Identification of Intron Retention in the Slc16a3 Gene Transcript Encoding the Transporter MCT4 in the Brain of Aged and Alzheimer-Disease Model (APPswePS1dE9) Mice
Highlights
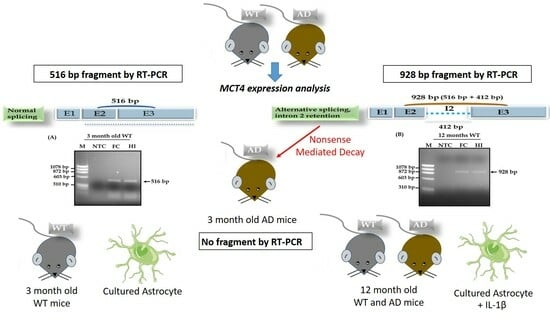
- We demonstrate the existence of an intron retention mechanism for the Slc16a3 gene transcript, which encodes the transporter MCT4 in brain cells.
- In addition, it is noteworthy that both in vitro and in vivo experiments showed the same splicing defect in MCT4 gene transcript expression.
- The occurrence of intron retention for the MCT4 transcript in astrocytes triggered by inflammatory signals may be a sign of cellular stress and contribute to its detection in normal and pathological aging.
- The early detection of intron 2 retention for the MCT4 transcript (and/or the lack of MCT4 transcript detection) could become useful as an early biomarker of Alzheimer’s disease (AD).
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Cultures
2.3. Lipopolysaccharide and Interleukin-1β Treatments
2.4. RNA Isolation
2.5. RT-PCR Amplification
2.6. DNA Sequencing of RT-PCR Products
2.7. Statistical Analysis
3. Results
3.1. Detection of Intron Retention in Aged Brain MCT4 mRNA
3.2. A Proposed Mechanism of Alternative RNA Splicing for the MCT4 Transporter in Aged Mice
3.3. Detection of Intron Retention in an Alzheimer Mouse Model
3.4. Detection of Non Intron Retention in 6-Month-Old Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Halestrap, A.P.; Meredith, D. The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pflugers Arch. 2004, 447, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Dimmer, K.S.; Friedrich, B.; Lang, F.; Deitmer, J.W.; Bröer, S. The low-affinity monocarboxylate transporter MCT4 is adapted to the export of lactate in highly glycolytic cells. Biochem. J. 2000, 350, 219–227. [Google Scholar] [CrossRef]
- Juel, C. Lactate-proton cotransport in skeletal muscle. Physiol. Rev. 1997, 77, 321–358. [Google Scholar] [CrossRef] [PubMed]
- Rafiki, A.; Boulland, J.L.; Halestrap, A.P.; Ottersen, O.P.; Bergersen, L. Highly differential expression of the monocarboxylate transporters MCT2 and MCT4 in the developing rat brain. Neuroscience 2003, 122, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Balmaceda-Aguilera, C.; Cortés-Campos, C.; Cifuentes, M.; Peruzzo, B.; Mack, L.; Tapia, J.C.; Oyarce, K.; Garcia, M.A.; Nualart, F. Glucose transporter 1 and monocarboxylate transporters 1, 2, and 4 localization within the glial cells of shark blood-brain-barriers. PLoS ONE 2012, 7, e32409. [Google Scholar] [CrossRef]
- Kong, L.; Wang, Z.; Liang, X.; Wang, Y.; Gao, L.; Ma, C. Monocarboxylate transporter 1 promotes classical microglial activation and pro-inflammatory effect via 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3. J. Neuroinflamm. 2019, 216, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Rosafio, K.; Pellerin, L. Oxygen tension controls the expression of the monocarboxylate transporter MCT4 in cultured mouse cortical astrocytes via a hypoxia-inducible factor-1α-mediated transcriptional regulation. Glia 2014, 62, 477–490. [Google Scholar] [CrossRef]
