Multi-Trait Exome-Wide Association Study of Back Pain-Related Phenotypes
Abstract
:1. Introduction
2. Materials and Methods
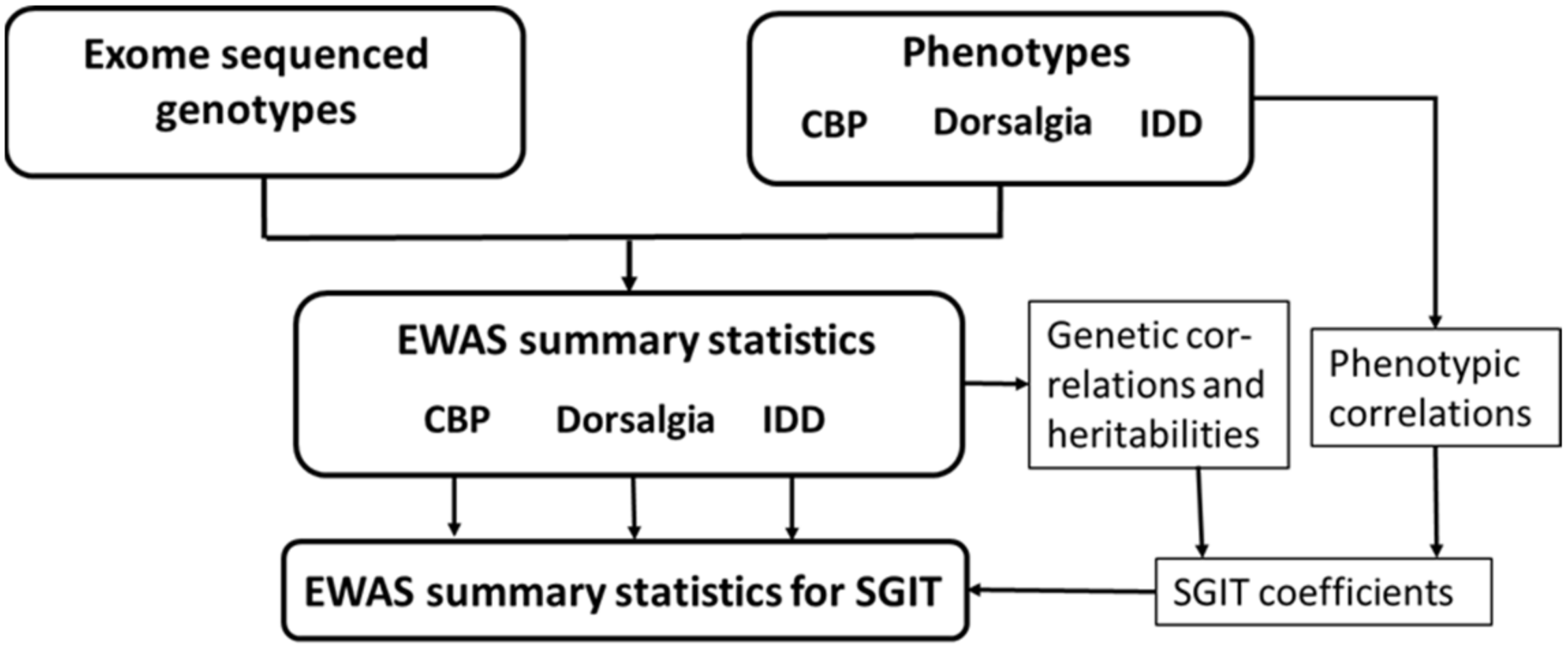
2.1. Overview of the Study Design
2.2. Data
2.3. EWAS of Original Traits
2.4. EWAS of Multi-Trait
2.5. Gene-Based Association Analyses
2.5.1. Matrices of Genotype Correlations
2.5.2. Variant Annotations
2.5.3. Collapsing
2.5.4. Methods of Gene-Based Analysis
2.6. Conditional Analysis
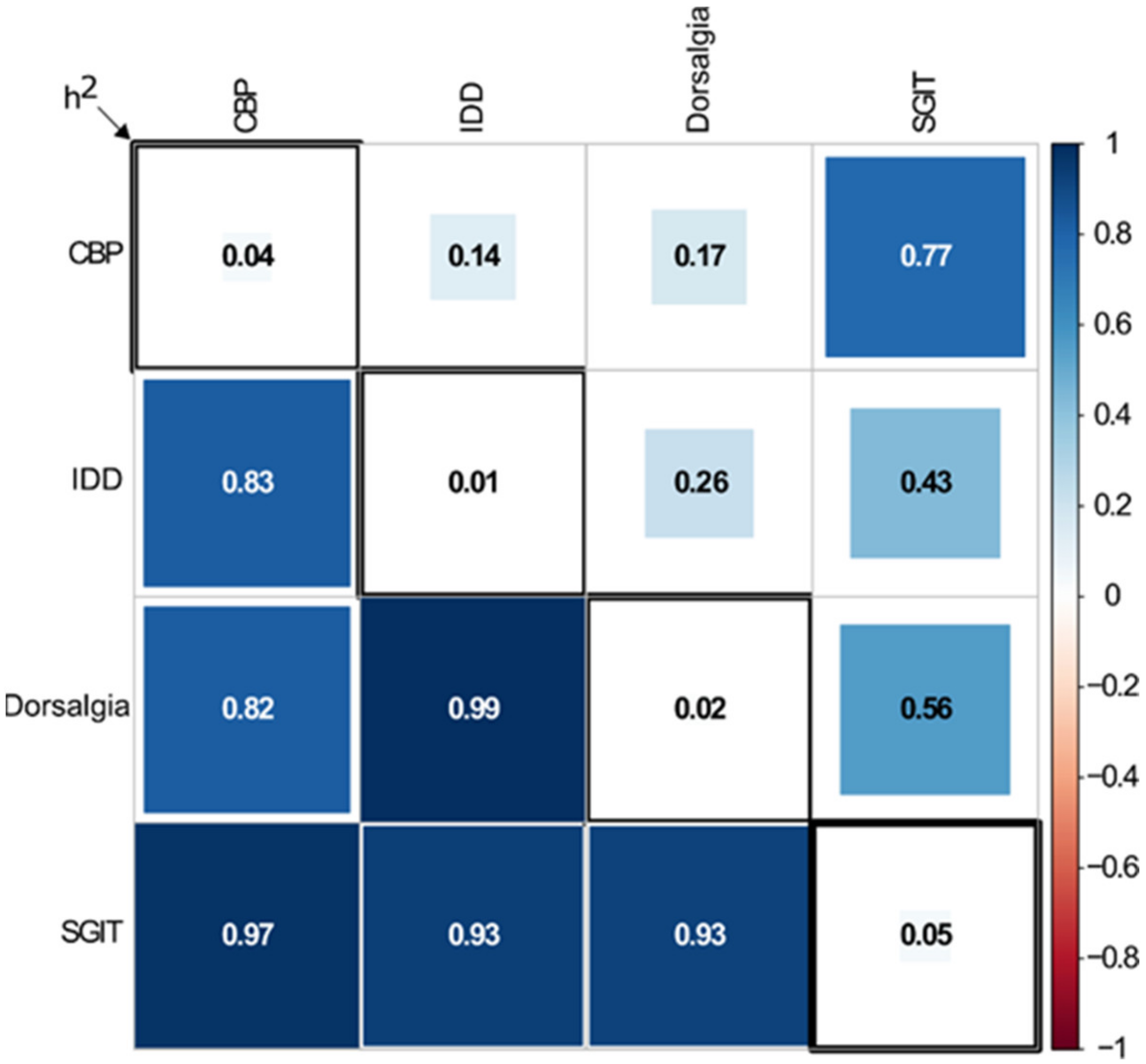
3. Results
3.1. Single Variants Association Analysis
3.2. Gene-Based Association Analysis
3.3. Conditional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoy, D.; March, L.; Brooks, P.; Blyth, F.; Woolf, A.; Bain, C.; Williams, G.; Smith, E.; Vos, T.; Barendregt, J.; et al. The global burden of low back pain: Estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis. 2014, 73, 968–974. [Google Scholar] [CrossRef]
- Manchikanti, L.; Singh, V.; Falco, F.J.; Benyamin, R.M.; Hirsch, J.A. Epidemiology of low back pain in adults. Neuromodulation 2014, 17 (Suppl. 2), 3–10. [Google Scholar] [CrossRef] [PubMed]
- Bortsov, A.V.; Parisien, M.; Khoury, S.; Martinsen, A.E.; Lie, M.U.; Heuch, I.; Hveem, K.; Zwart, J.A.; Winsvold, B.S.; Diatchenko, L. Brain-specific genes contribute to chronic but not to acute back pain. Pain. Rep. 2022, 7, e1018. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhou, H.; Luo, S.; Cai, X.; Ye, F.; He, Q.; Huang, C.; Zheng, X.; Li, Y.; Du, Z.; et al. Investigating the Causal Relationship Between Physical Activity and Chronic Back Pain: A Bidirectional Two-Sample Mendelian Randomization Study. Front. Genet. 2021, 12, 758639. [Google Scholar] [CrossRef] [PubMed]
- Johnston, K.J.A.; Adams, M.J.; Nicholl, B.I.; Ward, J.; Strawbridge, R.J.; Ferguson, A.; McIntosh, A.M.; Bailey, M.E.S.; Smith, D.J. Genome-wide association study of multisite chronic pain in UK Biobank. PLoS Genet. 2019, 15, e1008164. [Google Scholar] [CrossRef] [PubMed]
- Tsepilov, Y.A.; Freidin, M.B.; Shadrina, A.S.; Sharapov, S.Z.; Elgaeva, E.E.; Zundert, J.V.; Karssen Lcapital Es, C.; Suri, P.; Williams, F.M.K.; Aulchenko, Y.S. Analysis of genetically independent phenotypes identifies shared genetic factors associated with chronic musculoskeletal pain conditions. Commun. Biol. 2020, 3, 329. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, G.; Benonisdottir, S.; Sveinbjornsson, G.; Styrkarsdottir, U.; Thorleifsson, G.; Walters, G.B.; Bjornsson, A.; Olafsson, I.H.; Ulfarsson, E.; Vikingsson, A.; et al. Sequence variant at 8q24.21 associates with sciatica caused by lumbar disc herniation. Nat. Commun. 