martins, J.C. Towards a molecular understanding of the membrane altering properties of viscosin, a cyclic lipodepsipeptide. Biophys. J. 2022, 121, 162a. [Google Scholar] [CrossRef]
- Geudens, N.; Kovács, B.; Sinnaeve, D.; Oni, F.E.; Höfte, M.; Martins, J.C. Conformation and dynamics of the cyclic Lipopeptide Viscosinamide at the water-lipid interface. Molecules 2019, 24, 2257. [Google Scholar] [CrossRef]
- Cheung, D.L.; Samantray, S. Molecular dynamics simulation of protein biosurfactants. Colloids Interfaces 2018, 2, 39. [Google Scholar] [CrossRef]
- Euston, S.R. Molecular simulation of biosurfactants with relevance to food systems. Curr. Opin. Colloid Interface Sci. 2017, 28, 110–119. [Google Scholar] [CrossRef]
- Baccile, N.; Seyrig, C.; Poirier, A.; Alonso-de Castro, S.; Roelants, S.L.; Abel, S. Self-assembly, interfacial properties, interactions with macromolecules and molecular modelling and simulation of microbial bio-based amphiphiles (biosurfactants). A tutorial review. Green Chem. 2021, 23, 3842–3944. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).