- Sun, X.; Wang, M.; Wang, M.; Yao, L.; Li, X.; Dong, H.; Li, M.; Sun, T.; Liu, X.; Xu, Y. Role of Proton-coupled monocarboxylate transporters in cancer: From metabolic crosstalk to therapeutic potential. Front. Cell Dev. Biol. 2020, 8, 651. [Google Scholar] [CrossRef]
- Rosafio, K.; Castillo, X.; Hirt, L.; Pellerin, L. Cell-specific modulation of monocarboxylate transporter expression contributes to the metabolic reprograming taking place following cerebral ischemia. Neuroscience 2016, 317, 108–120. [Google Scholar] [CrossRef]
- Goyal, M.S.; Vlassenko, A.G.; Blazey, T.M.; Su, Y.; Couture, L.E.; Durbin, T.J.; Bateman, R.J.; Benzinger, T.L.; Morris, J.C.; Raichle, M.E. Loss of brain aerobic glycolysis in normal human aging. Cell Metab. 2017, 26, 353–360. [Google Scholar] [CrossRef]
- Vlassenko, A.G.; Gordon, B.A.; Goyal, M.S.; Su, Y.; Blazey, T.M.; Durbin, T.J.; Couture, L.E.; Christensen, J.J.; Jafri, H.; Morris, J.C.; et al. Aerobic glycolysis and tau deposition in preclinical Alzheimer’s disease. Neurobiol. Aging 2018, 67, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.S.; Gordon, B.A.; Couture, L.E.; Flores, S.; Xiong, C.; Morris, J.C.; Raichle, M.E.; Benzinger, T.L.; Vlassenko, A.G. Spatiotemporal relationship betwwen subthreshold amyloid accumulation and aerobic glycolysis in the human brain. Neurobiol. Aging 2020, 96, 165–175. [Google Scholar] [CrossRef]
- Zhang, M.; Cheng, X.; Dang, R.; Zhang, W.; Zhang, J.; Yao, Z. Lactate deficit in an Alzheimer disease mouse model: The relationship with neuronal damage. J. Neuropathol. Exp. Neurol. 2018, 77, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.; Zhang, X.; Gao, S.; Wang, P. Role of monocarboxylate transporter 4 in Alzheimer disease. Neurotoxicology 2020, 76, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Vérité, J.; Janet, T.; Chassaing, D.; Fauconneau, B.; Rabeony, H.; Page, G. Longitudinal chemokine profile expression in a blood-brain barrier model from Alzheimer transgenic versus wild-type mice. J. Neuroinflamm. 2018, 15, 182. [Google Scholar] [CrossRef]
- François, A.; Terro, F.; Janet, T.; Rioux Bilan, A.; Paccalin, M.; Page, G. Involvement of interleukin-1β in the autophagic process of microglia: Relevance to Alzheimer’s disease. J. Neuroinflamm. 2013, 10, 151. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Wang, B.B.; Brendel, V. Genome wide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar] [CrossRef]
- Ghigna, C.; Valacca, C.; Biamonti, G. Alternative Splicing and Tumor Progression. Curr. Genom. 2008, 9, 556–570. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Sakabe, N.J.; de Souza, S.J. Sequence features responsible for intron retention in human. BMC Genom. 2007, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Vaquero-Garcia, J.; Barrera, A.; Gazzara, M.R.; Gonzales-Vallinas, J.; Lahens, N.F.; Hogenesch, J.B.; Lynch, K.W.; Barash, Y. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife 2016, 5, e11752. [Google Scholar] [CrossRef] [PubMed]
- Kervestin, S.; Jacobson, A. NMD: A multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 2012, 13, 700–712. [Google Scholar] [CrossRef]