2017, 8, 14265. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, G.; Stefansdottir, L.; Thorleifsson, G.; Sulem, P.; Norland, K.; Ferkingstad, E.; Oddsson, A.; Zink, F.; Lund, S.H.; Nawaz, M.S.; et al. Rare SLC13A1 variants associate with intervertebral disc disorder highlighting role of sulfate in disc pathology. Nat. Commun. 2022, 13, 634. [Google Scholar] [CrossRef] [PubMed]
- Belonogova, N.M.; Kirichenko, A.V.; Freidin, M.B.; Williams, F.M.K.; Suri, P.; Aulchenko, Y.S.; Axenovich, T.I.; Tsepilov, Y.A. Noncoding rare variants in PANX3 are associated with chronic back pain. Pain 2023, 164, 864–869. [Google Scholar] [CrossRef]
- Ou-Yang, D.C.; Kleck, C.J.; Ackert-Bicknell, C.L. Genetics of Intervertebral Disc Degeneration. Curr. Osteoporos. Rep. 2023, 21, 56–64. [Google Scholar] [CrossRef]
- Suri, P.; Palmer, M.R.; Tsepilov, Y.A.; Freidin, M.B.; Boer, C.G.; Yau, M.S.; Evans, D.S.; Gelemanovic, A.; Bartz, T.M.; Nethander, M.; et al. Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. PLoS Genet. 2018, 14, e1007601. [Google Scholar] [CrossRef] [PubMed]
- Battie, M.C.; Videman, T.; Levalahti, E.; Gill, K.; Kaprio, J. Heritability of low back pain and the role of disc degeneration. Pain 2007, 131, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Hartvigsen, J.; Nielsen, J.; Kyvik, K.O.; Fejer, R.; Vach, W.; Iachine, I.; Leboeuf-Yde, C. Heritability of spinal pain and consequences of spinal pain: A comprehensive genetic epidemiologic analysis using a population-based sample of 15,328 twins ages 20–71 years. Arthritis Rheum. 2009, 61, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Mather, L.; Karkkainen, S.; Narusyte, J.; Ropponen, A.; Mittendorfer-Rutz, E.; Svedberg, P. Sick leave due to back pain, common mental disorders and disability pension: Common genetic liability. Eur. J. Pain. 2020, 24, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Nyman, T.; Mulder, M.; Iliadou, A.; Svartengren, M.; Wiktorin, C. High heritability for concurrent low back and neck-shoulder pain: A study of twins. Spine 2011, 36, E1469–E1476. [Google Scholar] [CrossRef] [PubMed]
- Freidin, M.B.; Tsepilov, Y.A.; Palmer, M.; Karssen, L.C.; Suri, P.; Aulchenko, Y.S.; Williams, F.M.K.; Group, C.M.W. Insight into the genetic architecture of back pain and its risk factors from a study of 509,000 individuals. Pain 2019, 160, 1361–1373. [Google Scholar] [CrossRef] [PubMed]