- Shoemaker, C.J.; Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012, 19, 594–601. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Bennett, E.J. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell. Biol. 2014, 204, 467–476. [Google Scholar] [CrossRef]
- Green, R.E.; Lewis, B.P.; Hillman, R.T.; Blanchette, M.; Lareau, L.F.; Garnett, A.T.; Rio, D.C.; Brenner, S.E. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 2003, 19 (Suppl. S1), 118–121. [Google Scholar] [CrossRef]
- Ameur, A.; Zaghlool, A.; Halvardson, J.; Wetterbom, A.; Gyllensten, U.; Cavelier, L.; Feuk, L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat. Struct. Mol. Biol. 2011, 18, 1435–1440. [Google Scholar] [CrossRef]
- Monteuuis, G.; Schmitz, U.; Petrova, V.; Kearney, P.S.; Rasko, J.E. Holding on to Junk Bonds: Intron Retention in Cancer and Therapy. Cancer Res. 2021, 81, 779–789. [Google Scholar] [CrossRef]
- He, F.; Peltz, S.W.; Donahue, J.L.; Rosbash, M.; Jacobson, A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc. Natl. Acad. Sci. USA 1993, 90, 7034–7038. [Google Scholar] [CrossRef] [PubMed]
- Pulak, R.; Anderson, P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993, 7, 1885–1897. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Hopfler, M.; Ayatollahi, Z.; Barta, A.; Kalyna, M. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res. 2015, 25, 995–1007. [Google Scholar] [CrossRef] [PubMed]
- Adusumalli, S.; Ngian, Z.-K.; Lin, W.-Q.; Benoukraf, T.; Ong, C.-T. Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell 2019, 18, e12928. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, M.; Howell, P.; Dutta, S.; Heintz, C.; Mair, W.B. Alternative splicing in aging and longevity. Hum. Genet. 2020, 139, 357–369. [Google Scholar] [CrossRef]
- Ong, C.-T.; Adusumalli, A. Increased intron retention is linked to Alzheimer’s disease. Neural Regen. Res. 2020, 15, 259–260. [Google Scholar] [CrossRef]
- Ngian, Z.-K.; Tan, Y.-Y.; Choo, C.-T.; Lin, W.-Q.; Leow, C.-Y.; Mah, S.-J.; Lai, M.K.-P.; Chen, C.L.-H.; Ong, C.-T. Truncated Tau caused by intron retention is enriched in Alzheimer’s disease cortex and exhibits altered biochemical properties. Proc. Natl. Acad. Sci. USA 2022, 119, e2204179119. [Google Scholar] [CrossRef]
- Giannopoulou, A.; Konstantakou, E.G.; Veletzas, A.D.; Avgeris, S.N.; Avgeris, M.; Papandreou, N.C.; Zoi, I.; Filippa, V.; Katarachia, S.; Lampidonis, A.D.; et al. Gene-specific intron retention serves as molecular signature that distinguishes melanoma from non-melanoma cancer cells in Greek patients. Int. J. Mol. Sci. 2019, 20, 937. [Google Scholar] [CrossRef]
- Braunschweig, U.; Barbosa-Morais, N.L.; Pan, Q.; Nachman, E.N.; Alipanahi, B.; Gonatopoulos-Pournatzis, T.; Frey, B.; Irimia, M.; Blencowe, B.J. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014, 24, 1774–1786. [Google Scholar] [CrossRef]
- Jacob, A.G.; Smith, C.W.J. Intron retention as a component of regulated gene expression programs. Hum. Genet. 2017, 136, 1043–1057. [Google Scholar] [CrossRef]
- Yao, J.; Ding, D.; Li, X.; Shen, T.; Fu, H.; Zhong, H.; Wei, G.; Ni, T. Prevalent intron retention fine-tunes gene expression and contributes to cellular senescence. Aging Cell 2020, 19, e13276. [Google Scholar] [CrossRef] [PubMed]