- Wainschtein, P.; Jain, D.; Zheng, Z.; Group, T.O.A.W.; Consortium, N.T.-O.f.P.M.; Cupples, L.A.; Shadyab, A.H.; McKnight, B.; Shoemaker, B.M.; Mitchell, B.D.; et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat. Genet. 2022, 54, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Ao, X.; Parisien, M.; Zidan, M.; Grant, A.V.; Martinsen, A.E.; Winsvold, B.S.; Diatchenko, L. Rare variant analyses in large-scale cohorts identified SLC13A1 associated with chronic pain. Pain 2023, 164, 1841–1851. [Google Scholar] [CrossRef]
- Svishcheva, G.R.; Tiys, E.S.; Elgaeva, E.E.; Feoktistova, S.G.; Timmers, P.; Sharapov, S.Z.; Axenovich, T.I.; Tsepilov, Y.A. A Novel Framework for Analysis of the Shared Genetic Background of Correlated Traits. Genes 2022, 13, 1694. [Google Scholar] [CrossRef]
- Jiang, L.; Zheng, Z.; Fang, H.; Yang, J. A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 2021, 53, 1616–1621. [Google Scholar] [CrossRef]
- Stephens, M. A unified framework for association analysis with multiple related phenotypes. PLoS ONE 2013, 8, e65245. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genom. Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Van Hout, C.V.; Tachmazidou, I.; Backman, J.D.; Hoffman, J.D.; Liu, D.; Pandey, A.K.; Gonzaga-Jauregui, C.; Khalid, S.; Ye, B.; Banerjee, N.; et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 2020, 586, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Bi, W.; Zhao, Z.; Dey, K.K.; Jagadeesh, K.A.; Karczewski, K.J.; Daly, M.J.; Neale, B.M.; Lee, S. SAIGE-GENE+ improves the efficiency and accuracy of set-based rare variant association tests. Nat. Genet. 2022, 54, 1466–1469. [Google Scholar] [CrossRef]
- Belonogova, N.M.; Svishcheva, G.R.; Kirichenko, A.V.; Zorkoltseva, I.V.; Tsepilov, Y.A.; Axenovich, T.I. sumSTAAR: A flexible framework for gene-based association studies using GWAS summary statistics. PLoS Comput. Biol. 2022, 18, e1010172. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Wu, M.C.; Lin, X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics 2012, 13, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Abbott, D. A principal components regression approach to multilocus genetic association studies. Genet. Epidemiol. 2008, 32, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Svishcheva, G.R.; Belonogova, N.M.; Zorkoltseva, I.V.; Kirichenko, A.V.; Axenovich, T.I. Gene-based association tests using GWAS summary statistics. Bioinformatics 2019, 35, 3701–3708. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, S.; Li, Z.; Morrison, A.C.; Boerwinkle, E.; Lin, X. ACAT: A Fast and Powerful p Value Combination Method for Rare-Variant Analysis in Sequencing Studies. Am. J. Hum. Genet. 2019, 104, 410–421. [Google Scholar] [CrossRef]
- Yang, J.; Ferreira, T.; Morris, A.P.; Medland, S.E.; Genetic Investigation of ANthropometric Traits (GIANT) Consortium, DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; Weedon, M.N.; et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012, 44, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Rahman, M.S.; Winsvold, B.S.; Chavez Chavez, S.O.; Borte, S.; Tsepilov, Y.A.; Sharapov, S.Z.; Pain, H.A.-I.; Aulchenko, Y.S.; Hagen, K.; Fors, E.A.; et al. Genome-wide association study identifies RNF123 locus as associated with chronic widespread musculoskeletal pain. Ann. Rheum. Dis. 2021, 80, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Langford, R.; Hurrion, E.; Dawson, P.A. Genetics and pathophysiology of mammalian sulfate biology. J. Genet. Genom. 2017, 44, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Gatchel, R.J.; McGeary, D.D.; McGeary, C.A.; Lippe, B. Interdisciplinary chronic pain management: Past, present, and future. Am. Psychol. 2014, 69, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Gatchel, R.J.; Peng, Y.B.; Peters, M.L.; Fuchs, P.N.; Turk, D.C. The biopsychosocial approach to chronic pain: Scientific advances and future directions. Psychol. Bull. 2007, 133, 581–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Fan, B.; Dai, Q.; Xu, X.; Jiang, P.; Zhu, L.; Dai, H.; Yao, Z.; Xu, Z.; Liu, X. Fascin-1 Contributes to Neuropathic Pain by Promoting Inflammation in Rat Spinal Cord. Neurochem. Res. 2018, 43, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cheng, L.; Tian, D.; Li, Z.; Yao, F.; Luo, Y.; Liu, Y.; Zhu, Z.; Zheng, M.; Jing, J. Fascin-1 is Highly Expressed Specifically in Microglia After Spinal Cord Injury and Regulates Microglial Migration. Front. Pharmacol. 2021, 12, 729524. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef]
- Koskeridis, F.; Evangelou, E.; Said, S.; Boyle, J.J.; Elliott, P.; Dehghan, A.; Tzoulaki, I. Pleiotropic genetic architecture and novel loci for C-reactive protein levels. Nat. Commun. 2022, 13, 6939. [Google Scholar] [CrossRef]
- Rong, K.; Liang, Z.; Xiang, W.; Wang, Z.; Wen, F.; Lu, L. IL1R2 polymorphisms and their interaction are associated with osteoporosis susceptibility in the Chinese Han population. Int. J. Immunogenet. 2021, 48, 510–525. [Google Scholar] [CrossRef]