- Mauger, O.; Lemoine, F.; Scheiffele, P. Targeted Intron Retention and Excision for RapidGene Regulation in Response to Neuronal Activity. Neuron 2016, 92, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.Y.; Xue, H.; Wu, R.; Fazli, L.; Lin, D.; Collins, C.C.; Gleave, M.E.; Gout, P.W.; Wang, Y. The MCT4 Gene: A Novel, Potential Target for Therapy of Advanced Prostate Cancer. Clin. Cancer Res. 2016, 22, 2721–2733. [Google Scholar] [CrossRef]
- Figley, C.R. Lactate transport and metabolism in the human brain: Implications for the astrocyte-neuron lactate shuttle hypothesis. J. Neurosci. 2011, 31, 4768–4770. [Google Scholar] [CrossRef] [PubMed]
- Neher, J.J.; Neniskyte, U.; Zhao, J.W.; Bal-Price, A.; Tolkovsky, A.M.; Brown, G.C. Inhibition of microglial phagocytosis is sufficient to prevent inflammatory neuronal death. J. Immunol. 2011, 186, 4973–4983. [Google Scholar] [CrossRef]
- Brown, G.C. The endotoxin hypothesis of neurodegeneration. J. Neuroinflamm. 2019, 16, 180. [Google Scholar] [CrossRef]
- Shen, Y.; Qin, H.; Chen, J.; Mou, L.; He, Y.; Yan, Y.; Zhou, H.; Lv, Y.; Chen, Z.; Wang, J.; et al. Postnatal activation of TLR4 in astrocytes promotes excitatory synaptogenesis in hippocampal neurons. J. Cell Biol. 2016, 215, 719–734. [Google Scholar] [CrossRef]
- Hua, L.L.; Zhao, M.L.; Cosenza, M.; Kim, M.O.; Huang, H.; Tanowitz, H.B.; Brosnan, C.F.; Lee, S.C. Role of mitogen-activated protein kinases in inducible nitric oxide synthase and TNF expression in human fetal astrocytes. J. Neuroimmunol. 2002, 126, 180–189. [Google Scholar] [CrossRef]
- Guo, X.; Harada, C.; Namekata, K.; Matsuzawa, A.; Camps, M.; Ji, H.; Swinnen, D.; Jorand-Lebrun, C.; Muzerelle, M.; Vitte, P.-A.; et al. Regulation of the severity of neuroinflammation and demyelination by TLR-ASK1-p38 pathway. EMBO Mol. Med. 2010, 2, 504–515. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
EL-Seedy, A.; Pellerin, L.; Page, G.; Ladeveze, V. Identification of Intron Retention in the Slc16a3 Gene Transcript Encoding the Transporter MCT4 in the Brain of Aged and Alzheimer-Disease Model (APPswePS1dE9) Mice. Genes 2023, 14, 1949. https://doi.org/10.3390/genes14101949
EL-Seedy A, Pellerin L, Page G, Ladeveze V. Identification of Intron Retention in the Slc16a3 Gene Transcript Encoding the Transporter MCT4 in the Brain of Aged and Alzheimer-Disease Model (APPswePS1dE9) Mice. Genes. 2023; 14(10):1949. https://doi.org/10.3390/genes14101949
Chicago/Turabian StyleEL-Seedy, Ayman, Luc Pellerin, Guylène Page, and Veronique Ladeveze. 2023. "Identification of Intron Retention in the Slc16a3 Gene Transcript Encoding the Transporter MCT4 in the Brain of Aged and Alzheimer-Disease Model (APPswePS1dE9) Mice" Genes 14, no. 10: 1949. https://doi.org/10.3390/genes14101949
APA StyleEL-Seedy, A., Pellerin, L., Page, G., & Ladeveze, V. (2023). Identification of Intron Retention in the Slc16a3 Gene Transcript Encoding the Transporter MCT4 in the Brain of Aged and Alzheimer-Disease Model (APPswePS1dE9) Mice. Genes, 14(10), 1949. https://doi.org/10.3390/genes14101949