- Hendrickx, G.; Boudin, E.; Van Hul, W. A look behind the scenes: The risk and pathogenesis of primary osteoporosis. Nat. Rev. Rheumatol. 2015, 11, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Australo-Anglo-American Spondyloarthritis, C.; Reveille, J.D.; Sims, A.M.; Danoy, P.; Evans, D.M.; Leo, P.; Pointon, J.J.; Jin, R.; Zhou, X.; Bradbury, L.A.; et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 2010, 42, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Itai, T.; Wang, Z.; Nishimura, G.; Ohashi, H.; Guo, L.; Wakano, Y.; Sugiura, T.; Hayakawa, H.; Okada, M.; Saisu, T.; et al. De novo heterozygous variants in KIF5B cause kyphomelic dysplasia. Clin. Genet. 2022, 102, 3–11. [Google Scholar] [CrossRef]
- Koivunen, P.; Tiainen, P.; Hyvarinen, J.; Williams, K.E.; Sormunen, R.; Klaus, S.J.; Kivirikko, K.I.; Myllyharju, J. An endoplasmic reticulum transmembrane prolyl 4-hydroxylase is induced by hypoxia and acts on hypoxia-inducible factor α. J. Biol. Chem. 2007, 282, 30544–30552. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Li, J.; Yu, Z.; Dang, X.; Wang, K. The hypoxia-inducible factor pathway, prolyl hydroxylase domain protein inhibitors, and their roles in bone repair and regeneration. Biomed. Res. Int. 2014, 2014, 239356. [Google Scholar] [CrossRef] [PubMed]
- Danis, A. Mechanism of bone lengthening by the Ilizarov technique. Bull. Mem. Acad. R Med. Belg. 2001, 156, 107–112. [Google Scholar] [PubMed]
- Kraatari-Tiri, M.; Soikkonen, L.; Myllykoski, M.; Jamshidi, Y.; Karimiani, E.G.; Komulainen-Ebrahim, J.; Kallankari, H.; Mignot, C.; Depienne, C.; Keren, B.; et al. HIDEA syndrome is caused by biallelic, pathogenic, rare or founder P4HTM variants impacting the active site or the overall stability of the P4H-TM protein. Clin. Genet. 2022, 102, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Rahikkala, E.; Myllykoski, M.; Hinttala, R.; Vieira, P.; Nayebzadeh, N.; Weiss, S.; Plomp, A.S.; Bittner, R.E.; Kurki, M.I.; Kuismin, O.; et al. Biallelic loss-of-function P4HTM gene variants cause hypotonia, hypoventilation, intellectual disability, dysautonomia, epilepsy, and eye abnormalities (HIDEA syndrome). Genet. Med. 2019, 21, 2355–2363. [Google Scholar] [CrossRef]
- Grotzinger, A.D.; Rhemtulla, M.; de Vlaming, R.; Ritchie, S.J.; Mallard, T.T.; Hill, W.D.; Marioni, R.E.; McIntosh, A.M.; Koellinger, P.D.; Harden, K.P. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat. Hum. Behav. 2019, 3, 513–525. [Google Scholar] [CrossRef]
| Trait | Variant Annotation | Gene | #MAC a >10 | #MAC a ≤ 10 | SKATO b | PCA b | ACAT-O b |
|---|---|---|---|---|---|---|---|
| CBP c | LoF + missense | P4HTM | 29 | 149 | 6.82 × 10−6 | 6.35 × 10−3 | 1.36 × 10−5 |
| LoF + protein coding | P4HTM | 45 | 203 | 1.45 × 10−7 | 9.95 × 10−3 | 2.89 × 10−7 | |
| LoF + missense | SLC13A1 | 34 | 227 | 1.56 × 10−3 | 1.25 × 10−6 | 2.50 × 10−6 | |
| LoF + protein coding | KIF5B | 33 | 302 | 3.91 × 10−3 | 9.74 × 10−6 | 1.94 × 10−5 | |
| LoF + missense | TIMM44 | 19 | 134 | 1.82 × 10−1 | 5.90 × 10−6 | 1.18 × 10−5 | |
| Dorsalgia | All intragenic | RPL37 | 27 | 166 | 1.07 × 10−5 | 4.28 × 10−2 | 2.14 × 10−5 |
| LoF + missense | AVPR1A | 12 | 174 | 8.95 × 10−6 | 2.24 × 10−4 | 1.72 × 10−5 | |
| IDD d | LoF + missense | RNF115 | 12 | 105 | 1.04 × 10−5 | 1.85 × 10−3 | 2.07 × 10−5 |
| LoF + protein coding | KIAA2012 | 85 | 518 | 3.99 × 10−6 | 5.75 × 10−2 | 7.99 × 10−6 | |
| All intragenic | LETMD1 | 37 | 334 | 5.15 × 10−6 | 5.78 × 10−4 | 1.02 × 10−5 | |
| SGIT e | LoF | FSCN3 | 5 | 10 | 2.94 × 10−7 | 6.44 × 10−6 | 5.62 × 10−7 |
| LoF burden f | IL1R2 | 29 g | 1.14 × 10−5 h |
| Annotation | CBP | Dorsalgia | IDD | SGIT |
|---|---|---|---|---|
| LoF | 6.4 × 10−5 | 7.1 × 10−4 | 5.1 × 10−2 | 5.6 × 10−7 |
| LoF + missense | 1.3 × 10−3 | 1.6 × 10−2 | 6.9 × 10−2 | 9.2 × 10−5 |
| LoF + protein coding | 7.2 × 10−3 | 5.7 × 10−2 | 1.7 × 10−2 | 3.3 × 10−4 |
| All intragenic | 3.8 × 10−3 | 1.5 × 10−1 | 7.4 × 10−1 | 1.0 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zorkoltseva, I.V.; Elgaeva, E.E.; Belonogova, N.M.; Kirichenko, A.V.; Svishcheva, G.R.; Freidin, M.B.; Williams, F.M.K.; Suri, P.; Tsepilov, Y.A.; Axenovich, T.I. Multi-Trait Exome-Wide Association Study of Back Pain-Related Phenotypes. Genes 2023, 14, 1962. https://doi.org/10.3390/genes14101962
Zorkoltseva IV, Elgaeva EE, Belonogova NM, Kirichenko AV, Svishcheva GR, Freidin MB, Williams FMK, Suri P, Tsepilov YA, Axenovich TI. Multi-Trait Exome-Wide Association Study of Back Pain-Related Phenotypes. Genes. 2023; 14(10):1962. https://doi.org/10.3390/genes14101962
Chicago/Turabian StyleZorkoltseva, Irina V., Elizaveta E. Elgaeva, Nadezhda M. Belonogova, Anatoliy V. Kirichenko, Gulnara R. Svishcheva, Maxim B. Freidin, Frances M. K. Williams, Pradeep Suri, Yakov A. Tsepilov, and Tatiana I. Axenovich. 2023. "Multi-Trait Exome-Wide Association Study of Back Pain-Related Phenotypes" Genes 14, no. 10: 1962. https://doi.org/10.3390/genes14101962






